Virus-Induced Gene Silencing in Chrysanthemum seticuspe Using the Tomato Aspermy Virus Vector
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. TAV Vector
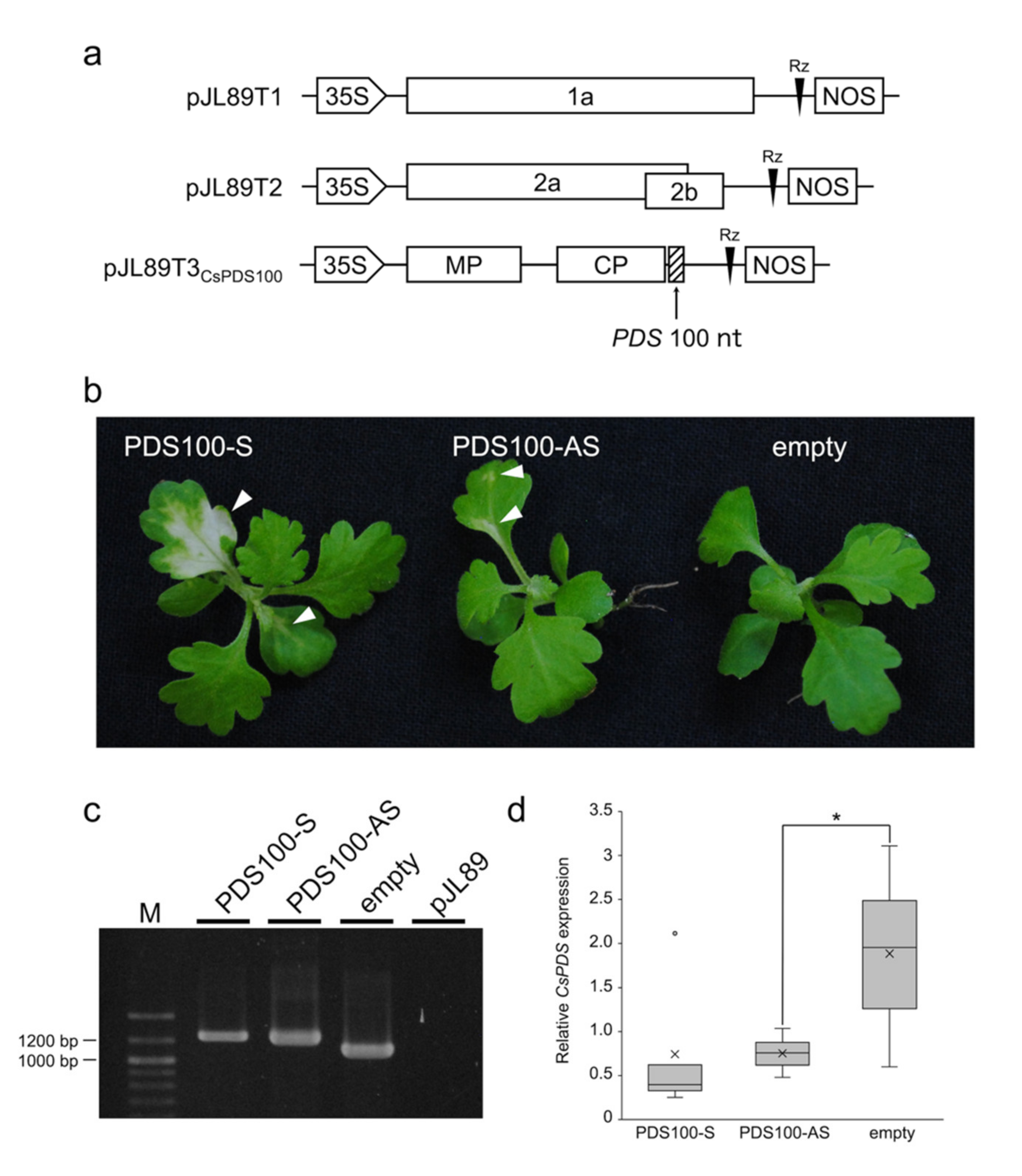
2.3. VIGS Constructs
2.4. Virus Inoculation
2.5. Detection of Viral CP by a Dot Immunobinding Assay (DIBA)
2.6. Detection of Viral RNA by RT–PCR and Gene Expression Analysis by RT–qPCR
3. Results
3.1. TAV Inoculation Method for C. seticuspe
3.2. Infectivity of 2b Mutants That Have Various VSR Activities to C. seticuspe
3.3. VIGS in C. seticuspe
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Funk, V.A.; Susanna, A.; Stuessy, T.F.; Robinson, H. Classification of Compositae Systematics, Evolution, and Biogeography of Compositae; Austria International Association for Plant Taxonomy-IAPT: Vienna, Austria, 2009. [Google Scholar]
- Anderson, N.O. Flower Breeding and Genetics: Issues, Challenges and Opportunities for the 21st Century; Springer Science & Business Media: New York, NY, USA, 2006. [Google Scholar]
- Sasaki, K.; Mitsuda, N.; Nashima, K.; Kishimoto, K.; Katayose, Y.; Kanamori, H.; Ohmiya, A. Generation of Expressed Sequence Tags for Discovery of Genes Responsible for Floral Traits of Chrysanthemum Morifolium by Next-Generation Sequencing Technology. BMC Genom. 2017, 18, 683. [Google Scholar]
- Dowrick, G.J.; El-Bayoumi, A. The Origin of New Forms of the Garden Chrysanthemum. Euphytica 1966, 15, 32–38. [Google Scholar] [CrossRef]
- Nakano, M.; Hirakawa, H.; Fukai, E.; Toyoda, A.; Kajitani, R.; Minakuchi, Y.; Itoh, T.; Higuchi, Y.; Kozuka, T.; Bono, H.; et al. A Chromosome-Level Genome Sequence of Chrysanthemum Seticuspe, a Model Species for Hexaploid Cultivated Chrysanthemum. Commun. Biol. 2021, 4, 1167. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, H.; Sumitomo, K.; Hisamatsu, T.; Nagano, S.; Shirasawa, K.; Higuchi, Y.; Kusaba, M.; Koshioka, M.; Nakano, Y.; Yagi, M.; et al. De Novo Whole-Genome Assembly in Chrysanthemum Seticuspe, a Model Species of Chrysanthemums, and Its Application to Genetic and Gene Discovery Analysis. DNA Res. 2019, 26, 195–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dommes, A.B.; Gross, T.; Herbert, D.B.; Kivivirta, K.I.; Becker, A. Virus-Induced Gene Silencing: Empowering Genetics in Non-Model Organisms. J. Exp. Bot. 2019, 70, 757–770. [Google Scholar] [CrossRef] [PubMed]
- Baulcombe, D. RNA Silencing in Plants. Nature 2004, 431, 356–363. [Google Scholar] [CrossRef]
- Burch-Smith, T.M.; Anderson, J.C.; Martin, G.B.; Dinesh-Kumar, S.P. Applications and Advantages of Virus-Induced Gene Silencing for Gene Function Studies in Plants. Plant J. 2004, 39, 734–746. [Google Scholar] [CrossRef]
- Song, A.; You, Y.; Chen, F.; Li, P.; Jiang, J.; Chen, S. A Multiplex RT-PCR for Rapid and Simultaneous Detection of Viruses and Viroids in Chrysanthemum. Lett. Appl. Microbiol. 2013, 56, 8–13. [Google Scholar] [CrossRef]
- Kumar, S.; Khan, M.S.; Raj, S.K.; Sharma, A.K. Elimination of Mixed Infection of Cucumber Mosaic and Tomato Aspermy Virus from Chrysanthemum Morifolium Ramat. Cv. Pooja by Shoot Meristem Culture. Sci. Hortic. 2009, 119, 108–112. [Google Scholar] [CrossRef]
- Matsuura, S.; Hoshino, S.; Hayashi, H.; Kohguchi, T.; Hagiwara, K.; Omura, T. Effects of Latent Infection of Stock Plants and Abundance of Thrips on the Occurrence of Tomato Spotted Wilt Virus in Chrysanthemum Fields. J. Gen. Plant Pathol. 2002, 68, 99–102. [Google Scholar] [CrossRef]
- Murai, H.; Atsumaru, K.; Mochizuki, T. Effect of mutations in the 2b protein of tomato aspermy virus on RNA silencing suppressor activity, virulence, and virus-induced gene silencing. Arch. Virol. 2022, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Palukaitis, P.; García-Arenal, F. Cucumoviruses. Adv. Virus Res. 2003, 62, 241–323. [Google Scholar] [CrossRef] [PubMed]
- Hayes, R.J.; Buck, K.W. Complete Replication of a Eukaryotic Virus RNA in Vitro by a Purified RNA-Dependent RNA Polymerase. Cell 1990, 63, 363–368. [Google Scholar] [CrossRef]
- Ding, S.-W.; Li, W.-X.; Symons, R.H. A Novel Naturally Occurring Hybrid Gene Encoded by a Plant RNA Virus Facilitates Long Distance Virus Movement. EMBO J. 1995, 14, 5762–5772. [Google Scholar] [CrossRef] [PubMed]
- Schwinghamer, M.W.; Symons, R.H. Fractionation of Cucumber Mosaic Virus RNA and Its Translation in a Wheat Embryo Cell-Free System. Virology 1975, 63, 252–262. [Google Scholar] [CrossRef]
- Nakano, M.; Taniguchi, K.; Masuda, Y.; Kozuka, T.; Aruga, Y.; Han, J.; Motohara, K.; Nakata, M.; Sumitomo, K.; Hisamatsu, T.; et al. A Pure Line Derived from a Self-Compatible Chrysanthemum Seticuspe Mutant as a Model Strain in the Genus Chrysanthemum. Plant Sci. 2019, 287, 110174. [Google Scholar] [CrossRef]
- Oda, A.; Narumi, T.; Li, T.; Kando, T.; Higuchi, Y.; Sumitomo, K.; Fukai, S.; Hisamatsu, T. CsFTL3, a Chrysanthemum FLOWERING LOCUS T-like Gene, Is a Key Regulator of Photoperiodic Flowering in Chrysanthemums. J. Exp. Bot. 2012, 63, 1461–1477. [Google Scholar] [CrossRef] [Green Version]
- Feng, M.; Zhang, H.; Pan, Y.; Hu, Y.; Chen, J.; Zuo, D.; Jiang, T. Complete Nucleotide Sequence of Strawberry Vein Banding Virus Chinese Isolate and Infectivity of Its Full-Length DNA Clone. Virol. J. 2016, 13, 164. [Google Scholar] [CrossRef] [Green Version]
- Yan, H.; Fu, D.; Zhu, B.; Liu, H.; Shen, X.; Luo, Y. Sprout Vacuum-Infiltration: A Simple and Efficient Agroinoculation Method for Virus-Induced Gene Silencing in Diverse Solanaceous Species. Plant Cell Rep. 2012, 31, 1713–1722. [Google Scholar] [CrossRef]
- Senthil-Kumar, M.; Hema, R.; Anand, A.; Kang, L.; Udayakumar, M.; Mysore, K.S. A Systematic Study to Determine the Extent of Gene Silencing in Nicotiana Benthamiana and Other Solanaceae Species When Heterologous Gene Sequences Are Used for Virus-Induced Gene Silencing. New Phytol. 2007, 176, 782–791. [Google Scholar] [CrossRef]
- Osaka, M.; Itabashi, T.; Chiba, N.; Sumitomo, K.; Matsushita, Y. The Effects on Adventitious Root Formation Caused by Chrysanthemum Stunt Viroid in Chrysanthemum Morifolium and C. Seticuspe. J. Phytopathol. 2021, 169, 710–715. [Google Scholar] [CrossRef]
- Zhou, Y.; Deng, Y.; Liu, D.; Wang, H.; Zhang, X.; Liu, T.; Wang, J.; Li, Y.; Ou, L.; Liu, F.; et al. Promoting Virus-Induced Gene Silencing of Pepper Genes by a Heterologous Viral Silencing Suppressor. Plant Biotechnol. J. 2021, 19, 2398–2400. [Google Scholar] [CrossRef] [PubMed]
- Goodin, M.M.; Zaitlin, D.; Naidu, R.A.; Lommel, S.A. Nicotiana Benthamiana: Its History and Future as a Model for Plant-Pathogen Interactions. Mol. Plant Microbe Interact. 2008, 21, 1015–1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaffar, F.Y.; Koch, A. Catch Me If You Can! RNA Silencing-Based Improvement of Antiviral Plant Immunity. Viruses 2019, 11, 673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Killiny, N.; Nehela, Y.; Hijaz, F.; Ben-Mahmoud, S.K.; Hajeri, S.; Gowda, S. Citrus Tristeza Virus-Based Induced Gene Silencing of Phytoene Desaturase Is More Efficient When Antisense Orientation Is Used. Plant Biotechnol. Rep. 2019, 13, 179–192. [Google Scholar] [CrossRef]
- Lacomme, C.; Hrubikova, K.; Hein, I. Enhancement of Virus-Induced Gene Silencing through Viral-Based Production of Inverted-Repeats. Plant J. 2003, 34, 543–553. [Google Scholar] [CrossRef]
- Zhang, C.; Bradshaw, J.D.; Whitham, S.A.; Hill, J.H. The Development of an Efficient Multipurpose Bean Pod Mottle Virus Viral Vector Set for Foreign Gene Expression and RNA Silencing. Plant Physiol. 2010, 153, 52–65. [Google Scholar] [CrossRef] [Green Version]
- Liu, E.; Page, J.E. Optimized CDNA Libraries for Virus-Induced Gene Silencing (VIGS) Using Tobacco Rattle Virus. Plant Methods 2008, 4, 5. [Google Scholar] [CrossRef] [Green Version]
- Yamagishi, M.; Masuta, C.; Suzuki, M.; Netsu, O. Peanut Stunt Virus-Induced Gene Silencing in White Lupin (Lupinus Albus). Plant Biotechnol. 2015, 32, 181–191. [Google Scholar] [CrossRef] [Green Version]
- Peele, C.; Jordan, C.V.; Muangsan, N.; Turnage, M.; Egelkrout, E.; Eagle, P.; Hanley-Bowdoin, L.; Robertson, D. Silencing of a Meristematic Gene Using Geminivirus-Derived Vectors: Meristematic Gene Silencing Using Geminivirus-Derived Vectors. Plant J. 2001, 27, 357–366. [Google Scholar] [CrossRef] [Green Version]
- Hiriart, J.-B.; Lehto, K.; Tyystjärvi, E.; Junttila, T.; Aro, E.-M. Suppression of a Key Gene Involved in Chlorophyll Biosynthesis by Means of Virus-Inducing Gene Silencing. Plant Mol. Biol. 2002, 50, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Bruun-Rasmussen, M.; Madsen, C.T.; Jessing, S.; Albrechtsen, M. Stability of Barley Stripe Mosaic Virus–Induced Gene Silencing in Barley. Mol. Plant Microbe Interact. 2007, 20, 1323–1331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
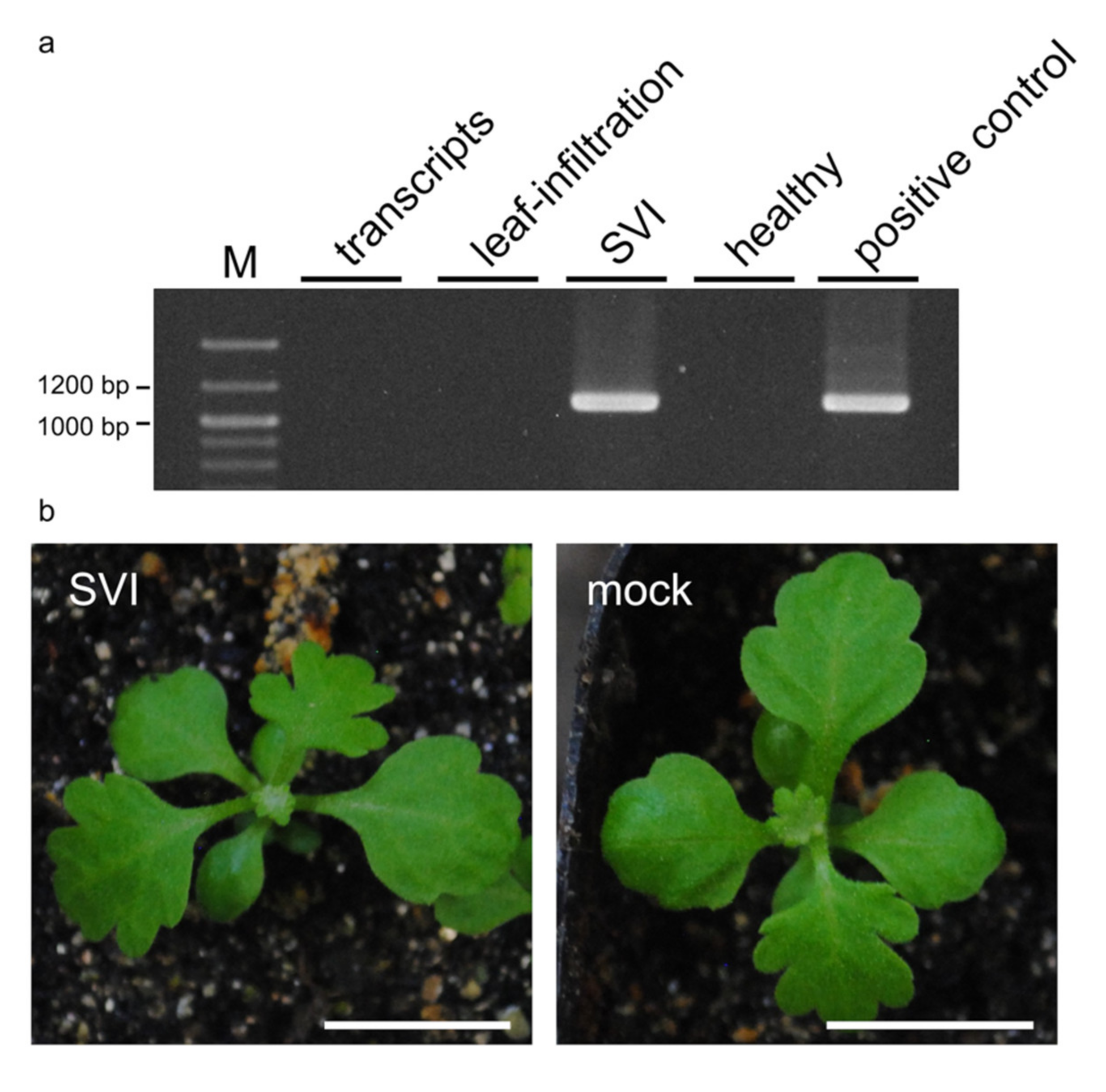
| Inoculation Method | No. of Inoculated C. seticuspe a | No. of TAV-Detected C. seticuspe b | Detection Percentage c |
|---|---|---|---|
| Transcripts | 12 | 0 | 0% |
| Leaf infiltration | 12 | 0 | 0% |
| Sprout vacuum infiltration | 20 | 10 | 50% |
| 2b Variation | No. of Inoculated C. seticuspe a | No. of TAV-Detected C. seticuspe b | Detection Percentage c |
|---|---|---|---|
| wt | 20 | 10 | 50% |
| ΔC61 | 22 | 0 | 0% |
| ΔC23 | 19 | 0 | 0% |
| R46C | 20 | 3 | 15% |
| S4042A | 24 | 1 | 4.2% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murai, H.; Mochizuki, T. Virus-Induced Gene Silencing in Chrysanthemum seticuspe Using the Tomato Aspermy Virus Vector. Plants 2022, 11, 430. https://doi.org/10.3390/plants11030430
Murai H, Mochizuki T. Virus-Induced Gene Silencing in Chrysanthemum seticuspe Using the Tomato Aspermy Virus Vector. Plants. 2022; 11(3):430. https://doi.org/10.3390/plants11030430
Chicago/Turabian StyleMurai, Hirotomo, and Tomofumi Mochizuki. 2022. "Virus-Induced Gene Silencing in Chrysanthemum seticuspe Using the Tomato Aspermy Virus Vector" Plants 11, no. 3: 430. https://doi.org/10.3390/plants11030430
APA StyleMurai, H., & Mochizuki, T. (2022). Virus-Induced Gene Silencing in Chrysanthemum seticuspe Using the Tomato Aspermy Virus Vector. Plants, 11(3), 430. https://doi.org/10.3390/plants11030430






