Genome-Wide Association Study for Resistance to Tan Spot in Synthetic Hexaploid Wheat
Abstract
:1. Introduction
2. Results
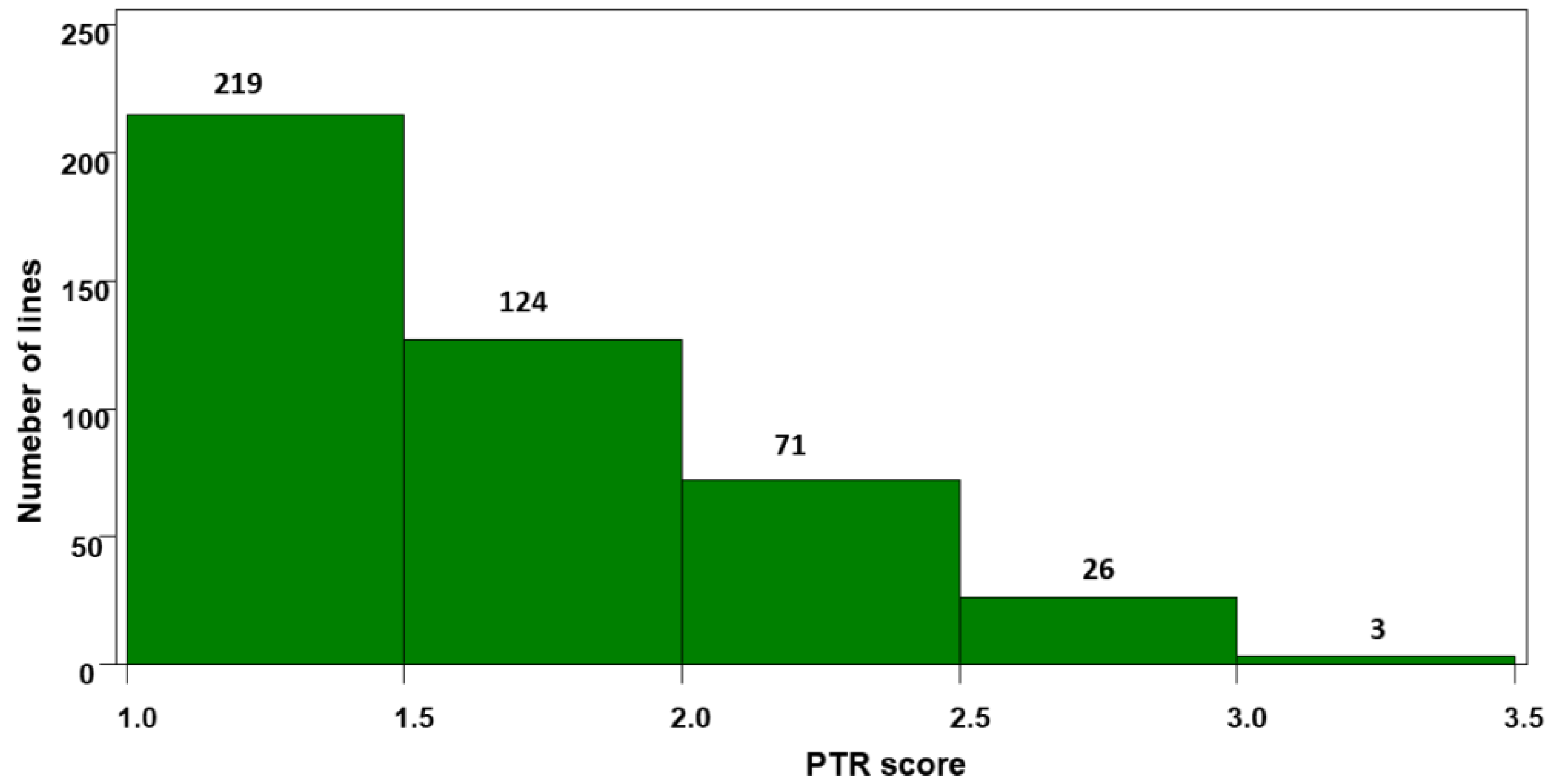
2.1. Resistance to Tan Spot at the Seedling Stage
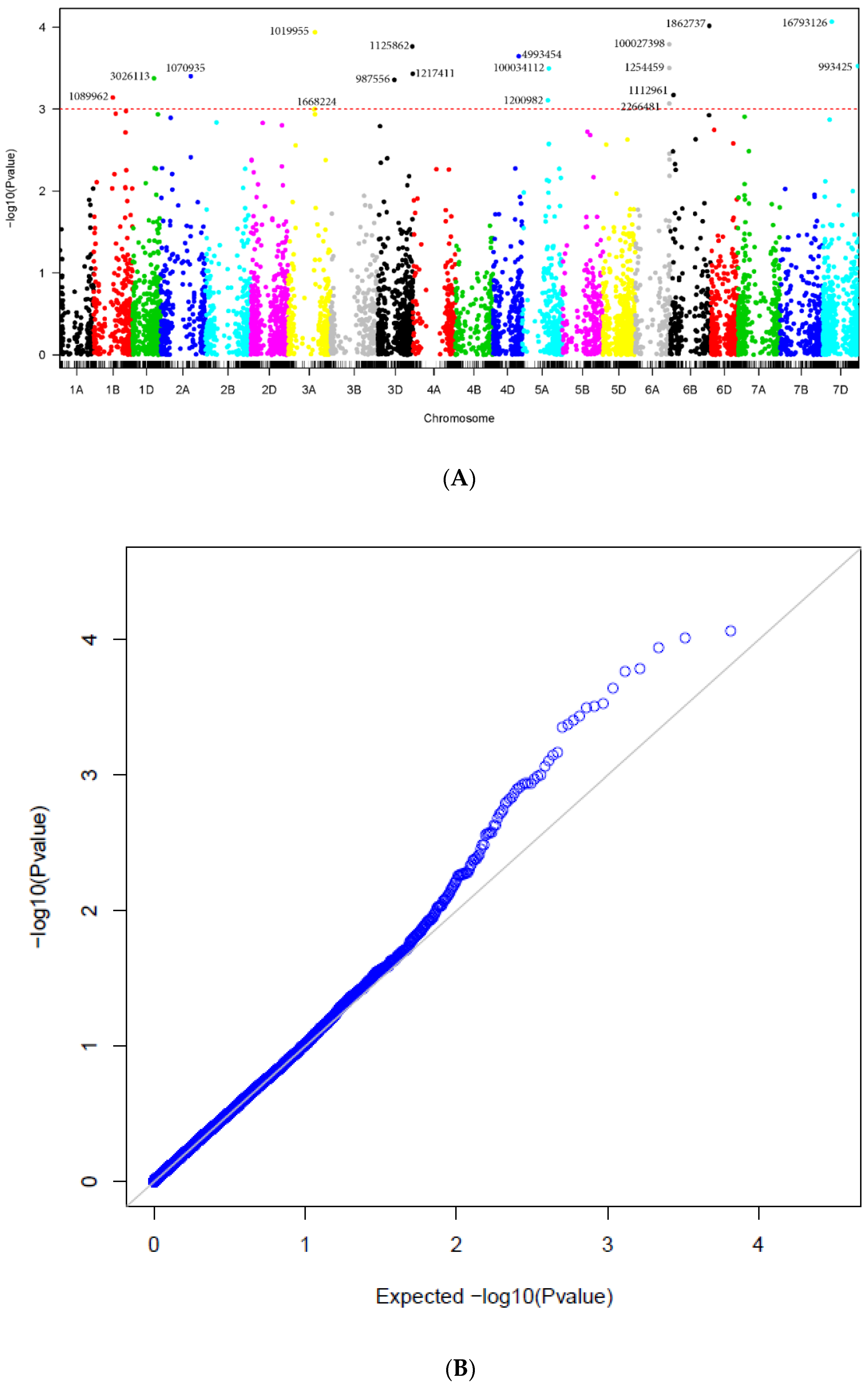
2.2. Genome-Wide Association Mapping under Different References Maps
2.3. Comparison of the Significant Markers across the Different Maps
2.4. Marker-Trait Associations and QTL for Tan Spot Resistance
3. Discussion
3.1. Tan Spot Resistance in SHW
3.2. Comparisons with Previous Studies
3.2.1. Significant Markers Found in the D- Genome Chromosomes
3.2.2. Significant Markers Found at the A and B Genome Chromosomes
3.3. Underlying Candidate Genes Based on Protein Annotation
4. Materials and Methods
4.1. Plant Material
4.2. Phenotypic Evaluation for Tan Spot
4.3. Plant Genotyping
4.4. Statistical Analysis and Genome-Wide Association Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board statement
Informed Consent statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singh, P.K.; Singh, R.P.; Duveiller, E.; Mergoum, M.; Adhikari, T.B.; Elias, E.M. Genetics of wheat-Pyrenophora tritici-repentis interactions. Euphytica 2009, 171, 1–13. [Google Scholar] [CrossRef]
- Gurung, S.; Mamidi, S.; Bonman, J.M.; Xiong, M.; Brown, G.; Adhikari, T.B. Genome wide association study reveals novel quantitative trait loci associated with resistance to multiple leaf spot diseases of spring wheat. PLoS ONE 2014, 9, e108179. [Google Scholar] [CrossRef] [Green Version]
- Oerke, E.C. Crop losses to pest. J. Agric. Sci. 2006, 144, 31–43. [Google Scholar] [CrossRef]
- Annone, J. Roya de la hoja en trigo. Importancia económica y estrategias para reducir los efectos sobre la producción. In Trigo Informe de Actualización Técnica N° 1; EEA INTA Ms Juárez: Córdoba, Argentina, 2006; p. 3. [Google Scholar]
- Manning, V.; Pandelova, I.; Dhillon, B.; Wilhelm, L.; Goodwin, S.; Berlin, A.M.; Figueroa, M.; Freitag, M.; Hane, J.K.; Henrissat, B.; et al. Comparative genomics of a plant-pathogenic fungus, Pyrenophora tritici-repentis, reveals transduplication and the impact of repeat elements on pathogenicity and population divergence. G3 2013, 3, 41–63. [Google Scholar] [CrossRef] [Green Version]
- Shabeer, A.; Bockus, W.W. Tan spot effects on yield and yield components relative to growth stage in winter wheat. Plant Dis. 1988, 72, 599–602. [Google Scholar] [CrossRef]
- Strelkov, S.E.; Lamari, L. Host-parasite interactions in tan spot (Pyrenophora tritici-repentis) of wheat. Can. J. Plant Pathol. 2003, 25, 339–349. [Google Scholar] [CrossRef]
- Tomas, A.; Bockus, W.W. Cultivar-specific toxicity of culture filtrates of Pyrenophora tritici-repentis. Phytopathology 1987, 77, 1337–1340. [Google Scholar] [CrossRef]
- Lamari, L.; Bernier, C.C. Evaluation of wheat lines and cultivars to tan spot [Pyrenophora tritici-repentis] based on lesion type. Can. J. Plant Pathol. 1989, 11, 49–56. [Google Scholar] [CrossRef]
- Lamari, L.; Strelkov, S.E.; Yahyaoui, A.; Orabi, J.; Smith, R.B. The identification of two new races of Pyrenophora tritici-repentis from the host center of diversity confirms a one-to-one relationship in tan spot of wheat. Phytopathology 2003, 93, 391–396. [Google Scholar] [CrossRef] [Green Version]
- Trethowan, R.M.; van Ginkel, M. Synthetic wheat—An emerging genetic resource. In Wheat-Science and Trade; Carver, B., Ed.; Wiley-Blackwell: Ames, IA, USA, 2009; pp. 369–386. [Google Scholar]
- Siedler, H.; Obst, A.; Hsam, S.L.K.; Zeller, F.J. Evaluation for resistance to Pyrenophora tritici-repentis in Aegilops tauschii Coss. and synthetic hexaploid wheat SHW. Genet. Resour. Crop Evol. 1994, 41, 27–34. [Google Scholar] [CrossRef]
- Zhu, C.; Gore, M.; Buckler, E.S.; Yu, J. Status and prospects of association mapping in plants. Plant Genome 2008, 1, 5–20. [Google Scholar] [CrossRef]
- Waugh, R.; Jannink, J.L.; Muehlbauer, G.J.; Ramsay, L. The emergence of whole genome association scans in barley. Curr. Opin. Plant Biol. 2009, 12, 218–222. [Google Scholar] [CrossRef] [PubMed]
- Rafalski, J.A. Association genetics in crop improvement. Curr. Opin. Plant Biol. 2010, 13, 174–180. [Google Scholar] [CrossRef]
- Hamblin, M.T.; Buckler, E.S.; Jannink, J.L. Population genetics of genomics-based crop improvement methods. Trends Genet. 2011, 27, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Gurung, S.; Mamidi, S.; Bonman, J.M.; Jackson, E.W.; del Rio, L.E.; Acevedo, M.; Mergoum, M.; Adhikari, T.B. Identification of novel genomic regions associated with resistance to Pyrenophora tritici-repentis races 1 and 5 in spring wheat using association analysis. Theor. Appl. Genet. 2011, 123, 1029–1041. [Google Scholar] [CrossRef]
- Patel, J.S.; Mamidi, S.; Bonman, J.M.; Adhikari, T.B. Identification of QTL in spring wheat associated with resistance to a novel isolate of Pyrenophora tritici-repentis. Crop Sci. 2013, 53, 842–852. [Google Scholar] [CrossRef]
- Kollers, S.; Rodemann, B.; Ling, J.; Korzun, V.; Ebmeyer, E.; Argillier, O.; Hinze, M.; Plieske, J.; Kulosa, D.; Ganal, M.W.; et al. Genome-wide association mapping of tan spot resistance (Pyrenophora tritici-repentis) in European winter wheat. Mol. Breed. 2014, 34, 363–371. [Google Scholar] [CrossRef]
- Liu, Z.; El-Basyoni, I.; Kariyawasam, G.; Zhang, G.; Fritz, A.; Hansen, J.M.; Marias, F.; Fiskop, A.; Chao, S.; Akhunov, E.; et al. Evaluation and association mapping of resistance to tan spot and Stagonospora nodorum blotch in adapted winter wheat germplasm. Plant Dis. 2015, 99, 1333–1341. [Google Scholar] [CrossRef] [Green Version]
- Dinglasan, E.G.; Singh, D.; Shankar, M.; Afanasenko, O.; Platz, G.; Godwin, I.D.; Voss-Fels, K.P.; Hickey, L.T. Discovering new alleles for yellow spot resistance in the Vavilov wheat collection. Theor. Appl. Genet. 2019, 132, 149–162. [Google Scholar] [CrossRef]
- Singh, P.K.; Crossa, J.; Duveiller, E.; Singh, R.P.; Djurle, A. Association mapping for resistance to tan spot induced by Pyrenophora tritici-repentis race 1 in CIMMYTs historical bread wheat set. Euphytica 2016, 207, 515–525. [Google Scholar] [CrossRef]
- Juliana, P.; Singh, R.P.; Singh, P.K.; Poland, J.A.; Bergstrom, G.C.; Huerta-Espino, J.; Bhavani, S.; Crossa, J.; Sorrells, M.E. Genome-wide association mapping for resistance to leaf rust, stripe rust and tan spot in wheat reveals potential candidate genes. Theor. Appl. Genet. 2018, 131, 1405–1422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phuke, R.M.; He, X.; Juliana, P.; Bishnoi, S.K.; Singh, G.P.; Kabir, M.R.; Roy, K.K.; Joshi, A.K.; Singh, R.P.; Singh, P.K. Association mapping of seedling resistance to tan spot (Pyrenophora tritici-repentis race 1) in CIMMYT and South Asian wheat germplasm. Front. Plant Sci. 2020, 11, 1309. [Google Scholar] [CrossRef]
- Faris, J.D.; Friesen, T.L. Identification of quantitative trait loci for race-nonspecific resistance to tan spot of wheat. Theor. Appl. Genet. 2005, 111, 386–392. [Google Scholar] [CrossRef]
- Bhatta, M.; Morgounov, A.; Vikas Belamkar, V.; Wegulo, S.N.; Dababat, A.A.; Erginbas-Orakci, G.; El Bouhssini, M.; Gautam, P.; Poland, J.; Akci, N.; et al. Genome-wide association study for multiple biotic stress resistance in synthetic hexaploid wheat. Int. J. Mol. Sci. 2019, 20, 3667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tadesse, W.; Schmolke, M.; Hsam, S.L.K.; Mohler, V.; Wenzel, G.; Zeller, F.J. Molecular mapping to tan spot (Pyrenophora tritici-repentis race 1) in synthetic wheat lines. Theor. Appl. Genet. 2007, 114, 855–862. [Google Scholar] [CrossRef]
- Kokhmetova, A.; Sehgal, D.; Ali, S.; Atishova, M.; Kumarbayeva, M.; Irina Leonova, I.; Dreisigacker, S. Genome-wide association study of tan spot resistance in a hexaploid wheat collection from Kazakhstan. Front. Genet. 2021, 11, 581214. [Google Scholar] [CrossRef] [PubMed]
- Kalia, B.; Bockus, W.W.; Singh, S.; Tiwari, V.K.; Gill, B.S. Mapping of quantitative trait loci for resistance to race 1 of Pyrenophora tritici-repentis in synthetic hexaploid wheat. Plant Breed. 2018, 137, 313–319. [Google Scholar] [CrossRef]
- Chu, C.G.; Friesen, T.L.; Xu, S.S.; Faris, J.D. Identification of novel tan spot resistance loci beyond the known host-selective toxin insensitivity genes in wheat. Theor. Appl. Genet. 2008, 117, 873–881. [Google Scholar] [CrossRef]
- Zhang, X.; Gonzalez-Carranza, Z.H.; Zhang, S.; Miao, Y.; Liu, C.; Roberts, J.A. F-Box proteins in plants. Annu. Plant Rev. 2019, 2, 1–21. [Google Scholar] [CrossRef]
- Durst, F.; Benveniste, I. Cytochrome P450 in Plants. In Cytochrome P450; Schenkman, J.B., Griem, H., Eds.; Springer: Berlin/Heidelberg, Germany, 1993. [Google Scholar]
- Jun, X.; Xin-Yu, W.; Wang-Zhen, G. The cytochrome P450 superfamily: Key players in plant development and defense. J. Integr. Agric. 2015, 14, 1673–1686. [Google Scholar]
- Gunupuru, L.R.; Arunachalam, C.; Keshav, B.M.; Kahla, A.; Perochon, A.; Jia, J.; Thapa, G.; Doohan, F.M. A wheat cytochrome P450 enhances both resistance to deoxynivalenol and grain yield. PLoS ONE 2018, 13, e0204992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dreisigacker, S.; Sehgal, D.; Reyes-Jaimez, A.E.; Luna-Garrido, B.; Muñoz-Zavala, S.; Núñez Ríos, C.; Mollins, J.; Mall, S. CIMMYT Wheat Molecular Genetics: Laboratory Protocols and Applications to Wheat Breeding; Version 1; CIMMYT: Texcoco, Mexico, 2016. [Google Scholar]
- Sansaloni, C.; Petroli, C.; Jaccoud, D.; Carling, J.; Detering, F. Diversity arrays technology (DArT) and next-generation sequencing combined: Genome wide, high throughput, highly informative genotyping for molecular breeding of Eucalyptus. BMC Proc. 2011, 5, 54. [Google Scholar] [CrossRef] [Green Version]
- Sansaloni, C.; Franco, J.; Santos, B.; Percival-Alwyn, L.; Singh, S.; Petroli, C.; Campos, J.; Dreher, K.; Payne, T.; Marshall, D.; et al. Diversity analysis of 80,000 wheat accessions reveals consequences and opportunities of selection footprints. Nat. Commun. 2020, 11, 4572. [Google Scholar] [CrossRef] [PubMed]
- The International Wheat Genome Sequencing Consortium (IWGSC); Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef] [Green Version]
- Maccaferri, M.; Sanguineti, M.C.; Mantovani, P.; Demontis, A.; Massi, A.; Ammar, K.; Kolmer, J.A.; Czembor, J.H.; Ezrati, S.; Tuberosa, R. Association mapping of leaf rust response in durum wheat. Mol. Breed. 2019, 26, 189–228. [Google Scholar] [CrossRef]
- Luo, M.-C.; Gu, Y.Q.; Puiu, D.; Wang, H.; Twardziok, S.O.; Deal, K.R.; Huo, N.; Zhu, T.; Wang, L.; Wang, Y.; et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 2017, 551, 498–502. [Google Scholar] [CrossRef]
- SAS Institute. SAS/STAT User’s Guide; Version 9.1; SAS Institute, Inc.: Cary, NC, USA, 2004. [Google Scholar]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Yu, J.; Pressoir, G.; Briggs, W.; Bi, I.V.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef]


| DW Parents | SHW | ||||
|---|---|---|---|---|---|
| Pedigree | Tan Spot Scores | Reaction Type | Number of SHW Progeny | Mean Tan Spot Scores | Mean Reaction Type |
| BOTNO | 4.3 | S | 1 | 2.2 | MR |
| SCAUP | 3.9 | S | 3 | 2.2 | MR |
| CROC_1 | 3.7 | S | 30 | 1.7 | MR |
| D67.2/PARANA 66.270 | 3.7 | S | 13 | 1.7 | MR |
| YAR | 3.7 | S | 4 | 1.4 | R |
| 68.111/RGB-U//WARD RESEL/3/STIL | 3.6 | S | 31 | 1.5 | R |
| DECOY 1 | 3.5 | MS | 30 | 2.1 | MR |
| SORA | 3.4 | MS | 14 | 1.6 | MR |
| 6973/WARD.7463//74110 | 3.3 | MS | 3 | 1.6 | MR |
| CPI8/GEDIZ/3/GOO//ALB/CRA | 3.3 | MS | 31 | 1.9 | MR |
| LCK59.61 | 3.2 | MS | 2 | 2.3 | MR |
| 68.111/RGB-U//WARD | 3.1 | MS | 7 | 1.6 | MR |
| CHEN_7 | 3.0 | MS | 1 | 1.2 | R |
| ALG86/4/FGO/PALES//MEXI_1/3/RUFF/FGO/5/ENTE | 2.9 | MS | 3 | 2 | MR |
| YAV_2/TEZ | 2.9 | MS | 12 | 1.6 | MR |
| LOCAL RED | 2.9 | MS | 7 | 2.2 | MR |
| TK SN1081 | 2.9 | MS | 3 | 1.2 | R |
| YARMUK | 2.8 | MS | 4 | 1.7 | MR |
| ROK/KML | 2.7 | MS | 4 | 2.2 | MR |
| STY,DR/CELTA//PALS/3/SRN_5 | 2.7 | MS | 2 | 1.5 | R |
| ALTAR 84 | 2.6 | MS | 20 | 1.6 | MR |
| ACONCHI 89 | 2.6 | MS | 4 | 1.5 | R |
| DVERD_2 | 2.5 | MR | 13 | 1.5 | R |
| FGO/USA2111 | 2.5 | MR | 1 | 1.1 | R |
| ARLIN_1 | 2.4 | MR | 13 | 1.5 | R |
| 68.111/RGB-U//WARD/3/FGO/4/RABI | 2.4 | MR | 31 | 1.5 | R |
| SCOT/MEXI_1 | 2.4 | MR | 1 | 1.8 | MR |
| GARZA/BOY | 2.3 | MR | 7 | 1.8 | MR |
| 68112/WARD | 2.3 | MR | 4 | 1.2 | R |
| LARU | 2.3 | MR | 4 | 1.1 | R |
| RASCON_37 | 2.2 | MR | 2 | 1.3 | R |
| KAPUDE_1 | 2.1 | MR | 1 | 1.9 | MR |
| CERCETA | 1.9 | MR | 54 | 1.6 | MR |
| RABI//GS/CRA | 1.6 | MR | 4 | 1.5 | R |
| SNIPE/YAV79//DACK/TEAL | 1.5 | R | 7 | 1.1 | R |
| FALCIN_1 | 1.5 | R | 5 | 1.9 | MR |
| SHAG_22 | 1.5 | R | 6 | 1.5 | R |
| GREEN_3 | 1.2 | R | 1 | 1 | R |
| GAN | 1.1 | R | 39 | 1.4 | R |
| SCOOP_1 | 1.1 | R | 3 | 1 | R |
| Erik (R check) | 1.0 | R | --- | --- | --- |
| Glenlea (S check) | 4.8 | S | --- | --- | --- |
| 6B-662 (MR check) | 2.0 | MR | --- | --- | --- |
| 6B-365 (MS check) | 3.1 | MS | --- | --- | --- |
| Chr | Marker ID | Allele ID | Genetic Position on Consensus Map (cM) | F Statistics | Prob. | Marker R2 | −log10 p-Value | Effect of Allele Substitution (Genotype Effect) |
|---|---|---|---|---|---|---|---|---|
| 1B | 987556 | 987556|F|0-61:G>A-61:G>A | 60.43 | 8.36 | 2.78 × 10−4 | 0.042 | 3.56 | −0.22 |
| 1B | 6045377 | 6045377|F|0-16:T>C-16:T>C | 51.29 | 8.06 | 3.71 × 10−4 | 0.040 | 3.43 | −0.10 |
| 1B | 1089962 | 1089962|F|0-56:C>T-56:C>T | 83.57 | 7.21 | 8.40 × 10−4 | 0.036 | 3.08 | −0.19 |
| 2A | 1070935 | 1070935|F|0-45:G>A-45:G>A | 68.84 | 7.48 | 6.46 × 10−4 | 0.038 | 3.19 | −0.28 |
| 4A | 4993454 | 4993454|F|0-12:T>C-12:T>C | 10.72 | 8.20 | 3.24 × 10−4 | 0.041 | 3.49 | −0.55 |
| 5A | 1200982 | 1200982|F|0-30:C>G-30:C>G | 47.79 | 7.68 | 5.36 × 10−4 | 0.038 | 3.27 | 0.05 |
| 5A | 4393896 | 4393896|F|0-34:T>C-34:T>C | 48.67 | 7.21 | 8.43 × 10−4 | 0.036 | 3.07 | −0.20 |
| 5B | 100034112 | 100034112|F|0-10:C>T-10:C>T | 39.26 | 7.80 | 4.77 × 10−4 | 0.039 | 3.32 | −0.14 |
| 6A | 1862737 | 1862737|F|0-44:C>G-44:C>G | 90.36 | 9.15 | 1.30 × 10−4 | 0.046 | 3.89 | −0.20 |
| 6A | 100027398 | 100027398|F|0-42:A>G-42:A>G | 77.32 | 8.21 | 3.20 × 10−4 | 0.041 | 3.49 | −0.15 |
| 6A | 5331622 | 5331622|F|0-5:A>G-5:A>G | 98.51 | 8.05 | 3.72 × 10−4 | 0.040 | 3.43 | −0.12 |
| 6A | 1254459 | 1254459|F|0-8:A>C-8:A>C | 94.09 | 7.35 | 7.36 × 10−4 | 0.037 | 3.13 | −0.22 |
| 6A | 4993056 | 4993056|F|0-26:A>T-26:A>T | 91.17 | 7.18 | 8.68 × 10−4 | 0.036 | 3.06 | −0.23 |
| 6B | 1019955 | 1019955|F|0-55:A>G-55:A>G | 46.69 | 8.82 | 1.79 × 10−4 | 0.044 | 3.75 | −0.44 |
| 7D | 991140 | 991140|F|0-11:G>C-11:G>C | 153.02 | 10.19 | 4.84 × 10−5 | 0.051 | 4.31 | −0.15 |
| 7D | 993425 | 993425|F|0-28:A>G-28:A>G | 168.74 | 8.35 | 2.81 × 10−4 | 0.041 | 3.55 | 0.59 |
| Chr | Marker | Allele ID | Pos | F Statistic | Prob. | Marker R2 | −log10 p-Value | Effect of Allele Substitution (Genotype Effect) |
|---|---|---|---|---|---|---|---|---|
| 1B | 1089962 | 1089962|F|0-56:C>T-56:C>T | 340462174 | 7.37 | 7.23 × 10−4 | 0.037 | 3.14 | −0.19 |
| 1D | 3026113 | 3026113|F|0-19:G>T-19:G>T | 375647840 | 7.92 | 4.22 × 10−4 | 0.040 | 3.37 | 0.16 |
| 2A | 1070935 | 1070935|F|0-45:G>A-45:G>A | 525822786 | 7.99 | 3.97 × 10−4 | 0.040 | 3.40 | −0.29 |
| 3A | 1019955 | 1019955|F|0-55:A>G-55:A>G | 474447292 | 9.28 | 1.16 × 10−4 | 0.046 | 3.94 | −0.44 |
| 3A | 1668224 | 1668224|F|0-18:T>C-18:T>C | 468520788 | 7.03 | 1.00 × 10−3 | 0.035 | 3.00 | −0.24 |
| 3D | 1125862 | 1125862|F|0-8:C>A-8:C>A | 603632716 | 8.86 | 1.72 × 10−4 | 0.044 | 3.76 | −0.13 |
| 3D | 1217411 | 1217411|F|0-6:C>T-6:C>T | 610566593 | 8.06 | 3.71 × 10−4 | 0.040 | 3.43 | −0.21 |
| 3D | 987556 | 987556|F|0-61:G>A-61:G>A | 288544777 | 7.88 | 4.41 × 10−4 | 0.039 | 3.36 | −0.21 |
| 4D | 4993454 | 4993454|F|0-12:T>C-12:T>C | 449396486 | 8.57 | 2.26 × 10−4 | 0.043 | 3.64 | −0.56 |
| 5A | 100034112 | 100034112|F|0-10:C>T-10:C>T | 471723681 | 8.21 | 3.20 × 10−4 | 0.041 | 3.50 | −0.15 |
| 5A | 1200982 | 1200982|F|0-30:C>G-30:C>G | 454770585 | 7.28 | 7.83 × 10−4 | 0.036 | 3.11 | −0.05 |
| 6A | 100027398 | 100027398|F|0-42:A>G-42:A>G | 601233092 | 8.92 | 1.62 × 10−4 | 0.045 | 3.79 | −0.15 |
| 6A | 1254459 | 1254459|F|0-8:A>C-8:A>C | 602989232 | 8.23 | 3.15 × 10−4 | 0.041 | 3.50 | −0.23 |
| 6A | 2266481 | 2266481|F|0-54:C>T-54:C>T | 602745555 | 7.19 | 8.56 × 10−4 | 0.036 | 3.07 | −0.21 |
| 6B | 1862737 | 1862737|F|0-44:C>G-44:C>G | 689032602 | 9.46 | 9.65 × 10−5 | 0.047 | 4.02 | −0.20 |
| 6B | 1112961 | 1112961|F|0-43:G>A-43:G>A | 62173247 | 7.44 | 6.75 × 10−4 | 0.037 | 3.17 | −0.13 |
| 7D | 16793126 | 16793126|F|0-15:G>T-15:G>T | 161842641 | 9.59 | 8.59 × 10−5 | 0.048 | 4.07 | 0.05 |
| 7D | 993425 | 993425|F|0-28:A>G-28:A>G | 620252466 | 8.28 | 3.00 × 10−4 | 0.041 | 3.52 | 0.61 |
| Chr | Marker | Allelle ID | Position | F Statistic | Prob. | Marker R2 | −log10 p-Value | Effect of Allele Substitution (Genotype Effect) |
|---|---|---|---|---|---|---|---|---|
| 1B | 1106306 | 1106306|F|0-31:A>G-31:A>G | 18733634 | 9.04 | 1.45 × 10−4 | 0.045 | 3.84 | −0.24 |
| 1B | 1089962 | 1089962|F|0-56:C>T-56:C>T | 333205076 | 8.01 | 3.89 × 10−4 | 0.040 | 3.41 | −0.20 |
| 1B | 3026113 | 3026113|F|0-19:G>T-19:G>T | 493514948 | 7.86 | 4.47 × 10−4 | 0.039 | 3.35 | 0.16 |
| 1B | 4909460 | 4909460|F|0-15:T>C-15:T>C | 551136407 | 7.33 | 7.45 × 10−4 | 0.037 | 3.13 | −0.17 |
| 2D | 1046601 | 1046601|F|0-37:C>G-37:C>G | 543349511 | 7.33 | 7.47 × 10−4 | 0.037 | 3.13 | −0.01 |
| 2D | 1217245 | 1217245|F|0-50:G>A-50:G>A | 49063764 | 7.27 | 7.90 × 10−4 | 0.036 | 3.10 | −0.15 |
| 3A | 1065211 | 1065211|F|0-46:G>A-46:G>A | 477078596 | 7.49 | 6.43 × 10−4 | 0.037 | 3.19 | −0.26 |
| 3A | 1125872 | 1125872|F|0-29:C>T-29:C>T | 141341740 | 7.14 | 9.00 × 10−4 | 0.036 | 3.05 | −0.27 |
| 4A | 1125862 | 1125862|F|0-8:C>A-8:C>A | 558758715 | 8.35 | 2.80 × 10−4 | 0.042 | 3.55 | −0.14 |
| 5A | 3064590 | 3064590|F|0-39:T>A-39:T>A | 433029624 | 7.17 | 8.76 × 10−4 | 0.036 | 3.06 | −0.22 |
| 6A | 100027398 | 100027398|F|0-42:A>G-42:A>G | 597038442 | 11.53 | 1.36 × 10−5 | 0.058 | 4.87 | −0.17 |
| 6A | 1254459 | 1254459|F|0-8:A>C-8:A>C | 598610204 | 8.69 | 2.01 × 10−4 | 0.043 | 3.70 | −0.23 |
| 7D | 16793126 | 16793126|F|0-15:G>T-15:G>T | 162738314 | 9.30 | 1.13 × 10−4 | 0.047 | 3.95 | 0.05 |
| Ch. | Marker ID | Consensus Map (cM) | Position (CS) | Position (Svevo) | Pos (Ae.t.) | Gene (s) | GWAS | p-Value | Marker R2 | −log10 p-Value |
|---|---|---|---|---|---|---|---|---|---|---|
| 1B | 1106306 | 1B-18733634 | - | Durum-tauschii (phy. pos) | 1.45 × 10−4 | 0.045 | 3.84 | |||
| 1B | 6045377 | 1B-51.3 | Bread wheat (genetic map) | 3.71 × 10−4 | 0.040 | 3.43 | ||||
| 1B | 1089962 | 1B-83.6 | 1B-340462174 | 1B-333205076 | Aestivum (genetic map) | 8.40 × 10−4 | 0.036 | 3.08 | ||
| - | Aestivum (phy. pos.) | 7.23 × 10−4 | 0.037 | 3.14 | ||||||
| - | Durum-tauschii (phy. pos) | 3.89 × 10−4 | 0.040 | 3.41 | ||||||
| 1B | 4909460 | 1B-558561647 | 1B-551136407 | - | Durum-tauschii (phy. pos) | 7.45 × 10−4 | 0.037 | 3.13 | ||
| 1D | 3026113 | 1D-375647840 | 1D-381593800 | - | Aestivum (phy. pos.) | 4.22 × 10−4 | 0.040 | 3.37 | ||
| AET1Gv20669700 | Durum-tauschii (phy. pos) | 4.47 × 10−4 | 0.039 | 3.35 | ||||||
| 2A | 1070935 | 2A-68.8 | Aestivum (genetic map) | 6.46 × 10−4 | 0.038 | 3.19 | ||||
| 2A-525822786 | 2A-519747584 | - | Aestivum (phy. pos.) | 3.97 × 10−4 | 0.040 | 3.40 | ||||
| 2D | 1217245 | 2D-48123061 | 2D-49063764 | - | Durum-tauschii (phy. pos) | 7.90 × 10−4 | 0.036 | 3.10 | ||
| 2D | 1046601 | 2D-544685083 | 2D-543349511 | TraesCS2D02G432700 | Durum-tauschii (phy. pos) | 7.47 × 10−4 | 0.037 | 3.13 | ||
| 3A | 1125872 | 3A-135590641 | 3A-141341769 | - | Durum-tauschii (phy. pos) | 9.00 × 10−4 | 0.036 | 3.05 | ||
| 3A | 1668224 | 3A-468520788 | 3A-471432162 | - | Aestivum (phy. pos.) | 1.00 × 10−3 | 0.035 | 3.00 | ||
| 3A or 6B | 1019955 | 6B-46.7 | 3A-474447292, 6B-665557108 | 3A-477078694 | Aestivum (genetic map) | 1.79 × 10−4 | 0.044 | 3.75 | ||
| - | Aestivum (phy. pos.) | 1.16 × 10−4 | 0.046 | 3.94 | ||||||
| 3A | 1065211 | 3A-474447226 | 3A-477078596 | - | Durum-tauschii (phy. pos) | 6.43 × 10−4 | 0.037 | 3.19 | ||
| 3D | 987556 | 1B-60.4 | Aestivum (genetic map) | 2.78 × 10−4 | 0.042 | 3.56 | ||||
| 3D-288544838 | 3D-295969303 | - | Aestivum (phy. pos.) | 4.41 × 10−4 | 0.039 | 3.36 | ||||
| 3D | 1125862 | 3D-603632716 | 3D-614682837 | - | Aestivum (phy. pos.) | 1.72 × 10−4 | 0.044 | 3.76 | ||
| - | Durum-tauschii (phy. pos) | 2.80 × 10−4 | 0.042 | 3.55 | ||||||
| 3D | 1217411 | 3D-610566592 | 3D-622597928 | - | Aestivum (phy. pos.) | 3.71 × 10−4 | 0.040 | 3.43 | ||
| 4B or 4D | 4993454 | 4A-10.7 | 4B-561892901, 4D-449396542 | 4B-566325530 | 4D-455660733 | Aestivum (genetic map) | 3.24 × 10−4 | 0.041 | 3.49 | |
| - | Aestivum (phy. pos.) | 2.26 × 10−4 | 0.043 | 3.64 | ||||||
| 5A | 4393896 | 5A-48.7 | Aestivum (genetic map) | 8.43 × 10−4 | 0.036 | 3.07 | ||||
| 5A | 1200982 | 5A-47.8 | 5A-454770615 | 5A-416482338 | Aestivum (genetic map) | 5.36 × 10−4 | 0.038 | 3.27 | ||
| TraesCS5A02G238600 TRITD5Av1G148960 | Aestivum (phy. pos.) | 7.83 × 10−4 | 0.036 | 3.11 | ||||||
| 5A | 100034112 | 5B-39.3 | 5A-471723681 | 5A-433814227 | Aestivum (genetic map) | 4.77 × 10−4 | 0.039 | 3.32 | ||
| - | Aestivum (phy. pos.) | 3.20 × 10−4 | 0.041 | 3.50 | ||||||
| 5A | 3064590 | 5A-470186523 | 5A:433029663 | TraesCS5A02G254500 TRITD5Av1G155700 | Durum-tauschii (phy. pos) | 8.76 × 10−4 | 0.036 | 3.06 | ||
| 6A | 1862737 | 6A-90.4 | 6A-599622814 | 6A-595687891 | Aestivum (genetic map) | 1.30 × 10−4 | 0.046 | 3.89 | ||
| TraesCS6A02G378800, TRITD6Av1G217060 | Aestivum (phy. pos.) | 9.65 × 10−5 | 0.047 | 4.02 | ||||||
| 6A | 100027398 | 6A-77.3 | 6A-601233092 | 6A-597038469 | Aestivum (genetic map) | 3.20 × 10−4 | 0.041 | 3.49 | ||
| TraesCS6A02G381900 | Aestivum (phy. pos.) | 1.62 × 10−4 | 0.045 | 3.79 | ||||||
| TRITD6Av1G217800 | Durum-tauschii (phy. pos) | 1.36 × 10−5 | 0.058 | 4.87 | ||||||
| 6A | 1254459 | 6A-94.1 | 6A-602989232 | 6A-598610265 | Aestivum (genetic map) | 7.36 × 10−4 | 0.037 | 3.13 | ||
| - | Aestivum (phy. pos.) | 3.15 × 10−4 | 0.041 | 3.50 | ||||||
| - | Durum-tauschii (phy. pos) | 2.01 × 10−4 | 0.043 | 3.70 | ||||||
| 6A | 2266481 | 6A-602745555 | 6A-598380242 | TraesCS6A02G384200 | Aestivum (phy. pos.) | 8.56 × 10−4 | 0.036 | 3.07 | ||
| 6A | 4993056 | 6A-91.2 | Aestivum (genetic map) | 8.68 × 10−4 | 0.036 | 3.06 | ||||
| 6A | 5331622 | 6A-98.6 | Aestivum (genetic map) | 3.72 × 10−4 | 0.040 | 3.43 | ||||
| 6B | 1112961 | 6B-62173280 | 6B-59030547 | - | Aestivum (phy. pos.) | 6.75 × 10−4 | 0.037 | 3.17 | ||
| 7D | 16793126 | 7D-161842695 | 7D-162738368 | TraesCS7D02G203900 | Aestivum (phy. pos.) | 8.59 × 10−5 | 0.048 | 4.07 | ||
| AET7Gv20511100 AET7Gv20511200 | Durum-tauschii (phy. pos) | 1.13 × 10−4 | 0.047 | 3.95 | ||||||
| 7D | 991140 | 7D-153.0 | Aestivum (genetic map) | 4.84 × 10−5 | 0.051 | 4.31 | ||||
| 7D | 993425 | 7D-168.7 | 7D-620252508 | 7D-625050620 | Aestivum (genetic map) | 2.81 × 10−4 | 0.042 | 3.55 | ||
| TraesCS7D02G524200 AET7Gv21298500 | Aestivum (phy. pos.) | 3.00 × 10−4 | 0.041 | 3.52 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lozano-Ramírez, N.; Dreisigacker, S.; Sansaloni, C.P.; He, X.; Islas, S.S.; Pérez-Rodríguez, P.; Carballo, A.C.; Nava-Díaz, C.; Kishii, M.; Singh, P.K. Genome-Wide Association Study for Resistance to Tan Spot in Synthetic Hexaploid Wheat. Plants 2022, 11, 433. https://doi.org/10.3390/plants11030433
Lozano-Ramírez N, Dreisigacker S, Sansaloni CP, He X, Islas SS, Pérez-Rodríguez P, Carballo AC, Nava-Díaz C, Kishii M, Singh PK. Genome-Wide Association Study for Resistance to Tan Spot in Synthetic Hexaploid Wheat. Plants. 2022; 11(3):433. https://doi.org/10.3390/plants11030433
Chicago/Turabian StyleLozano-Ramírez, Nerida, Susanne Dreisigacker, Carolina P. Sansaloni, Xinyao He, Sergio Sandoval Islas, Paulino Pérez-Rodríguez, Aquiles Carballo Carballo, Cristian Nava-Díaz, Masahiro Kishii, and Pawan K. Singh. 2022. "Genome-Wide Association Study for Resistance to Tan Spot in Synthetic Hexaploid Wheat" Plants 11, no. 3: 433. https://doi.org/10.3390/plants11030433
APA StyleLozano-Ramírez, N., Dreisigacker, S., Sansaloni, C. P., He, X., Islas, S. S., Pérez-Rodríguez, P., Carballo, A. C., Nava-Díaz, C., Kishii, M., & Singh, P. K. (2022). Genome-Wide Association Study for Resistance to Tan Spot in Synthetic Hexaploid Wheat. Plants, 11(3), 433. https://doi.org/10.3390/plants11030433






