Proteomic Changes in Response to Colorless nonripening Mutation during Tomato Fruit Ripening
Abstract
1. Introduction
2. Results
2.1. The Biochemical Characteristics of Cnr Fruit
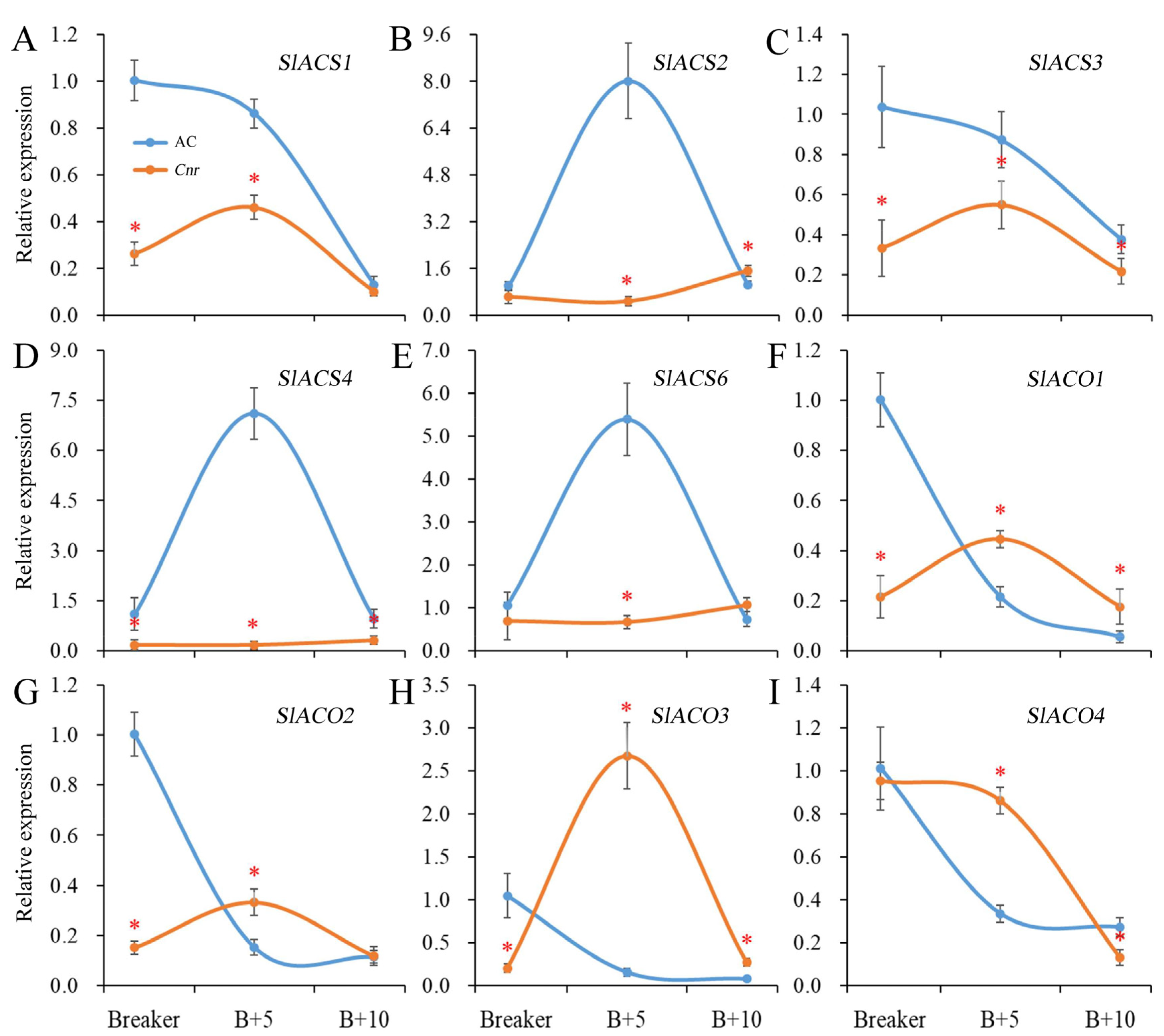
2.2. Expression of Genes Involved in Ethylene Biosynthesis in Cnr Fruit
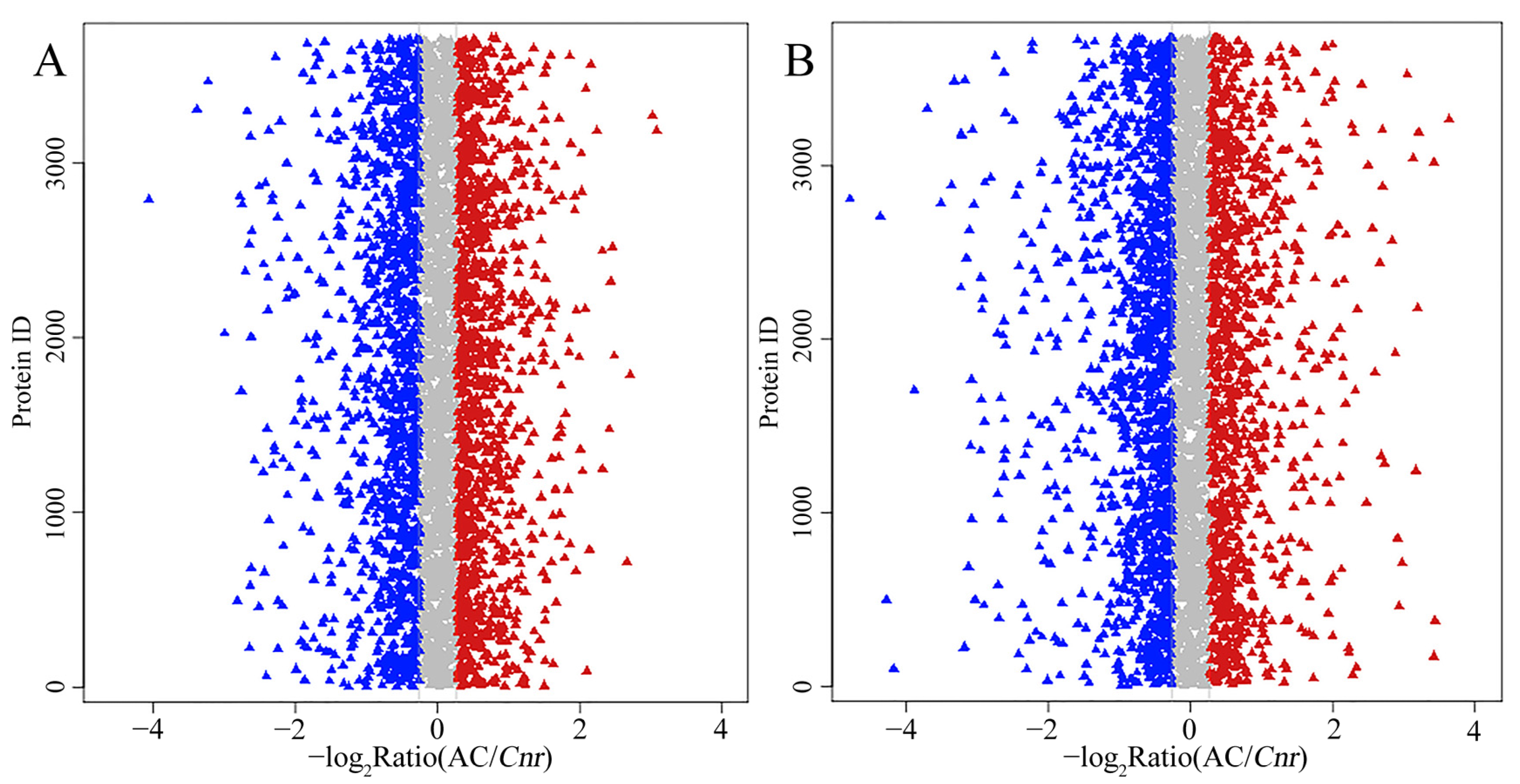
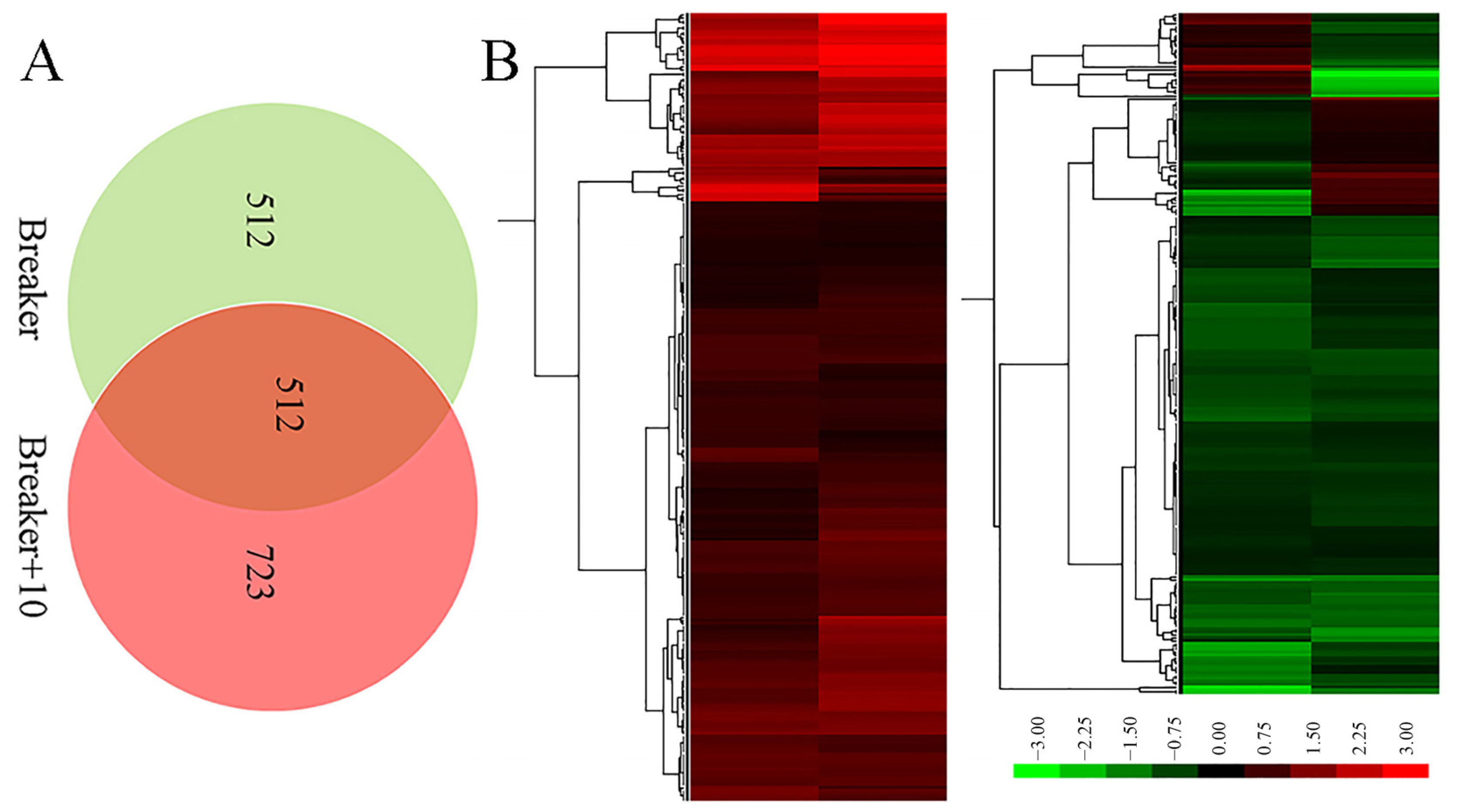
2.3. A Global View of iTRAQ Analysis
2.4. Function Classification of DEGs
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Determination of Biochemical Parameters
4.3. Relative Expression Analysis of Genes Involved in Ethylene Biosynthesis
4.4. Comparative Proteomics Analysis
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- The Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Pesaresi, P.; Mizzotti, C.; Colombo, M.; Masiero, S. Genetic regulation and structural changes during tomato fruit development and ripening. Front. Plant Sci. 2014, 5, 124. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, Y.; Nicolas, P.; Fernandez-Pozo, N.; Ma, Q.Y.; Evanich, D.J.; Shi, Y.N.; Xu, Y.M.; Zheng, Y.; Snyder, S.I.; Martin, L.B.B.; et al. High-resolution spatiotemporal transcriptome mapping of tomato fruit development and ripening. Nat. Commun. 2018, 9, 364. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.L.; Fei, Z.J.; Chen, Y.R.; Zheng, Y.; Huang, M.Y.; Vrebalov, J.; McQuinn, R.; Gapper, N.; Liu, B.; Xiang, J.; et al. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 2013, 31, 2. [Google Scholar] [CrossRef]
- Karlova, R.; Chapman, N.; David, K.; Angenent, G.C.; Seymour, G.B.; De-Maagd, R.A. Transcriptional control of fleshy fruit development and ripening. J. Exp. Bot. 2014, 65, 4527–4541. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Wang, W.H.; Cai, J.H.; Zhang, Y.R.; Qing, G.Z.; Tian, S.P. Tomato nuclear proteome reveals the involvement of specific E2 ubiquitin-conjugating enzymes in fruit ripening. Genome Biol. 2014, 15, 548. [Google Scholar] [CrossRef]
- Li, C.X.; Hou, X.M.; Qi, N.N.; Liu, H.W.; Li, Y.H.; Huang, D.J.; Wang, C.L. Insight into ripening-associated transcription factors in tomato: A review. Sci. Hortic.-Amst. 2021, 288, 110363. [Google Scholar] [CrossRef]
- Klee, H.J.; Giovannoni, J.J. Genetics and control of tomato fruit ripening and quality attributes. Annu. Rev. Genet. 2011, 45, 41–59. [Google Scholar] [CrossRef]
- Liu, R.E.; How-Kit, A.; Stammitti, L.; Teyssier, E.; Rolin, D.; Mortain-Bertrand, A.; Halle, S.; Liu, M.C.; Kong, J.H.; Wu, C.Q.; et al. A DEMETER-like DNA demethylase governs tomato fruit ripening. Proc. Natl. Acad. Sci. USA 2015, 112, 34. [Google Scholar] [CrossRef]
- Cedillo-Jimenez, C.A.; Feregrino-Perez, A.A.; Guevara-González, R.G.; Cruz-Hernández, A. MicroRNA regulation during the tomato fruit development and ripening: A review. Sci. Hortic. 2020, 270, 109435. [Google Scholar] [CrossRef]
- Lai, T.F.; Wang, X.H.; Ye, B.S.; Jin, M.F.; Chen, W.W.; Wang, Y.; Zhou, Y.Y.; Blanks, A.M.; Gu, M.; Zhang, P.C.; et al. Molecular and functional characterization of the SBP-box transcription factor SlSPL-CNR in tomato fruit ripening and cell death. J. Exp. Bot. 2020, 30, 2995–3011. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.L.; Mu, J.L.; Grierson, D.; Wang, Y.X.; Gao, L.P.; Zhao, X.Y.; Zhu, B.Z.; Luo, Y.B.; Shi, K.; Wang, Q.; et al. Noncoding RNAs: Functional regulatory factors in tomato fruit ripening. Theor. Appl. Genet. 2020, 133, 1753–1762. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.C.; Liu, X.C.; Jiang, G.X.; Li, Z.W.; Song, Y.B.; Zhang, D.D.; Jiang, Y.M.; Duan, X.W. SlJMJ7 orchestrates tomato fruit ripening via crosstalk between H3K4me3 and DML2-mediated DNA demethylation. N. Phytol. 2022, 233, 1202–1219. [Google Scholar] [CrossRef]
- Kou, X.H.; Zhou, J.Q.; Wu, C.E.; Yang, S.; Liu, Y.F.; Chai, L.P.; Xue, Z.H. The interplay between ABA/ethylene and NAC TFs in tomato fruit ripening: A review. Plant Mol. Biol. 2021, 106, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Chen, Y.; Liu, Z.Y.; Liu, Z.Q.; Shu, P.; Wang, R.C.; Hao, Y.W.; Su, D.; Pirrello, J.; Liu, Y.S.; et al. SlERF.F12 modulates the transition to ripening in tomato fruit by recruiting the co-repressor TOPLESS and histone deacetylases to repress key ripening genes. Plant Cell 2022, 34, 1250–1272. [Google Scholar] [CrossRef]
- Guo, J.E. Histone deacetylase gene SlHDT1 regulates tomato fruit ripening by affecting carotenoid accumulation and ethylene biosynthesis. Plant Sci. 2022, 318, 111235. [Google Scholar] [CrossRef]
- Wang, R.F.; Angenent, G.C.; Seymour, G.; DeMaagd, R.A. Revisiting the role of master regulators in tomato ripening. Trends Plant Sci. 2020, 25, 291–301. [Google Scholar] [CrossRef]
- Gupta, A.; Upadhyay, R.K.; Prabhakar, R.; Tiwari, N.; Garg, R.; Sane, V.A.; Sane, A.P. SlDREB3, a negative regulator of ABA responses, controls seed germination, fruit size and the onset of ripening in tomato. Plant Sci. 2022, 319, 111249. [Google Scholar] [CrossRef]
- Huang, W.; Hu, N.; Xiao, Z.; Qiu, Y.P.; Yang, Y.; Yang, J.; Mao, X.; Wang, Y.C.; Wang, Y.C.; Li, Z.G.; et al. A molecular framework of ethylene-mediated fruit growth and ripening processes in tomato. Plant Cell 2022, 34, 3280–3300. [Google Scholar] [CrossRef]
- Klein, J.; Saedler, H.; Huijser, P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 1996, 250, 7–16. [Google Scholar]
- Salinas, M.; Xing, S.P.; Höhmann, S.; Berndtgen, R.; Huijser, P. Genomic organization, phylogenetic comparison and differential expression of the SBP-box family of transcription factors in tomato. Planta 2012, 235, 1171–1184. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, E.M.; Bovy, A.; Manning, K.; Harrison, L.; Andrews, J.; De-Silva, J.; Tucker, G.A.; Seymour, G.B. Effect of the Colorless non-ripening mutation on cell wall biochemistry and gene expression during tomato fruit development and ripening. Plant Physiol. 2004, 136, 4184–4197. [Google Scholar] [CrossRef] [PubMed]
- Manning, K.; Tör, M.; Poole, M.; Hong, Y.G.; Thompson, A.J.; King, G.J.; Giovannoni, J.J.; Seymour, G.B. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 2006, 38, 8. [Google Scholar] [CrossRef] [PubMed]
- Lai, T.F.; Wang, Y.; Zhou, T.; Mei, F.L.; Zhang, P.C.; Zhou, Y.Y.; Shi, N.N.; Hong, Y.G. Virus-induced LeSPL-CNR silencing inhibits fruit ripening in tomato. J. Agr. Sci. 2015, 7, 184–195. [Google Scholar] [CrossRef][Green Version]
- Gao, Y.; Zhu, N.; Zhu, X.F.; Wu, M.; Jiang, C.Z.; Grierson, D.; Luo, Y.B.; Shen, W.; Zhong, S.L.; Fu, D.Q.; et al. Diversity and redundancy of the ripening regulatory networks revealed by the fruitENCODE and the new CRISPR/Cas9 CNR and NOR mutants. Hortic. Res. 2019, 6, 39. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.W.; Zhu, H.H.; Wang, J.Y.; Han, G.H.; Huang, R.N.; Hong, Y.G.; Yang, J.L. Comparative physiological and transcriptomic analyses reveal altered Fe-deficiency responses in tomato epimutant Coloress non-ripening. Front. Plant Sci. 2022, 12, 796893. [Google Scholar] [CrossRef]
- Zhu, H.H.; Wang, J.Y.; Jiang, D.; Hong, Y.G.; Xu, J.M.; Zheng, S.J.; Yang, J.L. The miR157-SPL-CNR module acts upstream of bHLH101 to negatively regulate iron deficiency responses in tomato. J. Integr. Plant Biol. 2022, 64, 1059–1075. [Google Scholar] [CrossRef]
- Chen, W.W.; Kong, J.H.; Lai, T.F.; Manning, K.; Wu, C.Q.; Wang, Y.; Qin, C.; Li, B.; Yu, Z.M.; Zhang, X.; et al. Tuning LeSPL-CNR expression by SlymiR157 affects tomato fruit ripening. Sci. Rep. 2015, 5, 7852. [Google Scholar] [CrossRef]
- Chen, W.W.; Kong, J.H.; Qin, C.; Yu, S.; Tan, J.J.; Chen, Y.R.; Wu, C.Q.; Wang, H.; Shi, Y.; Li, C.Y.; et al. Requirement of CHROMOMETHYLASE3 for somatic inheritance of the spontaneous tomato epimutation Colourless Non-Ripening. Sci. Rep. 2015, 5, 9192. [Google Scholar] [CrossRef]
- Huan, C.; Jiang, L.; An, X.J.; Kang, R.Y.; Yu, M.L.; Ma, R.J. Potential role of glutathione peroxidase gene family in peach fruit ripening under combined postharvest treatment with heat and 1-MCP. Postharvest Biol. Technol. 2016, 111, 175–184. [Google Scholar] [CrossRef]
- Yao, M.M.; Ge, W.Y.; Zhou, Q.; Zhou, X.; Luo, M.L.; Zhao, Y.B.; Wei, B.D. Exogenous glutathione alleviates chilling injury in postharvest bell pepper by modulating the ascorbate-glutathione (AsA-GSH) cycle. Food Chem. 2021, 352, 129458. [Google Scholar] [CrossRef]
- Zhou, Y.; Huang, X.G.; Li, R.S.; Lin, H.S.; Huang, Y.W.; Zhang, T.; Mo, Y.X.; Liu, K.D. Transcriptome and biochemical analyses of glutathione-dependent regulation of tomato fruit ripening. J. Plant Interact. 2022, 17, 537–547. [Google Scholar] [CrossRef]
- Alexander, L.; Grierson, D. Ethylene biosynthesis and action in tomato: A model for climacteric fruit ripening. J. Exp. Bot. 2002, 53, 2039–2055. [Google Scholar] [CrossRef]
- Liu, L.H.; Liu, H.R.; Li, S.; Zhang, X.; Zhang, M.; Zhu, N.; Dufresne, C.P.; Chen, S.X.; Wang, Q.M. Regulation of BZR1 in fruit ripening revealed by iTRAQ proteomics analysis. Sci. Rep. 2016, 6, 33635. [Google Scholar] [CrossRef]
- Giovannoni, J. Molecular biology of fruit maturation and ripening. Annu. Rev. Plant Biol. 2001, 52, 725–749. [Google Scholar] [CrossRef]
- Houben, M.; de Poel, V. 1-aminocyclopropane-1-carboxylic acid oxidase (ACO): The enzyme that makes the plant hormone ethylene. Front. Plant Sci. 2019, 10, 695. [Google Scholar] [CrossRef]
- Katz, J.J.; Norris, J.R.; Shipman, L.L.; Thurnauer, M.C.; Wasielewski, M.R. Chlorophyll function in the photosynthetic reaction center. Ann. Rev. Biophys. Bioeng. 1978, 7, 393–434. [Google Scholar] [CrossRef] [PubMed]
- Llorente, B.; D’Andrea, L.; Ruiz-Sola, M.A.; Botterweg, E.; Pulido, P.; Andilla, J.; Loza-Alvarez, P.; Rodriguez-Concepcion, M. Tomato fruit carotenoid biosynthesis is adjusted to actual ripening progression by a light-dependent mechanism. Plant J. 2016, 85, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Slimestad, R.; Fossen, T.; Verheul, M.J. The flavonoids of tomatoes. J. Agric. Food Chem. 2008, 56, 2436–2441. [Google Scholar] [CrossRef]
- Agati, G.; Tattini, M. Multiple functional roles of flavonoids in photoprotection. N. Phytol. 2010, 186, 786–793. [Google Scholar] [CrossRef] [PubMed]
- Samkumar, A.; Karppinen, K.; McGhie, T.K.; Espley, R.V.; Martinussen, I.; Jaakola, L. Flavonoid biosynthesis is differentially altered in detached and attached ripening bilberries in response to spectral light quality. Front. Plant Sci. 2022, 13, 969934. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Fourdin, R.; Quadrado, M.; Dargel-Graffin, C.; Tolleter, D.; Macherel, D.; Mireau, H. Rerouting of ribosomal proteins into splicing in plant organelles. Proc. Natl. Acad. Sci. USA 2020, 24, 29979–29987. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.D.; Zhang, X.L.; Yang, Y.X.; Zhang, H.; Zhu, W.M.; Nie, W.F. The histone variant SlH2A.Z regulates carotenoid biosynthesis and gene expression during tomato fruit ripening. Hortic. Res. 2021, 8, 85. [Google Scholar] [CrossRef]
- Guo, J.E.; Hu, Z.L.; Li, F.F.; Zhang, L.C.; Xu, X.H.; Tang, B.Y. Silencing of histone deacetylase SlHDT3 delays fruit ripening and suppresses carotenoid accumulation in tomato. Plant Sci. 2017, 265, 29–38. [Google Scholar] [CrossRef]
- Guo, J.E.; Hu, Z.L.; Zhu, M.K.; Li, F.F.; Zhu, Z.G.; Lu, Y.; Chen, G.P. The tomato histone deacetylase SlHDA1 contributes to the repression of fruit ripening and carotenoid accumulation. Sci. Rep. 2017, 7, 7930. [Google Scholar] [CrossRef]
- Wang, J.F.; Li, G.B.; Li, C.X.; Zhang, C.L.; Cui, L.; Guo, A.; Wang, X.; Zheng, F.Y.; Zhang, D.L.; Larkin, R.M.; et al. NF-Y plays essential roles in flavonoid biosynthesis by modulating histone modifications in tomato. N. Phytol. 2021, 229, 3237–3252. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, G.Y.; Li, Y.S.; Zhang, J.; Shi, M.J.; Muhammad, T.; Liang, Y. Transcriptome profiling of tomato uncovers and involvement of cytochrome P450s and peroxidases in stigma color formation. Front. Plant Sci. 2017, 8, 897. [Google Scholar] [CrossRef]
- Yasumoto, S.; Seki, H.; Shimizu, Y.; Fukushima, E.O.; Muranaka, T. Functional characterization of CYP716 family P450 enzymes in triterpenoid biosynthesis in tomato. Front. Plant Sci. 2017, 8, 21. [Google Scholar] [CrossRef] [PubMed]
- Brekke, T.D.; Stroud, J.A.; Shaw, D.S.; Crawford, S.; Steele, K.A. QTL mapping in salad tomatoes. Euphytica 2019, 215, 115. [Google Scholar] [CrossRef]
- Wang, X.H.; Ye, B.S.; Kang, X.P.; Zhou, T.; Lai, T.F. Potato virus x-induced LeHB-1 silencing delays tomato fruit ripening. J. Am. Soc. Hortic. Sci. 2018, 143, 454–461. [Google Scholar] [CrossRef]
- Xu, F.; Yuan, Q.P.; Dong, H.R. Determination of lycopene and β-carotene by high-performance liquid chromatography using sudan I as internal standard. J. Chromatogr. B 2006, 838, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.Y.; Tang, C.Y. Determination of total phenolic and flavonoid contents in selected fruits and vegetables, as well as their stimulatory effects on mouse splenocyte proliferation. Food Chem. 2006, 01, 140–147. [Google Scholar] [CrossRef]
- Zhang, H.B.; Jordheim, M.; Lewis, D.H.; Arathoon, S.; Andersen, Ø.M.; Davies, K.M. Anthocyanins and their differential accumulation in the floral and vegetative tissues of a shrub species (Rhabdothamnus solandri A. Cunn). Sci. Hortic. 2014, 165, 29–35. [Google Scholar] [CrossRef]
- Wang, Q.; Lai, T.F.; Qin, G.Z.; Tian, S.P. Response of jujube fruits to exogenous oxalic acid treatment based on proteomic analysis. Plant Cell Physiol. 2009, 50, 230–242. [Google Scholar] [CrossRef]
- Lai, T.F.; Wang, Y.; Bai, X.L.; Qi, Q.Q.; Xu, M.J.; Zhou, T. Dissecting inhibitory effect of boric acid on virulence and patulin production of Penicillium expasum. Postharvest Biol. Technol. 2016, 117, 187–196. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Kristensen, D.M.; Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Microbial genome analysis: The COG approach. Brief. Bioinform. 2019, 20, 1063–1070. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, T.; Li, R.; Yu, Q.; Wang, J.; Pan, J.; Lai, T. Proteomic Changes in Response to Colorless nonripening Mutation during Tomato Fruit Ripening. Plants 2022, 11, 3570. https://doi.org/10.3390/plants11243570
Zhou T, Li R, Yu Q, Wang J, Pan J, Lai T. Proteomic Changes in Response to Colorless nonripening Mutation during Tomato Fruit Ripening. Plants. 2022; 11(24):3570. https://doi.org/10.3390/plants11243570
Chicago/Turabian StyleZhou, Ting, Ran Li, Qinru Yu, Jingjing Wang, Jingjing Pan, and Tongfei Lai. 2022. "Proteomic Changes in Response to Colorless nonripening Mutation during Tomato Fruit Ripening" Plants 11, no. 24: 3570. https://doi.org/10.3390/plants11243570
APA StyleZhou, T., Li, R., Yu, Q., Wang, J., Pan, J., & Lai, T. (2022). Proteomic Changes in Response to Colorless nonripening Mutation during Tomato Fruit Ripening. Plants, 11(24), 3570. https://doi.org/10.3390/plants11243570




