Mutation in the Endo-β-1,4-glucanase (KORRIGAN) Is Responsible for Thick Leaf Phenotype in Sorghum
Abstract
1. Introduction
2. Results
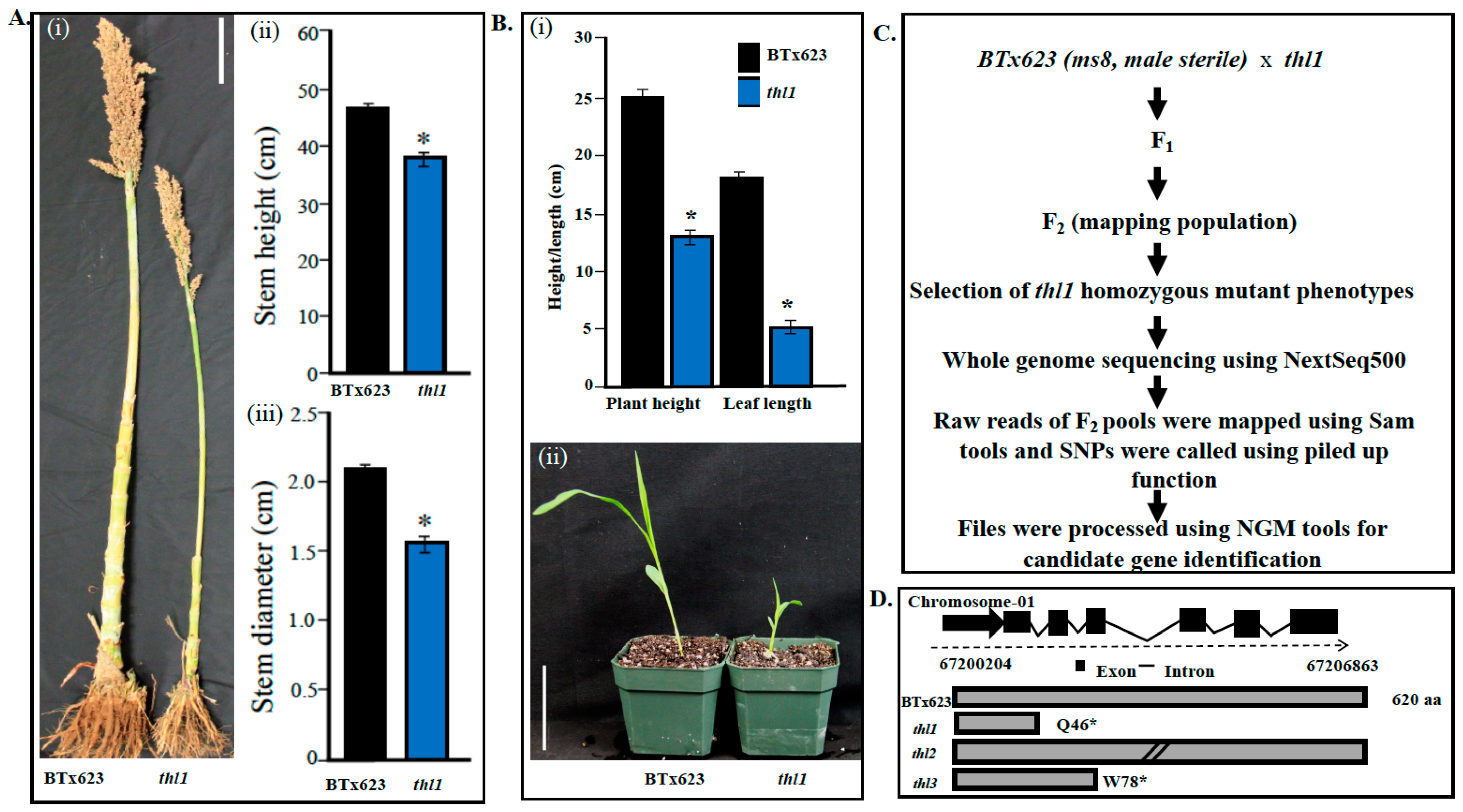
2.1. Identification and Characterization of thl Mutants
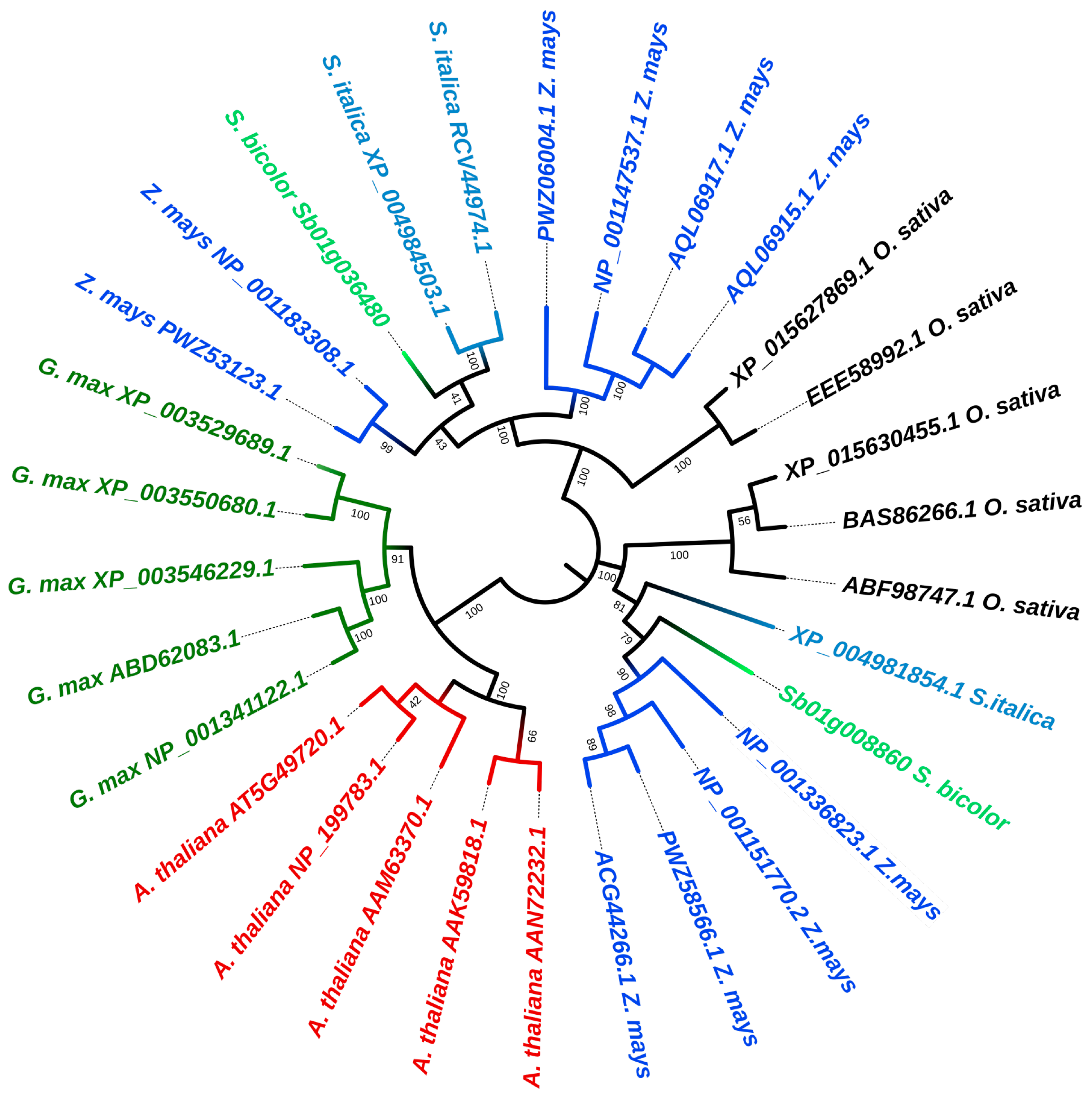
2.2. Phylogenetic Analysis Showed a Highly Conserved Nature of the KORRIGAN Proteins
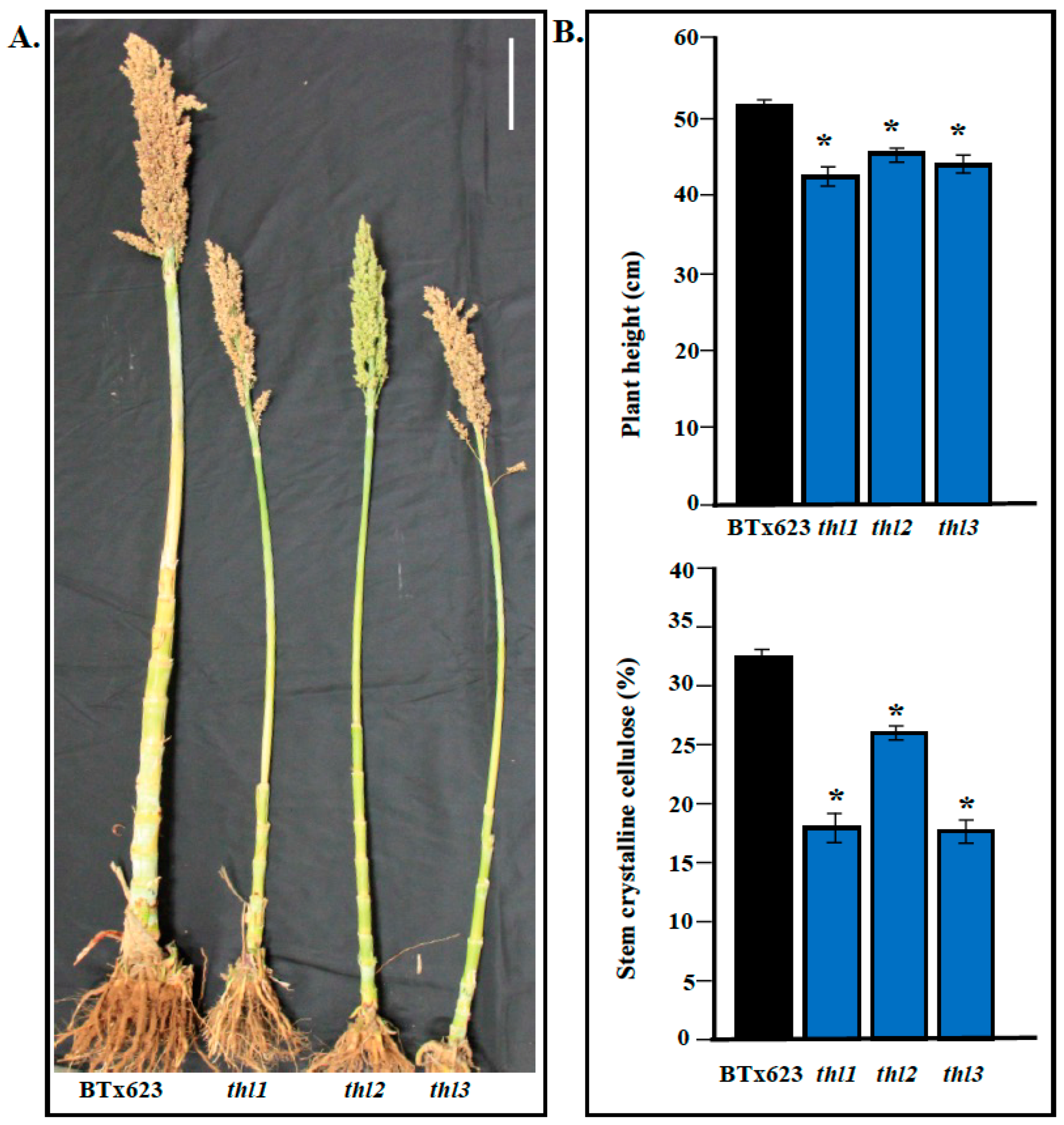
2.3. Identification and Characterization of Additional thl Mutants
2.4. Mutation in THL Resulted in Reduced Stem Crystalline Cellulose Content
2.5. Cell Elongation Is Compromised in thl Mutants
2.6. Arabidopsis KOR Mutant (rsw2-1) Phenotype Is Restored by SbKOR1
3. Discussion
4. Materials and Methods
4.1. Developing Mapping Population and Identification of thl Locus
4.2. Plant Growth Conditions, Phenotyping, and Preparation of Samples for Estimation of Crystalline Cellulose Content
4.3. Seedling Growth Conditions and Phenotyping under Light and Dark Conditions
4.4. Histochemical Analysis of Leaf Cross Sections
4.5. Arabidopsis Transformation
4.6. RNA Extraction from Plant Tissue, cDNA Synthesis, and Gene Amplification
4.7. Identification of Sorghum Homologs
4.8. Phylogenetic Tree Construction and Sequence Alignment
4.9. Cloning, Overexpression, and Complementation of SbKOR1
4.10. Plant Growth and Agrobacterium Mediated Plant Transformation
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Houston, K.; Tucker, M.R.; Chowdhury, J.; Shirley, N.; Little, A. The Plant Cell Wall: A Complex and Dynamic Structure as Revealed by the Responses of Genes under Stress Conditions. Front. Plant Sci. 2016, 7, 984. [Google Scholar] [CrossRef] [PubMed]
- Zuo, J.; Niu, Q.W.; Nishizawa, N.; Wu, Y.; Kost, B.; Chua, N.H. KORRIGAN, an Arabidopsis endo-1,4-beta-glucanase, localizes to the cell plate by polarized targeting and is essential for cytokinesis. Plant Cell 2000, 12, 1137–1152. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.M. The Biosynthesis of Cellulose. J. Macromol. Sci. Part A 1996, 33, 1345–1373. [Google Scholar] [CrossRef]
- Somerville, C. Cellulose Synthesis in Higher Plants. Annu. Rev. Cell Dev. Biol. 2006, 22, 53–78. [Google Scholar] [CrossRef]
- Vandavasi, V.G.; Putnam, D.K.; Zhang, Q.; Petridis, L.; Heller, W.T.; Nixon, B.T.; Haigler, C.H.; Kalluri, U.; Coates, L.; Langan, P.; et al. A Structural Study of CESA1 Catalytic Domain of Arabidopsis Cellulose Synthesis Complex: Evidence for CESA Trimers. Plant Physiol. 2016, 170, 123–135. [Google Scholar] [CrossRef]
- McNamara, J.T.; Morgan, J.L.W.; Zimmer, J. A molecular description of cellulose biosynthesis. Annu. Rev. Biochem. 2015, 84, 895–921. [Google Scholar] [CrossRef]
- Saxena, I.M.; Brown, R.M., Jr. Cellulose Biosynthesis: Current Views and Evolving Concepts. Ann. Bot. 2005, 96, 9–21. [Google Scholar] [CrossRef]
- McFarlane, H.E.; Döring, A.; Persson, S. The cell biology of cellulose synthesis. Annu. Rev. Plant Biol. 2014, 65, 69–94. [Google Scholar] [CrossRef]
- Hill, J.L., Jr.; Hammudi, M.B.; Tien, M. The Arabidopsis cellulose synthase complex: A proposed hexamer of CESA trimers in an equimolar stoichiometry. Plant Cell 2014, 26, 4834–4842. [Google Scholar] [CrossRef]
- Goubet, F.; Misrahi, A.; Park, S.K.; Zhang, Z.; Twell, D.; Dupree, P. AtCSLA7, a cellulose synthase-like putative glycosyltransferase, is important for pollen tube growth and embryogenesis in Arabidopsis. Plant Physiol. 2003, 131, 547–557. [Google Scholar] [CrossRef]
- Giddings, T.H., Jr.; Brower, D.L.; Staehelin, L.A. Visualization of particle complexes in the plasma membrane of Micrasterias denticulata associated with the formation of cellulose fibrils in primary and secondary cell walls. J. Cell Biol. 1980, 84, 327–339. [Google Scholar] [CrossRef] [PubMed]
- Kimura, S.; Laosinchai, W.; Itoh, T.; Cui, X.; Linder, C.R.; Brown, R.M., Jr. Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant Vigna angularis. Plant Cell 1999, 11, 2075–2086. [Google Scholar] [CrossRef] [PubMed]
- Bashline, L.; Li, S.; Anderson, C.T.; Lei, L.; Gu, Y. The endocytosis of cellulose synthase in Arabidopsis is dependent on μ2, a clathrin-mediated endocytosis adaptin. Plant Physiol. 2013, 163, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Nixon, B.T.; Mansouri, K.; Singh, A.; Du, J.; Davis, J.K.; Lee, J.-G.; Slabaugh, E.; Vandavasi, V.G.; O’Neill, H.; Roberts, E.M.; et al. Comparative Structural and Computational Analysis Supports Eighteen Cellulose Synthases in the Plant Cellulose Synthesis Complex. Sci. Rep. 2016, 6, 28696. [Google Scholar] [CrossRef] [PubMed]
- Kubicki, J.D.; Yang, H.; Sawada, D.; O’Neill, H.; Oehme, D.; Cosgrove, D. The Shape of Native Plant Cellulose Microfibrils. Sci. Rep. 2018, 8, 13983. [Google Scholar] [CrossRef]
- Richmond, T.A.; Somerville, C.R. The Cellulose Synthase Superfamily. Plant Physiol. 2000, 124, 495–498. [Google Scholar] [CrossRef]
- Persson, S.; Wei, H.; Milne, J.; Page, G.P.; Somerville, C.R. Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc. Natl. Acad. Sci. USA 2005, 102, 8633–8638. [Google Scholar] [CrossRef]
- Stork, J.; Harris, D.; Griffiths, J.; Williams, B.; Beisson, F.; Li-Beisson, Y.; Mendu, V.; Haughn, G.; DeBolt, S. CELLULOSE SYNTHASE9 Serves a Nonredundant Role in Secondary Cell Wall Synthesis in Arabidopsis Epidermal Testa Cells. Plant Physiol. 2010, 153, 580–589. [Google Scholar] [CrossRef]
- Mendu, V.; Griffiths, J.S.; Persson, S.; Stork, J.; Downie, A.B.; Voiniciuc, C.; Haughn, G.W.; DeBolt, S. Subfunctionalization of cellulose synthases in seed coat epidermal cells mediates secondary radial wall synthesis and mucilage attachment. Plant Physiol. 2011, 157, 441–453. [Google Scholar] [CrossRef]
- Polko, J.K.; Kieber, J.J. The Regulation of Cellulose Biosynthesis in Plants. Plant Cell 2019, 31, 282–296. [Google Scholar] [CrossRef]
- Nicol, F.; His, I.; Jauneau, A.; Vernhettes, S.; Canut, H.; Höfte, H. A plasma membrane-bound putative endo-1, 4-β-d-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 1998, 17, 5563–5576. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Sun, J.; Li, L. PtrCel9A6, an endo-1,4-β-glucanase, is required for cell wall formation during xylem differentiation in populus. Mol. Plant 2013, 6, 1904–1917. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Kato, T.; Kakegawa, K.; Ishii, T.; Liu, Y.-G.; Awano, T.; Takabe, K.; Nishiyama, Y.; Kuga, S.; Sato, S. Role of the putative membrane-bound endo-1, 4-β-glucanase KORRIGAN in cell elongation and cellulose synthesis in Arabidopsis thaliana. Plant Cell Physiol. 2001, 42, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Vain, T.; Crowell, E.F.; Timpano, H.; Biot, E.; Desprez, T.; Mansoori, N.; Trindade, L.M.; Pagant, S.; Robert, S.; Höfte, H.; et al. The Cellulase KORRIGAN Is Part of the Cellulose Synthase Complex. Plant Physiol. 2014, 165, 1521. [Google Scholar] [CrossRef] [PubMed]
- Mansoori, N.; Timmers, J.; Desprez, T.; Kamei, C.L.A.; Dees, D.C.T.; Vincken, J.-P.; Visser, R.G.F.; Höfte, H.; Vernhettes, S.; Trindade, L.M. KORRIGAN1 Interacts Specifically with Integral Components of the Cellulose Synthase Machinery. PLoS ONE 2014, 9, e112387. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Escalante, L.; Zhao, C.; Shukle, R.; Stuart, J. BSA-Seq Discovery and Functional Analysis of Candidate Hessian Fly (Mayetiola destructor) Avirulence Genes. Front. Plant Sci. 2020, 11, 956. [Google Scholar] [CrossRef]
- Mølhøj, M.; Jørgensen, B.; Ulvskov, P.; Borkhardt, B. Two Arabidopsis thaliana genes, KOR2 and KOR3, which encode membrane-anchored endo-1,4-beta-D-glucanases, are differentially expressed in developing leaf trichomes and their support cells. Plant Mol. Biol. 2001, 46, 263–275. [Google Scholar] [CrossRef]
- Lane, D.R.; Wiedemeier, A.; Peng, L.; Höfte, H.; Vernhettes, S.; Desprez, T.; Hocart, C.H.; Birch, R.J.; Baskin, T.I.; Burn, J.E.; et al. Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-beta-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol. 2001, 126, 278–288. [Google Scholar] [CrossRef]
- Williamson, R.E.; Burn, J.E.; Birch, R.; Baskin, T.I.; Arioli, T.; Betzner, A.S.; Cork, A. Morphology of rsw1, a cellulose-deficient mutant of Arabidopsis thaliana. Protoplasma 2001, 215, 116–127. [Google Scholar] [CrossRef]
- Gitz, D.C.; Liu-Gitz, L.; Xin, Z.; Baker, J.T.; Payton, P.; Lascano, R.J. Description of a Novel Allelic °∞Thick Leafed°± Mutant of Sorghum. Am. J. Plant Sci. 2017, 8, 10. [Google Scholar] [CrossRef][Green Version]
- Xin, Z.; Li Wang, M.; Barkley, N.A.; Burow, G.; Franks, C.; Pederson, G.; Burke, J. Applying genotyping (TILLING) and phenotyping analyses to elucidate gene function in a chemically induced sorghum mutant population. BMC Plant Biol. 2008, 8, 103. [Google Scholar] [CrossRef] [PubMed]
- Xin, Z.; Huang, J.; Smith, A.R.; Chen, J.; Burke, J.; Sattler, S.E.; Zhao, D. Morphological Characterization of a New and Easily Recognizable Nuclear Male Sterile Mutant of Sorghum (Sorghum bicolor). PLoS ONE 2017, 12, e0165195. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Burow, G.; Gladman, N.; Acosta-Martinez, V.; Chen, J.; Burke, J.; Ware, D.; Xin, Z. Efficient Identification of Causal Mutations through Sequencing of Bulked F (2) from Two Allelic Bloomless Mutants of Sorghum bicolor. Front. Plant Sci. 2017, 8, 2267. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lu, Z.; Regulski, M.; Jiao, Y.; Chen, J.; Ware, D.; Xin, Z. BSAseq: An interactive and integrated web-based workflow for identification of causal mutations in bulked F2 populations. Bioinformatics 2020, 37, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Lewandowska, M.A. The missing puzzle piece: Splicing mutations. Int. J. Clin. Exp. Pathol. 2013, 6, 2675–2682. [Google Scholar]
- Derbyshire, P.; Findlay, K.; McCann, M.C.; Roberts, K. Cell elongation in Arabidopsis hypocotyls involves dynamic changes in cell wall thickness. J. Exp. Bot. 2007, 58, 2079–2089. [Google Scholar] [CrossRef]
- Maloney, V.J.; Samuels, A.L.; Mansfield, S.D. The endo-1,4-β-glucanase Korrigan exhibits functional conservation between gymnosperms and angiosperms and is required for proper cell wall formation in gymnosperms. New Phytol. 2012, 193, 1076–1087. [Google Scholar] [CrossRef]
- Thu, S.W.; Rai, K.M.; Sandhu, D.; Rajangam, A.; Balasubramanian, V.K.; Palmer, R.G.; Mendu, V. Mutation in a PHD-finger protein MS4 causes male sterility in soybean. BMC Plant Biol. 2019, 19, 378. [Google Scholar] [CrossRef]
- Brummell, D.A.; Catala, C.; Lashbrook, C.C.; Bennett, A.B. A membrane-anchored E-type endo-1, 4-β-glucanase is localized on Golgi and plasma membranes of higher plants. Proc. Natl. Acad. Sci. USA 1997, 94, 4794–4799. [Google Scholar] [CrossRef]
- Brummell, D.A.; Lashbrook, C.C.; Bennett, A.B. Plant Endo-1, 4-β-D-glucanases: Structure, Properties, and Physiological Function; ACS Publications: Washington, DC, USA, 1994. [Google Scholar]
- Kalluri, U.C.; Payyavula, R.S.; Labbé, J.L.; Engle, N.; Bali, G.; Jawdy, S.S.; Sykes, R.W.; Davis, M.; Ragauskas, A.; Tuskan, G.A.; et al. Down-Regulation of KORRIGAN-Like Endo-β-1,4-Glucanase Genes Impacts Carbon Partitioning, Mycorrhizal Colonization and Biomass Production in Populus. Front. Plant Sci. 2016, 7, 1455. [Google Scholar] [CrossRef]
- Urbanowicz, B.R.; Catalá, C.; Irwin, D.; Wilson, D.B.; Ripoll, D.R.; Rose, J.K.C. A Tomato Endo-β-1,4-glucanase, SlCel9C1, Represents a Distinct Subclass with a New Family of Carbohydrate Binding Modules (CBM49)*. J. Biol. Chem. 2007, 282, 12066–12074. [Google Scholar] [CrossRef] [PubMed]
- Maloney, V.J.; Mansfield, S.D. Characterization and varied expression of a membrane-bound endo-β-1, 4-glucanase in hybrid poplar. Plant Biotechnol. J. 2010, 8, 294–307. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Chen, H.; Sun, J.; Li, L. PtrKOR1 is required for secondary cell wall cellulose biosynthesis in Populus. Tree Physiol. 2013, 34, 1289–1300. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, D.; Kumar, A.; Reddy, M.S. Genetic transformation of endo-1,4-β-glucanase (Korrigan) for cellulose enhancement in Eucalyptus tereticornis. Plant Cell Tissue Organ Cult. (PCTOC) 2015, 122, 363–371. [Google Scholar] [CrossRef]
- Mølhøj, M.; Pagant, S.; Höfte, H. Towards understanding the role of membrane-bound endo-β-1, 4-glucanases in cellulose biosynthesis. Plant Cell Physiol. 2002, 43, 1399–1406. [Google Scholar] [CrossRef]
- Arioli, T.; Peng, L.; Betzner, A.S.; Burn, J.; Wittke, W.; Herth, W.; Camilleri, C.; Höfte, H.; Plazinski, J.; Birch, R.; et al. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 1998, 279, 717–720. [Google Scholar] [CrossRef]
- Xin, Z.; Gitz, D.; Burow, G.; Hayes, C.; Burke, J.J. Registration of Two Allelic Erect Leaf Mutants of Sorghum. J. Plant Regist. 2015, 9, 254–257. [Google Scholar] [CrossRef]
- Michelmore, R.W.; Paran, I.; Kesseli, R.V. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 1991, 88, 9828–9832. [Google Scholar] [CrossRef]
- Updegraff, D.M. Semimicro determination of cellulose inbiological materials. Anal. Biochem. 1969, 32, 420–424. [Google Scholar] [CrossRef]
- Yuan, N.; Balasubramanian, V.K.; Chopra, R.; Mendu, V. The Photoperiodic Flowering Time Regulator FKF1 Negatively Regulates Cellulose Biosynthesis. Plant Physiol. 2019, 180, 2240–2253. [Google Scholar] [CrossRef]
- Dampanaboina, L.; Yuan, N.; Mendu, V. Estimation of Crystalline Cellulose Content of Plant Biomass using the Updegraff Method. J. Vis. Exp. 2021, 171, e62031. [Google Scholar] [CrossRef] [PubMed]
- Bailey, R.W. The reaction of pentoses with anthrone. Biochem J. 1958, 68, 669–672. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Bent, A.F. Arabidopsis in Planta Transformation. Uses, Mechanisms, and Prospects for Transformation of Other Species. Plant Physiol. 2000, 124, 1540–1547. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mendu, L.; Jalathge, G.; Dhillon, K.K.; Singh, N.P.; Balasubramanian, V.K.; Fewou, R.; Gitz, D.C., III; Chen, J.; Xin, Z.; Mendu, V. Mutation in the Endo-β-1,4-glucanase (KORRIGAN) Is Responsible for Thick Leaf Phenotype in Sorghum. Plants 2022, 11, 3531. https://doi.org/10.3390/plants11243531
Mendu L, Jalathge G, Dhillon KK, Singh NP, Balasubramanian VK, Fewou R, Gitz DC III, Chen J, Xin Z, Mendu V. Mutation in the Endo-β-1,4-glucanase (KORRIGAN) Is Responsible for Thick Leaf Phenotype in Sorghum. Plants. 2022; 11(24):3531. https://doi.org/10.3390/plants11243531
Chicago/Turabian StyleMendu, Lavanya, Gayani Jalathge, Kamalpreet Kaur Dhillon, Nagendra Pratap Singh, Vimal Kumar Balasubramanian, Rebecca Fewou, Dennis C. Gitz, III, Junping Chen, Zhanguo Xin, and Venugopal Mendu. 2022. "Mutation in the Endo-β-1,4-glucanase (KORRIGAN) Is Responsible for Thick Leaf Phenotype in Sorghum" Plants 11, no. 24: 3531. https://doi.org/10.3390/plants11243531
APA StyleMendu, L., Jalathge, G., Dhillon, K. K., Singh, N. P., Balasubramanian, V. K., Fewou, R., Gitz, D. C., III, Chen, J., Xin, Z., & Mendu, V. (2022). Mutation in the Endo-β-1,4-glucanase (KORRIGAN) Is Responsible for Thick Leaf Phenotype in Sorghum. Plants, 11(24), 3531. https://doi.org/10.3390/plants11243531







