Abstract
Orthotrichoideae aggregates epiphytic mosses widespread throughout temperate regions and high tropical mountains of the world. Recently, important advances have been made in elucidating its phylogenetic relationships and evolutionary patterns. Fourteen genera are currently recognized within the subfamily, which are spread over two main tribes: Orthotricheae, comprising Orthotrichinae and Lewinskyinae, and Zygodonteae. Despite the progress, some groups have received little attention, as is the case of genus Codonoblepharon. Recent studies have suggested that this genus may represent a separate lineage from Zygodonteae, in which it traditionally has been considered. Although, none of the studies were conclusive as they did not include a representative sampling of the Codonoblepharon species. This work aims to evaluate the taxonomic position of Codonoblepharon and its phylogenetic relationships within Orthotrichoideae. For this purpose, we present an updated phylogenetic tree based on four different loci, one belonging to the nuclear genome (ITS2) and the rest to the plastid genome (rps4, trnG and trnL-F). The phylogenetic reconstruction recovers all samples of Codonoblepharon in a monophyletic group, sister to the rest of the subfamily, constituting a lineage independent of the two currently recognized tribes. For this reason, we propose the new tribe Codonoblepharonteae to accommodate Codonoblepharon.
1. Introduction
Mosses are the most diversified lineage among Bryophytes [1,2]. Recent years have witnessed important advances in the elucidation of the phylogenetic relationships among the major lineages of mosses, e.g., order level and above [3,4,5]. However, much remains to be clarified at lower levels, which is a key issue to establish a robust molecular-based classification, especially in the case of larger families.
Orthotrichaceae Arn. is the second most speciose family of mosses, with an estimated 900 species [6,7]. Most are epiphytic taxa, both from tropical and temperate environments, although in each of these major climatic regions one of the two subfamilies of the group predominates. Macromitrioideae Broth. are cladocarpous mosses with almost exclusively tropical distribution, whereas Orthotrichoideae Broth. includes acrocarpous mosses that inhabit temperate regions of both hemispheres and high tropical mountains [8]. Orthotrichoideae is better known, both in terms of specific diversity, e.g., [9] and phylogenetically, e.g., [6,10,11]. Recently, Draper et al. [12,13] have provided new and more complete insights in the phylogenetic framework of the subfamily, besides some of the evolutionary patterns underlying its complexity. According to these works, Orthotrichoideae is composed of fourteen genera. Ten of them are grouped into Orthotricheae, a tribe that in turn integrates two well differentiated lineages recognized as subtribes: (i) Orthotrichinae, that includes Orthotrichum s.str., the most diversified genus; (ii) and Lewinskyinae, which includes Lewinskya and Ulota, the other two major genera of the tribe. The other four recognized genera are currently integrated into Zygodonteae [12], with Zygodon s.str. as the most species-rich genus of this tribe (Table 1).

Table 1.
Classification of Orthotrichoideae according to Draper et al. [12,13].
The phylogenetic reconstruction obtained by Draper et al. [12] did not conclusively resolve the analysed representation of Zygodonteae, as it separated its components in two well-supported clades but for which no robust conclusions could be drawn about their sister relationships. The results showed that one of the genera, Codonoblepharon, could constitute a separate lineage from Zygodonteae. Nevertheless, the placing of that lineage in a polytomy, together with a clade containing the rest of the genera of Zygodonteae and a clade containing the genera of Orthotrichoideae, advised postponing the possible consequences for the systematic of the subfamily until obtaining robust evidence.
Codonoblepharon, a genus initially conceived as grouping very heterogeneous taxa [14], was later circumscribed by Goffinet and Vitt [8] to segregate a section from Zygodon (Sect. Bryoides Malta), mainly characterized by the absence of papillae in its leaf cells [15]. Thus conceived, Codonoblepharon contains about seven mainly tropical and southern hemisphere species [14,16], although there are discrepancies about the ascription of C. forsteri (Dicks.) Goffinet, the only one restricted to the northern hemisphere. While studies based on morphological or biogeographical evidence suggest the exclusion of this species from Codonoblepharon [8,14], others based on phylogenetic reconstructions with partial taxa representation [6,12] indicate that it should be recognized as part of this genus.
In the present work we evaluate the hypothesis, based on the results obtained by Draper et al. [12], that Codonoblepharon represents a lineage independent of Zygodonteae. Our main objective is to achieve a robust phylogenetic reconstruction that would unequivocally reflect the relationships of Codonoblepharon within the subfamily Orthotrichoideae. Besides, we pursue to obtain more information about the phylogenetic structure of Codonoblepharon and the taxonomic position of C. forsteri.
2. Results
The independent analysis of the four markers resulted in phylogenetic trees with congruent topologies, which allowed concatenation. As shown in Table 2, the concatenated matrix resulted in a total length of 2065 bp, of which 550 were parsimony informative. In addition, indel coding added 153 informative variable positions. All the analyses performed (indels treatment and phylogenetic reconstruction method) were congruent. The best resolved phylogeny was originated from the BI analysis of the concatenated matrix with coded indels (Figure 1).

Table 2.
Number of variable and informative positions for the concatenated matrix and for the four analysed markers.
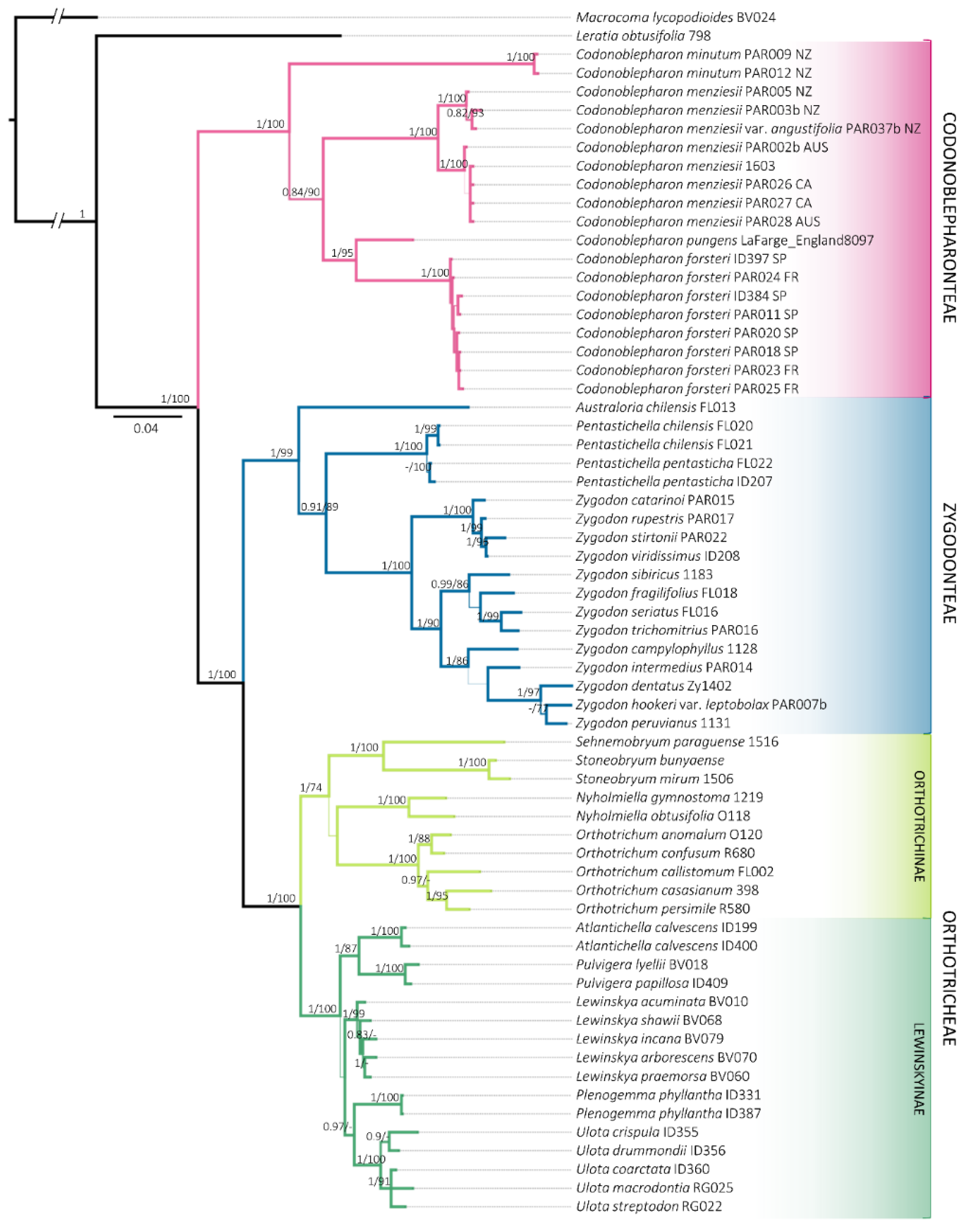
Figure 1.
Consensus phylogenetic tree obtained from the Bayesian Inference analysis with the combined matrix (ITS2, rps4, trnG, trnL-F and indels coded). Posterior probabilities greater than 0.8 for BI and Bootstrap values greater than 70% for ML are included. Sequences information is available in Table S1. The locality where the samples were collected is indicated for Codonoblepharonteae: Australia (AUS), California (CA), France (FR), New Zealand (NZ) and Spain (SP).
Representatives of Orthotrichoideae were resolved in three distinct clades. Zygodonteae, which until now had been treated as a single group, is divided in two maximally supported (1/100) lineages. On the one hand, Codonoblepharon appears as a monophyletic group (1/100), which is resolved as a sister clade to the rest of the subfamily. On the other hand, a lineage constituted of Pentastichella, Australoria and Zygodon is differentiated (1/99). This indicates that Zygodonteae s.l. represents a paraphyletic group, as was traditionally conceived. Conversely, Orthotricheae is grouped in a monophyletic clade (1/100), containing Lewinskyinae (1/100) and Orthotrichinae (1/74).
All the molecular markers used recover the samples of Codonoblepharon forsteri in a clade with maximal support that is nested within Codonoblepharon and sister to C. pungens (Müll.Hal.) A.Jaeger. This group is in turn sister to C. menziesii Schwägr., the type species of this genus.
3. Discussion
This work presents an updated molecular phylogeny with representation of all the accepted genera of the subfamily Orthotrichoideae based on Draper et al. [12], except for the recently described Rehubryum F.Lara, Garilleti and Draper [13] from Lewinskyinae. Special attention was paid to the tribe Zygodonteae and the genus Codonoblepharon. The variability and large number of analyses that have been conducted, consistent with each other, minimize the possibility of topological inconsistencies. The only analysed marker that has shown ambiguous alignments due to its high variability is nuclear ITS2. Nevertheless, the exclusion of these ambiguities did not affect phylogenetic results.
Orthotrichoideae is a taxonomically complex subfamily, which has led to numerous supraspecific reassignments. The species of Zygodon s.l., including the group with smooth leaf cells now segregated in the genus Codonoblepharon, have long been considered a large and important natural group. Unlike Orthotrichum s.l., the other traditional large genus of acrocarpous Orthotrichaceae, Zygodon s.l., includes mostly dioicous mosses, with sporophytes bearing a long seta and usually producing vegetative propagules on variably long and branched filamentous supports arising from the stem, never directly from the leaves, although propagules may be grouped in the leaf axils. Other distinguishing characteristics are the growth of the colonies forming lax turfs, rarely mats or cushions, the relatively small leaves with non or slightly recurved margins and little or no cell differentiation along the lamina, and the cucullate non-plicate calyptra, typically devoid of hairs. They typically grow on old or decaying bark of large trees or stumps. As in other groups of Orthotrichaceae, several species are saxicolous, either facultative or, more rarely, obligate [15,16] and [17] (pp. 15–135).
Malta [15], in the only world monograph on the genus, recognized a total of 77 Zygodon species grouped into four sections. Most of them (ca. 86%) belonged to the globally distributed section Euzygodon Müll.Hal. Section Stenomitrium Mitt. included a single species, with Andean and Patagonian distribution and deviant morphology due to its robust and creeping stems, leaves in pentastichous arrangement and dimorphic basal leaf cells. Section Obtusifolii Malta also contained a single species characterized by lingulate leaves with a rounded apex and papillose calyptra and a wide disjunct distribution including populations in Southeast Asia, Australasia, South America and Mexico. Finally, section Bryoides Malta included nine species, mainly distributed in the southern hemisphere and, furthermore, occurring in the north in some tropical localities and in Europe, characterized by smooth leaf cells. Malta [15] argued that this latter section represented a natural group that might merit subgeneric rank, as smooth cells are unusual among Orthotrichaceae. Malta’s taxonomic delimitation was basically followed by Calabrese [16], even though Goffinet and Vitt [8] had reinstated a few years earlier the genus Codonoblepharon for most of the species of section Bryoides and Bryomaltaea Goffinet to segregate Zygodon obtusifolius Hook (Table 3).

Table 3.
Evolution of the classification of Zygodon and related genera since Malta [15]. Genera not considered as belonging to tribe Zygodonteae in the respective study are marked with two asterisks (**).
Zygodon s.l. has been treated at various taxonomic ranks, including family (Zygodontaceae Schimp.) and subfamily (Zygodontoideae Broth.). These mosses have numerous characters shared with other Orthotrichaceae, but some deviate and more closely resemble representatives of other families, such as Ditrichaceae Limpr., Grimmiaceae Arn. or Pottiaceae Hampe [8,18,19], that are not phylogenetically close. In fact, Schimper [20] proposed the segregation of Zygodon as a family that he considered intermediate between Orthotrichaceae and Weissiaceae Schimp. (=Pottiaceae p.p.). Most of its components are currently included in Pottiaceae, and the proposal also included Amphidium Schimp., now placed in Amphidiaceae M.Stech. Among the characters that can be considered deviant are those that have been highlighted as being particularly significant [21]: the cucullate non-plicate calyptra and the orthotropic stems forming short turfs, not cushions or creeping mats as in other Orthotrichaceae. The current consideration of Zygodon s.l. as a tribe within Orthotrichoideae is relatively recent [8]. However, regardless of the taxonomic rank given to the group, it has traditionally been considered a fairly homogeneous entity, well differentiated from the rest of Orthotrichaceae. This idea that Zygodonteae is a natural and quite uniform group was only partially questioned in early molecular studies [6], which allowed the segregation of Bryomaltaea obtusifolia (Hook.) Goffinet as part of a phylogenetically distant lineage. However, this did not diminish the general consideration of Zygodonteae. Recently, however, Draper et al. [12] concluded that the genus Zygodon is a polyphyletic artificial group and their results supported the distinction of Pentastichella with the inclusion of Pleurorthotrichum Broth. and the establishment of the new genus Australoria (Table 1 and Table 3). Thus, Malta’s sections of Zygodon [15] are now treated as separate genera, Zygodon s.s. being currently restricted to the representatives of the section Euzygodon (Table 3).
The results of the present work support the distinction of Codonoblepharon as a separate lineage, as earlier suggested by Goffinet et al. [6] and Draper et al. [12]. Unequivocal evidence for this was obtained by inclusion in the analysis of a wide representation of the former Zygodonteae, including species with smooth leaf cells and a greater number of representatives with papillose leaf cells. The phylogenetic reconstruction obtained here places the monophyletic group constituted by Codonoblepharon as a sister group of the clade that includes all the other Orthotrichoideae. This suggests the need for recognition of this lineage as a separate tribe, which we propose to name Codonoblepharonteae; it is the third within the Orthotrichoideae, along with Zygodonteae and Orthotricheae. Thus, Codonoblepharon changes from being considered just a section of Zygodon to a major independent lineage among Orthotrichoideae. This taxonomic proposal is morphologically supported by smooth leaf cells, which is an exclusive character of Codonoblepharonteae within the subfamily. This classification is paralleled by Macromitrioideae, which contains a single genus characterized by entirely smooth leaf cells, Schlotheimia Brid., being also considered a separate tribe, Schlotheimieae Goffinet [8]. Future phylogenetic studies on Macromitrioideae may reveal the true relationships of this group with smooth cells within the subfamily.
The phylogenetic reconstruction (Figure 1) also shows that the lengths of the branches and the topology within Codonoblepharonteae are similar to what can be observed in other main lineages, such as Zygodonteae, which includes up to three genera. In contrast, in the new tribe all the terminals belong to a single genus, Codonoblepharon. This leads us to consider that the taxonomic diversity of the group is yet to be investigated and that the segregation of Codonoblepharon into several separate genera could be possible. Further in-depth studies are needed to unravel this possibly overlooked diversity, giving special attention to C. pungens, which, according to Malta [15], constitutes the systematic weak point of the group and to C. minutum (Müll.Hal. and Hampe) Matcham and O’Shea which in our reconstructions appears as the sister species of all the congeners included in the analysis. Further evidence of the possibly overlooked diversity among Codonoblepharonteae is the strong phylogenetic structure obtained for C. menziesii which could have considerable taxonomical and biogeographical significance. The New Zealand samples of C. menziesii are separated from the Australian and Californian ones, which could imply that they correspond to different taxa and might support the idea of Shevock [22] that the occurrence of C. menziesii in western North America is due to a recent introduction from Australia. Since Malta [15] already recognized a strong intraspecific morphological variability within C. menziesii, the current concept could hide a complex of species and calls for a deep integrative study.
Codonoblepharon forsteri is limited to Europe and northwestern Africa, making it the only representative of the genus with a Holarctic distribution [23]. Regarding its phylogenetic reconstruction, the samples are nested within the lineage of Codonoblepharonteae. Their segregation in a subclade together with C. pungens is significant as they are the only two autoicous species of the genus included in the analysis. In Orthotrichoideae, most genera are either dioicous or autoicous [12,13], which could support their segregation into a separate genus, although, once again, it is preferable to await the results of a more complete morphological and molecular study to resolve this in a robust and accurate manner.
4. Materials and Methods
4.1. Taxon Sampling
The final analyses were composed of 65 samples from 47 different taxa, out of which 94 sequences from 27 samples were newly obtained for this study. These include a representative from Australoria [A. chilensis (Calabrese and F.Lara) F.Lara, Garilleti and Draper], 4 taxa from Codonoblepharon [C. forsteri, C. menziesii, C. menziesii var. angustifolium (Malta) Matcham and O’Shea and C. minutum], the two known taxa from Pentastichella [P. chilensis (Broth.) F.Lara, Garilleti and Draper, and P. pentasticha (Mont.) Müll.Hal. ex Thér.], and 8 taxa from Zygodon [Z. catarinoi C.A.Garcia, F.Lara, Sérgio and Sim-Sim, Z. fragilifolius Broth. ex Malta, Z. hookeri var. leptobolax (Müll.Hal.) Calabrese, Z. intermedius Bruch and Schimp., Z. rupestris Schimp. ex Lorentz, Z. seriatus Thér. and Naveau, Z. stirtonii Schimp. and Z. trichomitrius Hook. and Wilson]. The information regarding these sequences is available in Table S1.
The identification of those samples was based on the analysis of microscopic characters of the leaves, stems, propagules, rhizoids, seta and capsules, according to the descriptions and taxonomical criteria of Lewinsky, Matcham and O’Shea, and Calabrese and Lara et al. [14,16,24] and [17] (pp. 17–27).
4.2. DNA Isolation and Amplification
Only the tip of a single gametophyte shoot from each sample was selected for DNA extraction to prevent contamination. The rest of the gametophyte, and the sporophyte if present, were preserved in a microscope slide fixed with glycerogelatin to allow identification revision. DNA was extracted using the standard DNeasy Plant Mini Kit protocol (QIAGEN). Nucleotide sequences were amplified by PCR from four genomic regions (Table 4): one of them from to the nuclear genome (ITS2) and the other three from to the plastid genome (rps4, trnG and trnL-F). PCRs were performed using Ready-To-Go™ PCR Beads (Amersham Pharmacia Biotech Inc.) in a final volume of 25 μL, initially with 2 μL of DNA, and in the case of subsequent PCR failure, with up to 10 μL. Amplification protocols are specified in Table 5. Amplification’s success was verified by electrophoresis and PCR products were purified using Exol/FastAP (Thermo Fisher Scientific, Spain) with 1 μL of Exonuclease and 4 μL of FastAP enzymes per tube, applying 37 °C and 85 °C for 15 min each. Finally, cleaned PCR products were sequenced by Macrogen. Two reads were obtained for each product, which were aligned using Geneious 2022.0.2.

Table 4.
Primers used for PCR amplification and sequencing.

Table 5.
PCR amplification protocols. Cycles are displayed in the first column of each region, heat and time in the second.
4.3. Molecular Analyses
The reconstruction of the phylogenetic relationships was conducted in a framework of the 65 aforementioned samples (Table S1). The genus Codonoblepharon was represented by 19 samples from 5 of the 8 accepted taxa. Outgroup were composed of 44 samples from 40 different taxa, representing 13 of the 14 genera currently accepted in the family [12,13]. Special attention was paid to Zygodonteae. In addition, based on the results of Goffinet et al. [6], Macrocoma lycopodioides (Schwägr.) Vitt was used to root the tree as a representative of Macromitrioideae, which was accompanied by Leratia obtusifolia (Hook.) Goffinet.
A matrix was constructed for each marker using PhyDE-1 0.9971 [31]. In order to eliminate uncertainties, 3′ and 5′ ends were trimmed in each matrix. Specifically, 99 bp in 3′ and 8 bp in 5′ for ITS2, 57 bp in 3′ and 10 bp in 5′ for rps4, 37 bp in 3′ and 63 bp in 5′ for trnG and 4 bp in 3′ and 18 bp in 5′ for trnL-F. The trimmed matrices were automatically aligned with MAFFT (Multiple Alignment using Fast Fourier Transform) using the EMBL-EBI multiple sequence alignment service [32]. Indels can sometimes lead to ambiguous alignments. Thus, each marker was analysed separately in three different ways: (1) indels considered as missing information, (2) indels coded as informative with the simple method of Simmons and Ochoterena [33] implemented in SeqState [34] and (3) removing divergent alignment zones using Gblocks 0.91b [35,36]. Gblocks values were modified, setting minimum length of a block to 5, allowing gap positions to “with half”, minimum number of sequences for a flank position to 25 and maximum number of contiguous non conserved positions to 10. The best evolutionary model and partition scheme were selected with PartitionFinder 2.1.1 [37,38,39] using the Bayesian information criterion (BIC).
Phylogenetic analyses were performed with BI using MrBayes Windows version 3.2.7a [40,41,42] and with ML using RAxML 8.2.12 [43] implemented in CIPRES [44]. BI analyses were run for 10,000,000 generations, saving trees and parameters every 1000. The initial 25% of the samples were not included in the consensus tree. Variable and informative positions were quantified with MEGA 11 [45,46]. Initially, each marker was analysed separately to detect possible incongruences, although the congruence of these markers in this group of mosses had already been verified in other studies, e.g., [12]. Once the congruence between the different resulting trees was visually confirmed, a combined matrix with the four markers was generated. The resulting phylogenetic trees were visualized and edited with FigTree 1.4.4 [47].
5. Conclusions
The present study has confirmed that the genus Codonoblepharon constitutes a separate lineage, which is resolved as sister from both Zygodonteae and Orthotricheae. This justifies the recognition of this lineage as an independent tribe, which we propose to name Codonoblepharonteae. Additionally, our results reveal that the diversity of the group is yet to be known, and that future integrative studies are necessary.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants11243557/s1, Table S1: Samples information. Those in italics were newly sequenced for this study.
Author Contributions
Conceptualization, F.L., I.D. and R.G.; morphological study, F.L. and P.A.-R.; DNA isolation and amplification, I.D., M.F. and P.A.-R.; phylogenetic analyses, I.D. and P.A.-R.; original draft preparation F.L., I.D. and P.A.-R.; writing—review and editing, F.L., I.D., P.A.-R., M.F. and R.G.; funding acquisition, F.L., I.D. and R.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Spanish Ministry of Economy, Industry and Competitiveness and the Spanish Research Agency of the Ministry of Science and Innovation (grant CGL2016-80772-P, PID202-115149GB-C21 and PID2020- 115149GB-C22), and by the European Union, Next Generation EU.
Data Availability Statement
Sequences used in this study were submitted to GenBank.
Acknowledgments
We are indebted to V. Huggonot, J. Brinda, A. Schäfer-Verwimp and D. Callaghan for kindly providing samples of Codonoblepharon. We also thank the referees for their help to improve the initial version of this manuscript.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Nomenclature
| Nomenclatural Changes: |
| Codonoblepharonteae F. Lara, Garilleti and Draper, tribe nova |
| TYPE: Codonoblepharon Schwägr. Species Muscorum Frondosorum, Supplementum Secundum 2: 142. pl. 137 p.p. 1824. |
References
- Goffinet, B.; Buck, W.R.; Shaw, A.J. Morphology, anatomy, and classification of the Bryophyta. In Bryophyte Biology, 2nd ed.; Goffinet, B., Shaw, A.J., Eds.; Camb: Cambridge, UK, 2009; pp. 55–138. [Google Scholar]
- Medina, N.G.; Draper, I.; Lara, F. Biogeography of Mosses and Allies: Does Size Matter? In Biogeography of Microscopic Organisms; Fontaneto, D., Ed.; Cambridge University Press: Cambridge, UK, 2011; pp. 209–233. [Google Scholar] [CrossRef]
- Chang, Y.; Graham, S.W. Inferring the Higher-Order Phylogeny of Mosses (Bryophyta) and Relatives Using a Large, Multigene Plastid Data Set. Am. J. Bot. 2011, 98, 839–849. [Google Scholar] [CrossRef]
- Cox, C.J.; Goffinet, B.; Wickett, N.J.; Boles, S.B.; Shaw, A.J. Moss Diversity: A Molecular Phylogenetic Analysis of Genera. Phytotaxa 2014, 9, 175–195. [Google Scholar] [CrossRef]
- Liu, Y.; Johnson, M.G.; Cox, C.J.; Medina, R.; Devos, N.; Vanderpoorten, A.; Hedenäs, L.; Bell, N.E.; Shevock, J.R.; Aguero, B.; et al. Resolution of the Ordinal Phylogeny of Mosses Using Targeted Exons from Organellar and Nuclear Genomes. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Goffinet, B.; Shaw, A.J.; Cox, C.J.; Wickett, N.J.; Boles, S.B. Phylogenetic Inferences in the Orthotrichoideae (Orthotrichaceae, Bryophyta) Based on Variation in Four Loci from All Genomes. Monogr. Syst. Bot. Missouri Bot. Gard. 2004, 98, 270–289. [Google Scholar]
- Frey, W.; Stech, M. Division of Bryophyta Schimp. (Musci, Mosses). In Syllabus of Plant Families. Adolf Engler’s Syllabus der Pflanzenfamilien, 13th edition. Part 3. Bryophytes and Seedless Vascular Plants; Frey, W., Ed.; Gebrüder Borntraeger: Berlin, Germany, 2009; pp. 116–257. [Google Scholar]
- Goffinet, B.; Vitt, D.H. Revised Generic Classification of the Orthotrichaceae Based on a Molecular Phylogeny and Comparative Morphology. In Bryology for the Twenty-first Century; Bates, J.W., Ashton, N.W., Duckett, J.G., Eds.; Maney Publishing and the British Bryological Society: Leeds, UK, 1998; pp. 143–160. [Google Scholar]
- Lara, F.; Garilleti, R.; Medina, R.; Mazimpaka, V. A New Key to the Genus Orthotrichum Hedw. in Europe and the Mediterranean Region. Cryptogamie Bryol. 2009, 30, 129–142. [Google Scholar]
- Sawicki, J.; Plášek, V.; Szczecińska, M. Molecular Data Do Not Support the Current Division of Orthotrichum (Bryophyta) Species with Immersed Stomata. J. Syst. Evol. 2012, 50, 12–24. [Google Scholar] [CrossRef]
- Lara, F.; Draper, I.; Flagmeier, M.; Calleja, J.A.; Mazimpaka, V.; Garilleti, R. Let’s Make Pulvigera Great Again: Re-Circumscription of a Misunderstood Group of Orthotrichaceae That Diversified in North America. Bot. J. Linn. Soc. 2020, 193, 180–206. [Google Scholar] [CrossRef]
- Draper, I.; Garilleti, R.; Calleja, J.A.; Flagmeier, M.; Mazimpaka, V.; Vigalondo, B.; Lara, F. Insights into the Evolutionary History of the Subfamily Orthotrichoideae (Orthotrichaceae, Bryophyta): New and Former Supra-Specific Taxa So Far Obscured by Prevailing Homoplasy. Front. Plant Sci. 2021, 12, 629035. [Google Scholar] [CrossRef]
- Draper, I.; Villaverde, T.; Garilleti, R.; Burleigh, J.G.; McDaniel, S.F.; Mazimpaka, V.; Calleja, J.A.; Lara, F. An NGS-Based Phylogeny of Orthotricheae (Orthotrichaceae, Bryophyta) With the Proposal of the New Genus Rehubryum from Zealandia. Front. Plant Sci. 2022, 13, 882960. [Google Scholar] [CrossRef]
- Matcham, H.W.; O’Shea, B.J. A Review of the Genus Codonoblepharon Schwägr. (Bryopsida: Orthotrichaceae). J. Bryol. 2005, 27, 129–135. [Google Scholar] [CrossRef]
- Malta, N. Die Gattung Zygodon Hook. et Tayl. Ein Monographischen Studie; Latvijas Universitates: Riga, Latvia, 1926. [Google Scholar]
- Calabrese, G.M. A Taxonomic Revision of Zygodon (Orthotrichaceae) in Southern South America. Bryologist 2006, 109, 453–509. [Google Scholar] [CrossRef]
- Lara, F.; Garilleti; Mazimpaka, V.; Guerra, J. Orthotrichaceae. In Flora Briofítica Ibérica, vol. V. Orthotrichales: Orthotrichaceae; Hedwigiales: Hedwigiaceae; Leucodontales: Fontinalaceae, Climaciaceae, Anomodontaceae, Cryphaeaceae, Leptodontaceae, Leucodontaceae, Neckeraceae; Hookeriales: Hypopterygiaceae, Hookeriaceae, Leucomiaceae, Pilotrichaceae; Guerra, J., Cano, M.J., Brugués, M., Eds.; Universidad de Murcia and Sociedad Española de Briología: Murcia, Spain, 2014; pp. 15–135. [Google Scholar]
- Goffinet, B. A Reconsideration of the Affinities of Kleioweisiopsis, Pleurozygodontopsis, Trigonodictyon, and the Microtheciellaceae (Bryopsida, Orthotrichales). J. Bryol. 1998, 20, 69–81. [Google Scholar] [CrossRef]
- Goffinet, B.; Bayer, R.J.; Vitt, D.H. Circumscription and Phylogeny of the Orthotrichales (Bryopsida) Inferred from RBCL Sequence Analyses. Am. J. Bot. 1998, 85, 1324–1337. [Google Scholar] [CrossRef] [PubMed]
- Schimper, W.P. Corollarium Bryologiae Europaeae; E. Schweizerbart: Stuttgart, Germany, 1856. [Google Scholar]
- Vitt, D.H. The Genera of Orthotrichaceae. Beih. Nova Hedwigia 1982, 71, 261–268. [Google Scholar]
- Shevock, J.R. Two Mosses New to North America with Southern Hemisphere Affinities. Evansia 2000, 17, 97–98. [Google Scholar] [CrossRef]
- Callaghan, D.A.; Aleffi, M.; Alegro, A.; Bisang, I.; Blockeel, T.L.; Collart, F.; Dragićević, S.; Draper, I.; Erdağ, A.; Erzberger, P.; et al. Global Geographical Range and Population Size of the Habitat Specialist Codonoblepharon forsteri (Dicks.) Goffinet in a Changing Climate. J. Bryol. 2022, 44, 35–50. [Google Scholar] [CrossRef]
- Lewinsky, J. Zygodon Hook. & Tayl. in Australasia: A Taxonomic Revision Including SEM-studies of Peristomes. Lindbergia 1989, 15, 109–139. [Google Scholar]
- Ziolkowski, P.A.; Sadowski, J. FISH-Mapping of RDNAs and Arabidopsis BACs on Pachytene Complements of Selected Brassicas. Genome 2002, 45, 189–197. [Google Scholar] [CrossRef]
- Nadot, S.; Bajon, R.; Lejeune, B. The Chloroplast Gene rps4 as a Tool for the Study of Poaceae Phylogeny. Plant Syst. Evol. 1994, 191, 27–38. [Google Scholar] [CrossRef]
- Souza-Chies, T.T.; Bittar, G.; Nadot, S.; Carter, L.; Besin, E.; Lejeune, B. Phylogenetic Analysis of Iridaceae with Parsimony and Distance Methods Using the Plastid Gene rps4. Plant Syst. Evol. 1997, 204, 109–123. [Google Scholar] [CrossRef]
- Vigalondo, B.; Lara, F.; Draper, I.; Valcárcel, V.; Garilleti, R.; Mazimpaka, V. Is It Really You, Orthotrichum acuminatum? Ascertaining a New Case of Intercontinental Disjunction in Mosses. Bot. J. Linn. Soc. 2016, 180, 30–49. [Google Scholar] [CrossRef]
- Werner, O.; Patiño, J.; González-Mancebo, J.M.; de Almeida Gabriel, R.M.; Ros, R.M. The Taxonomic Status and the Geographical Relationships of the Macaronesian Endemic Moss Fissidens luisieri (Fissidentaceae) Based on DNA Sequence Data. Bryologist 2009, 112, 315–324. [Google Scholar] [CrossRef]
- Pacak, A.; Szweykowska-Kulińska, Z. Molecular Data Concerning Alloploid Character and the Origin of Chloroplast and Mitochondrial Genomes in the Liverwort Pellia borealis. Plant Biotechnol. J. 2000, 2, 101–108. [Google Scholar]
- Müller, K.; Müller, J.; Neinhuis, C.; Quandt, D. PhyDE® (Phylogenetic Data Editor). 2006. Available online: http://www.phyde.de/ (accessed on 10 February 2022).
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI Search and Sequence Analysis Tools APIs in 2019. Nucleic Acids Res. 2019, 47, 636–641. [Google Scholar] [CrossRef] [PubMed]
- Simmons, M.P.; Ochoterena, H. Gaps as Characters in Sequence-Based Phylogenetic Analyses. Syst. Biol. 2000, 49, 369–381. [Google Scholar] [CrossRef]
- Müller, K. SeqState. Appl-Bioinformatics 2005, 4, 65–69. [Google Scholar] [CrossRef]
- Castresana, J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of Phylogenies after Removing Divergent and Ambiguously Aligned Blocks from Protein Sequence Alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Lanfear, R.; Calcott, B.; Ho, S.Y.W.; Guindon, S. PartitionFinder: Combined Selection of Partitioning Schemes and Substitution Models for Phylogenetic Analyses. Mol. Biol. Evol. 2012, 29, 1695–1701. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian Inference of Phylogenetic Trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian Phylogenetic Inference under Mixed Models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: Oxford, UK; New York, NY, USA, 2000. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree 1.4.4. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 3 June 2022).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).