Quercus ilex Phyllosphere Microbiome Environmental-Driven Structure and Composition Shifts in a Mediterranean Contex
Abstract
1. Introduction
2. Results
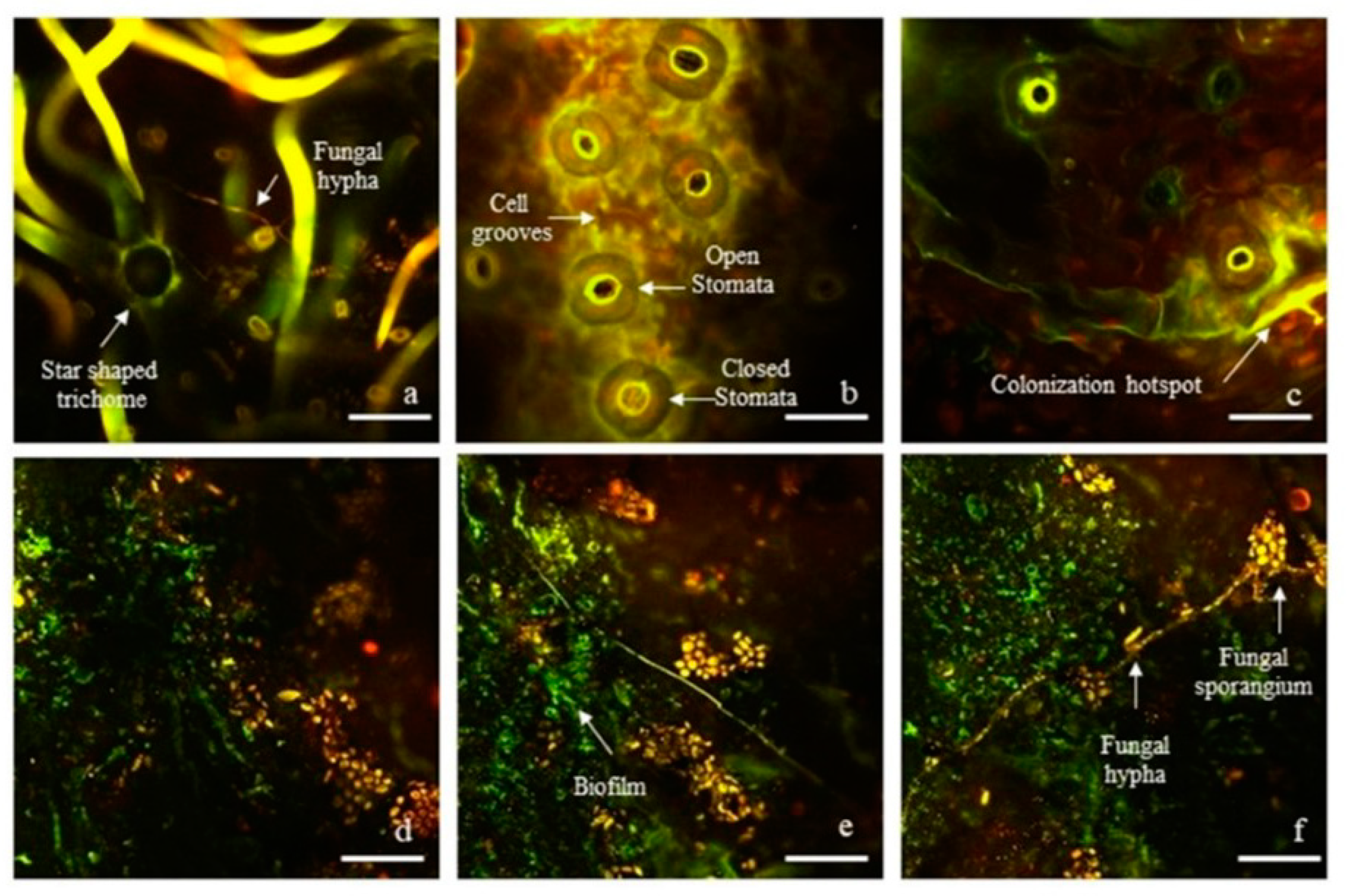
2.1. Visualization of Epiphytic Fungal and Bacterial Microbiome
2.2. Particulate Matter and PAHs Concentrations
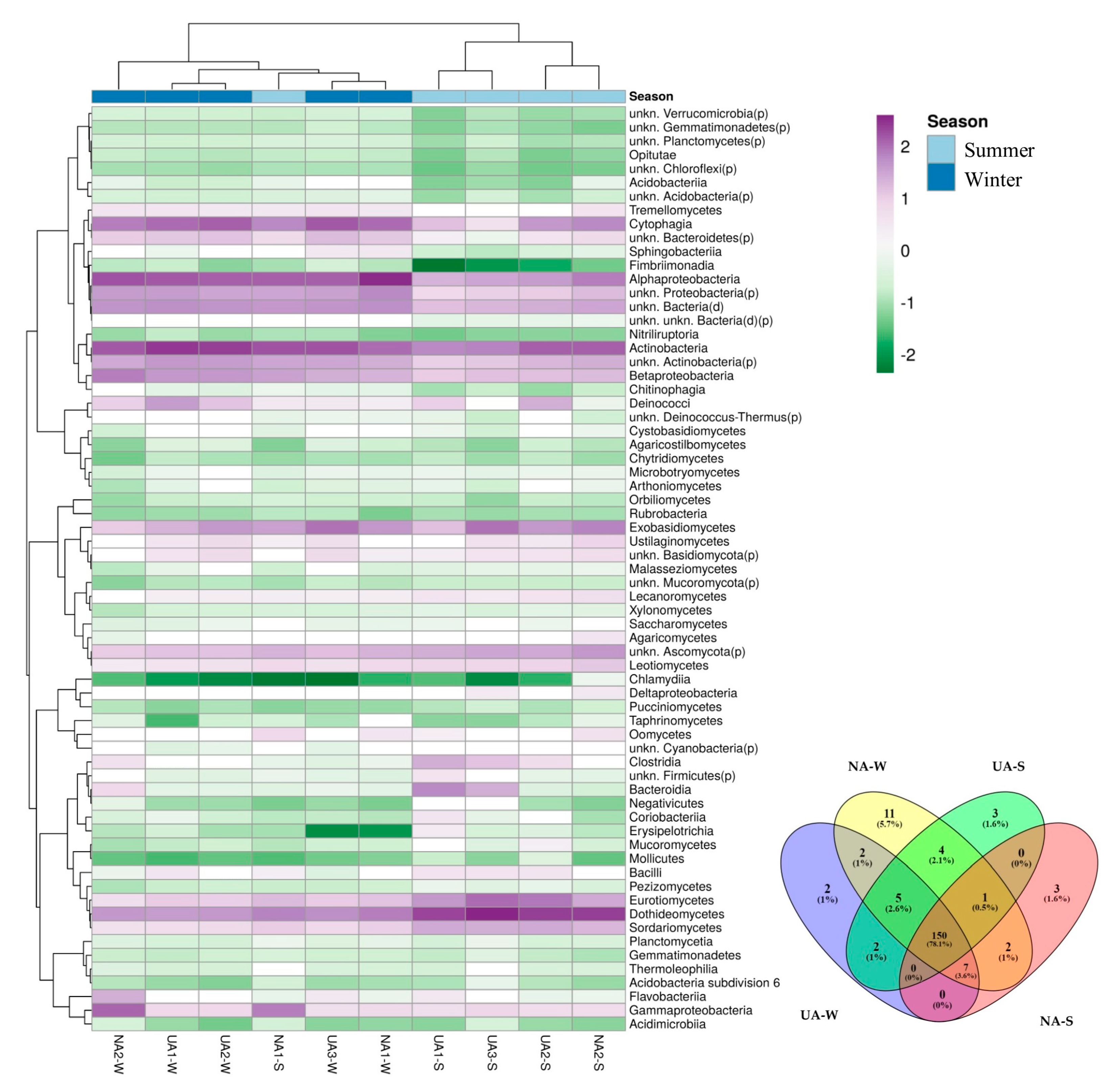
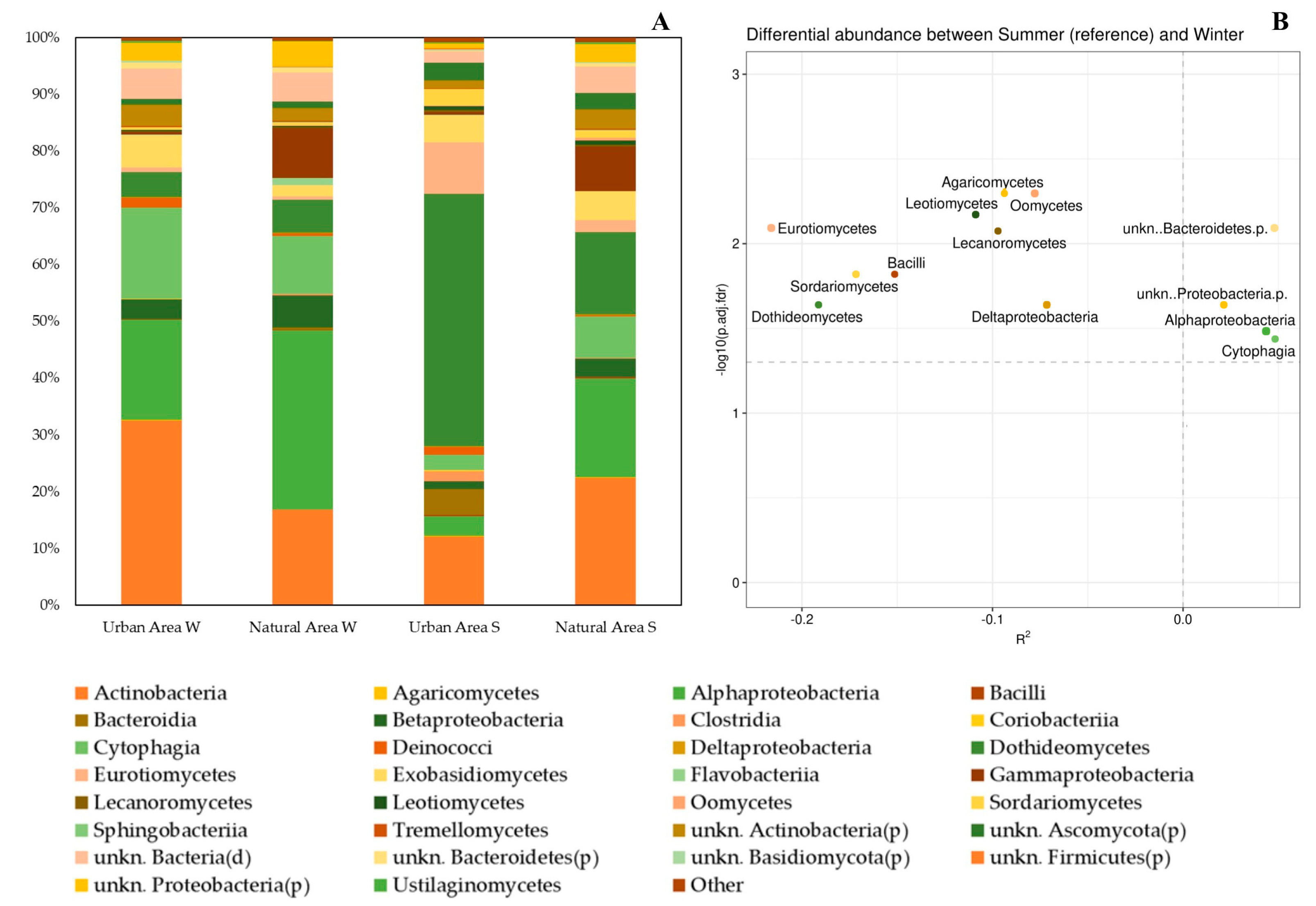
2.3. Relative Abundance of Phyla and Classes of Epiphytic Microorganisms
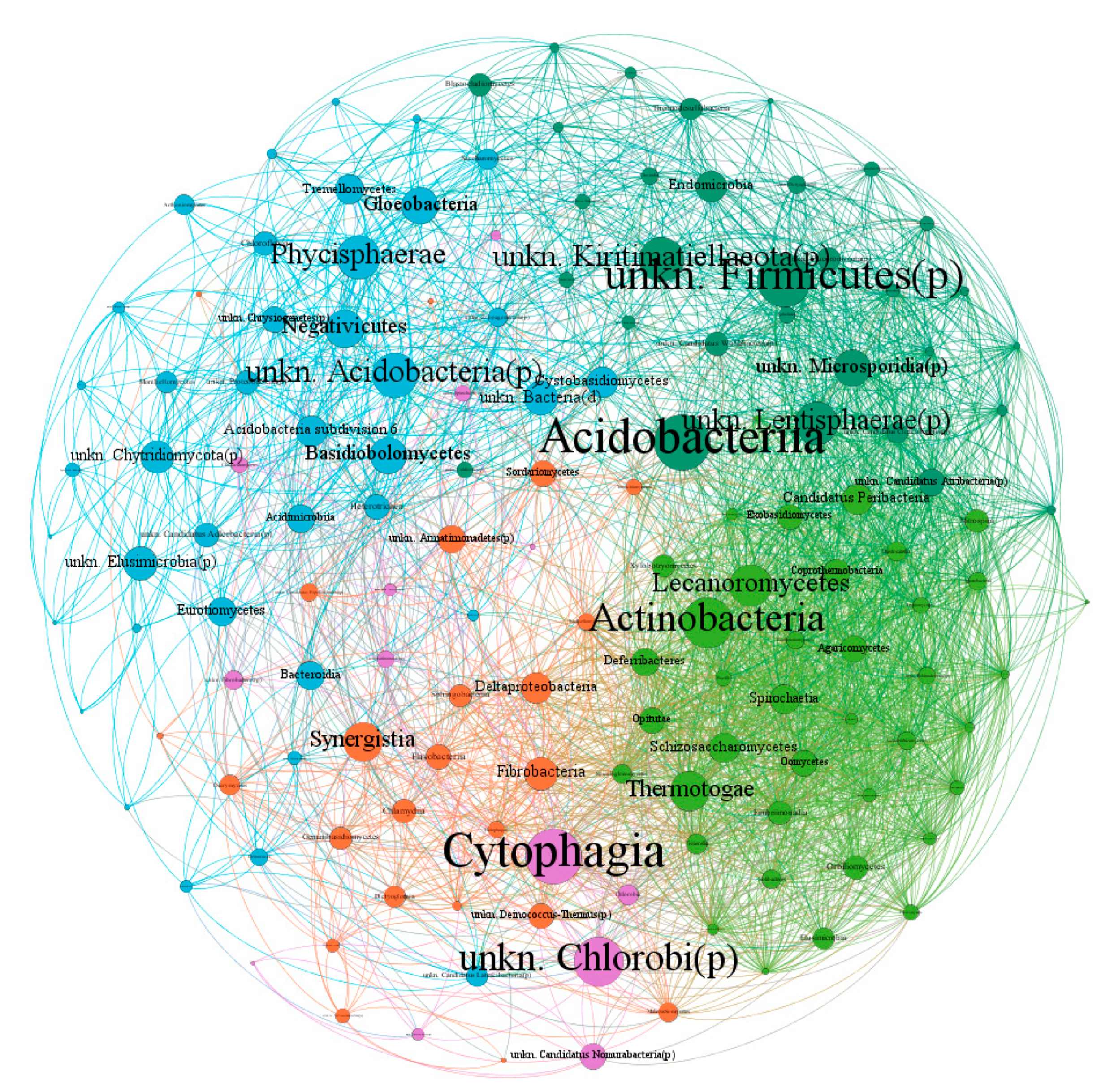
2.4. Epiphytic Microorganism Co-Occurrence Network
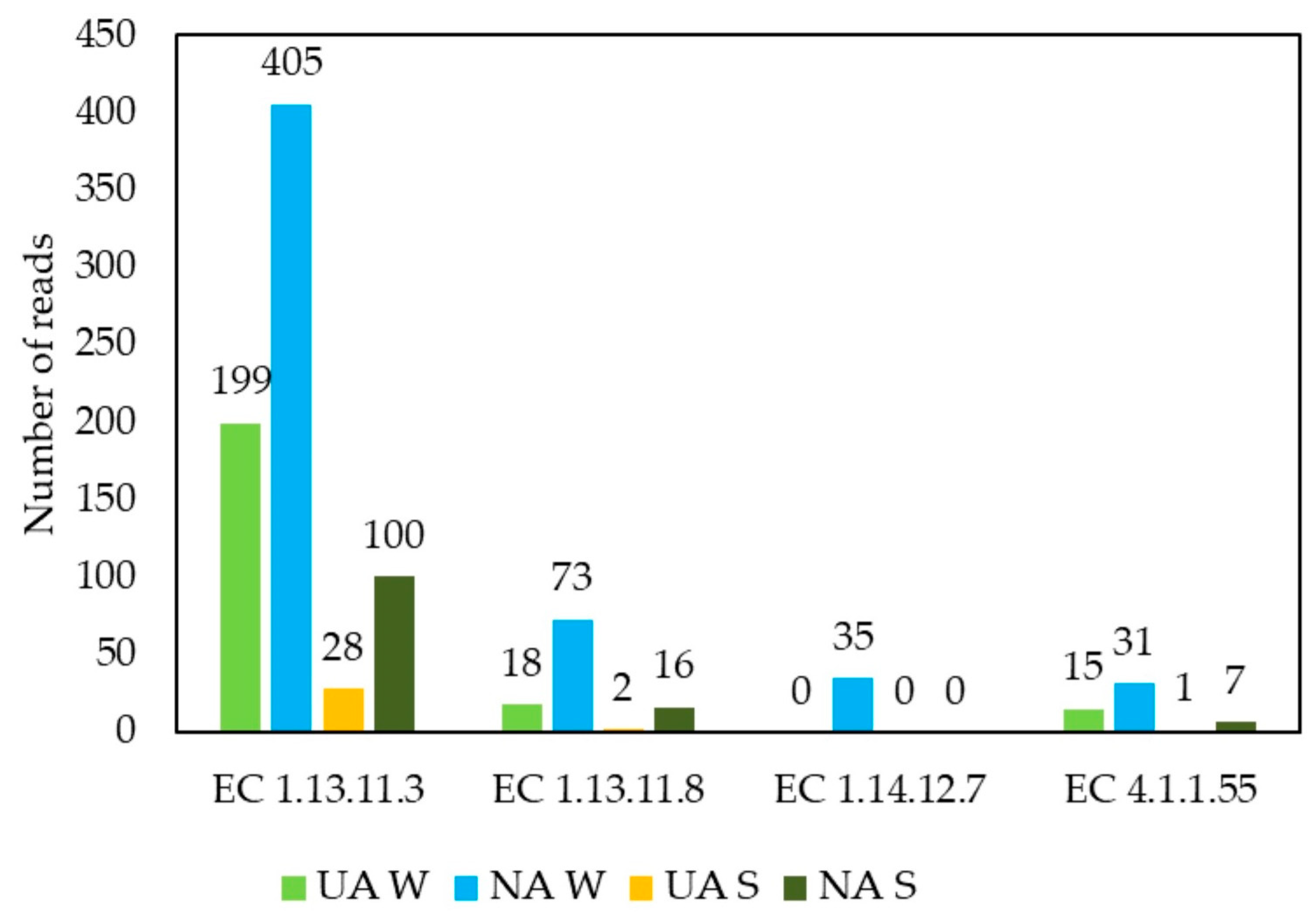
2.5. Functional Metagenomics
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Sites Description and Sample Collection
5.2. Two-Photon Excitation Microscopy (TPEM) Analyses
5.3. PM10 Extraction and Quantification on Leaf Surfaces
5.4. PAHs Associated to PM10 Chemical Analyses
5.5. DNA Extraction for Taxonomic Analysis and Functional Profiling
5.6. Metagenomic Analyses of Bacterial and Fungal Communities
5.7. Network Analysis of Phyllosphere Microorganisms
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jacques, M.; Kinkel, L.L.; Morris, C.E. Population sizes, immigration, and growth of epiphytic bacteria on leaves of different ages and positions of field-grown endive (Cichorium endivia var. latifolia). Appl. Environ. Microbiol. 1995, 61, 899–906. [Google Scholar] [CrossRef]
- Vorholt, J.A. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 2012, 10, 828–840. [Google Scholar] [CrossRef]
- Morris, C.E.; Kinkel, L.L. Fifty Years of Phyllosphere Microbiology: Significant contributions to research in related fields. In Phyllosphere Microbiology; Lindow, S.E., Hecht-Poinar, E.I., Elliot, V., Eds.; APS Press: St. Paul, MN, USA, 2002; pp. 365–375. ISBN 0890542864. [Google Scholar]
- Remus-Emsermann, M.N.P.; Schlechter, R.O. Phyllosphere microbiology: At the interface between microbial individuals and the plant host. New Phytol. 2018, 218, 1327–1333. [Google Scholar] [CrossRef]
- Müller, C.; Riederer, M. Plant surface properties in chemical ecology. J. Chem. Ecol. 2005, 31, 2621–2651. [Google Scholar] [CrossRef]
- Hirsch, A.M.; Fujishige, N.A. Commensalisms. In Encyclopedia of Ecology; Jørgensen, S.E., Fath, B.D., Eds.; Academic Press: Amsterdam, The Netherlands, 2008; pp. 679–683. ISBN 9780080454054. [Google Scholar] [CrossRef]
- Stone, B.W.; Weingarten, E.A.; Jackson, C.R. The role of the phyllosphere microbiome in plant health and function. Annu. Plant Rev. Online 2018, 1, 1–24. [Google Scholar] [CrossRef]
- Thapa, S.; Prasanna, R. Prospecting the characteristics and significance of the phyllosphere microbiome. Ann. Microbiol. 2018, 68, 229–245. [Google Scholar] [CrossRef]
- Yadav, R.K.; Karamanoli, K.; Vokou, D. Bacterial colonization of the phyllosphere of Mediterranean perennial species as influenced by leaf structural and chemical features. Microb. Ecol. 2005, 50, 185–196. [Google Scholar] [CrossRef]
- Schlechter, R.O.; Miebach, M.; Remus-Emsermann, M.N.P. Driving factors of epiphytic bacterial communities: A review. J. Adv. Res. 2019, 19, 57–65. [Google Scholar] [CrossRef]
- Jacques, M.A.; Morris, C.E. A review of issues related to the quantification of bacteria from the phyllosphere. FEMS Microbiol. Ecol. 1995, 18, 1–14. [Google Scholar] [CrossRef]
- Weyens, N.; Thijs, S.; Popek, R.; Witters, N.; Przybysz, A.; Espenshade, J.; Gawronska, H.; Vangronsveld, J.; Gawronski, S.W. The role of plant-microbe interactions and their exploitation for phytoremediation of air pollutants. Int. J. Mol. Sci. 2015, 16, 25576–25604. [Google Scholar] [CrossRef]
- Liu, J.; Song, M.; Wei, X.; Zhang, H.; Bai, Z.; Zhuang, X. Responses of phyllosphere microbiome to Ozone stress: Abundance, community compositions and functions. Microorganisms 2022, 10, 680. [Google Scholar] [CrossRef] [PubMed]
- Gandolfi, I.; Canedoli, C.; Imperato, V.; Tagliaferri, I.; Gkorezis, P.; Vangronsveld, J.; Padoa Schioppa, E.; Papacchini, M.; Bestetti, G.; Franzetti, A.; et al. Diversity and hydrocarbon-degrading potential of epiphytic microbial communities on Platanus × acerifolia leaves in an urban area. Environ. Pollut. 2017, 220, 650–658. [Google Scholar] [CrossRef] [PubMed]
- Grady, K.L.; Sorensen, J.W.; Stopnisek, N.; Guittar, J.; Shade, A. Assembly and seasonality of core phyllosphere microbiota on perennial biofuel crops. Nat. Commun. 2019, 10, 4135. [Google Scholar] [CrossRef] [PubMed]
- Franzetti, A.; Gandolfi, I.; Bestetti, G.; Padoa Schioppa, E.; Canedoli, C.; Brambilla, D.; Cappelletti, D.; Sebastiani, B.; Federici, E.; Papacchini, M.; et al. Plant-microorganisms interaction promotes removal of air pollutants in Milan (Italy) urban area. J. Hazard. Mater. 2020, 384, 121021. [Google Scholar] [CrossRef]
- Noble, A.S.; Noe, S.; Clearwater, M.J.; Lee, C.K. A core phyllosphere microbiome exists across distant populations of a tree species indigenous to New Zealand. PLoS ONE 2020, 15, e0237079. [Google Scholar] [CrossRef] [PubMed]
- Espenshade, J.; Thijs, S.; Gawronski, S.; Bové, H.; Weyens, N.; Vangronsveld, J. Influence of urbanization on epiphytic bacterial communities of the Platanus × hispanica tree leaves in a biennial study. Front. Microbiol. 2019, 10, 675. [Google Scholar] [CrossRef]
- Imperato, V.; Kowalkowski, L.; Portillo-Estrada, M.; Gawronski, S.W.; Vangronsveld, J.; Thijs, S. Characterisation of the Carpinus betulus L. phyllomicrobiome in urban and forest areas. Front. Microbiol. 2019, 10, 1110. [Google Scholar] [CrossRef]
- Stevens, V.; Thijs, S.; Vangronsveld, J. Diversity and plant growth-promoting potential of (un)culturable bacteria in the Hedera helix phylloplane. BMC Microbiol. 2021, 21, 66. [Google Scholar] [CrossRef]
- Sivakumar, N.; Sathishkumar, R.; Selvakumar, G.; Shyamkumar, R.; Arjunekumar, K. Phyllospheric microbiomes: Diversity, ecological significance, and biotechnological applications. In Plant Microbiomes for Sustainable Agriculture; Yadav, A., Singh, J., Rastegari, A., Yadav, N., Eds.; Springer: New York, NY, USA, 2020; Volume 25, pp. 113–172. ISBN 978-3-030-38452-4. [Google Scholar] [CrossRef]
- Imperato, V.; Portillo-Estrada, M.; Saran, A.; Thoonen, A.; Kowalkowski, Ł.; Gawronski, S.W.; Rineau, F.; Vangronsveld, J.; Thijs, S. Exploring the diversity and aromatic hydrocarbon degrading potential of epiphytic fungi on hornbeams from chronically polluted areas. J. Fungi 2021, 7, 972. [Google Scholar] [CrossRef]
- Vacher, C.; Hampe, A.; Porté, A.J.; Sauer, U.; Compant, S.; Morris, C.E. The phyllosphere: Microbial jungle at the plant-climate interface. Ann. Rev. Ecol. 2016, 47, 1–24. [Google Scholar] [CrossRef]
- Wuyts, K.; Smets, W.; Lebeer, S.; Samson, R. Green infrastructure and atmospheric pollution shape diversity and composition of phyllosphere bacterial communities in an urban landscape. FEMS Microbiol. Ecol. 2020, 96, fiz173. [Google Scholar] [CrossRef] [PubMed]
- Shakir, S.; Zaidi, S.S.; de Vries, F.T.; Mansoor, S. Plant genetic networks shaping phyllosphere microbial community. Trends Genet. 2021, 37, 306–316. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.G.; Xiong, C.; Wei, Z.; Chen, Q.L.; Ma, B.; Zhou, S.Y.; Tan, J.; Zhang, L.M.; Cui, H.L.; Duan, G.L.; et al. Impacts of global change on phyllosphere microbiome. New Phytol. 2022, 234, 1977–1986. [Google Scholar] [CrossRef] [PubMed]
- Whipps, J.M.; Hand, P.; Pink, D.; Bending, G.D. Phyllosphere microbiology with special reference to diversity and plant genotype. J. Appl. Microbiol. 2008, 105, 1744–1755. [Google Scholar] [CrossRef]
- Kim, M.; Singh, D.; Lai-Hoe, A.; Go, R.; Abdul Rahim, R.; Ainuddin, A.N.; Chun, J.; Adams, J.M. Distinctive phyllosphere bacterial communities in tropical trees. Microb. Ecol. 2012, 63, 674–681. [Google Scholar] [CrossRef]
- Yutthammo, C.; Thongthammachat, N.; Pinphanichakarn, P.; Luepromchai, E. Diversity and activity of PAH-degrading bacteria in the phyllosphere of ornamental plants. Microb. Ecol. 2010, 59, 357–368. [Google Scholar] [CrossRef]
- Roeland, S.; Moretti, M.; Amorim, J.H.; Branquinho, C.; Fares, S.; Morelli, F.; Niinemets, Ü.; Paoletti, E.; Pinho, P.; Sgrigna, G.; et al. Towards an Integrative Approach to Evaluate the Environmental Ecosystem Services Provided by Urban Forest. J. For. Res. 2019, 30, 1981–1996. [Google Scholar] [CrossRef]
- Sæbø, A.; Janhall, S.; Gawronski, S.W.; Hanslin, H.M. Urban forestry and pollution mitigation. In Routledge Handbook of Urban Forestry, 1st ed.; Ferrini, F., van den Konijnendijk Bosch, C.C., Fini, A., Eds.; Routledge: Abingdon, UK, 2017; ISBN 9781315627106. [Google Scholar] [CrossRef]
- Prigioniero, A.; Zuzolo, D.; Niinemets, Ü.; Guarino, C. Nature-based solutions as tools for air phytoremediation: A review of the current knowledge and gaps. Environ. Pollut. 2021, 277, 116817. [Google Scholar] [CrossRef]
- Dzierzanowski, K.; Popek, R.; Gawrońska, H.; Saebø, A.; Gawroński, S.W. Deposition of particulate matter of different size fractions on leaf surfaces and in waxes of urban forest species. Int. J. Phytoremediat. 2011, 13, 1037–1046. [Google Scholar] [CrossRef]
- Irga, P.J.; Burchett, M.D.; Torpy, F.R. Does urban forestry have a quantitative effect on ambient air quality in an urban environment? Atmos. Environ. 2015, 120, 173–181. [Google Scholar] [CrossRef]
- Sgrigna, G.; Sæbø, A.; Gawronski, S.; Popek, R.; Calfapietra, C. Particulate Matter deposition on Quercus ilex leaves in an industrial city of central Italy. Environ. Pollut. 2015, 197, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Leonard, R.J.; McArthur, C.; Hochuli, D.F. Particulate matter deposition on roadside plants and the importance of leaf trait combinations. Urban For. Urban Green. 2016, 20, 249–253. [Google Scholar] [CrossRef]
- Prigioniero, A.; Zuzolo, D.; Niinemets, Ü.; Postiglione, A.; Mercurio, M.; Izzo, F.; Trifuoggi, M.; Toscanesi, M.; Scarano, P.; Tartaglia, M.; et al. Particulate matter and polycyclic aromatic hydrocarbon uptake in relation to leaf surface functional traits in Mediterranean evergreens: Potentials for air phytoremediation. J. Hazard. Mater. 2022, 435, 129029. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Agrawal, M. Use of GLM approach to assess the responses of tropical trees to urban air pollution in relation to leaf functional traits and tree characteristics. Ecotoxicol. Env. Saf. 2018, 15, 42–54. [Google Scholar] [CrossRef]
- Sun, X.; Sun, M.; Chao, Y.; Wang, H.; Pan, H.; Yang, Q.; Cui, X.; Lou, Y.; Zhuge, Y. Alleviation of lead toxicity and phytostimulation in perennial ryegrass by the Pb-resistant fungus Trichoderma asperellum SD-5. Funct. Plant Biol. 2021, 48, 333–341. [Google Scholar] [CrossRef]
- Sánchez-López, A.S.; Carrillo-González, R.; del González-Chávez, C.A.; Rosas-Saito, G.H.; Vangronsveld, J. Phytobarriers: Plants capture particles containing potentially toxic elements originating from mine tailings in semiarid regions. Environ. Pollut. 2015, 205, 33–42. [Google Scholar] [CrossRef]
- Grote, R.; Samson, R.; Alonso, R.; Amorim, J.H.; Cariñanos, P.; Churkina, G.; le Fares, S.; Thiec, D.; Niinemets, Ü.; Mikkelsen, T.N.; et al. Functional traits of urban trees: Air pollution mitigation potential. Front. Ecol. Environ. 2016, 14, 543–550. [Google Scholar] [CrossRef]
- Meyer, K.M.; Leveau, J.H.J. Microbiology of the phyllosphere: A playground for testing ecological concepts. Oecologia 2012, 168, 621–629. [Google Scholar] [CrossRef]
- Vargas, C.; Pérez-Esteban, J.; Escolástico, C.; Masaguer, A.; Moliner, A. Phytoremediation of Cu and Zn by vetiver grass in mine soils amended with humic acids. Environ. Sci. Pollut. Res. 2016, 23, 13521–13530. [Google Scholar] [CrossRef]
- Agler, M.T.; Ruhe, J.; Kroll, S.; Morhenn, C.; Kim, S.T.; Weigel, D.; Kemen, E.M. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 2016, 14, e1002352. [Google Scholar] [CrossRef]
- Hassani, M.A.; Durán, P.; Hacquard, S. Microbial interactions within the plant holobiont. Microbiome 2018, 6, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Barroso-Bergadà, D.; Pauvert, C.; Vallance, J.; Delière, L.; Bohan, D.A.; Buée, M.; Vacher, C. Microbial networks inferred from environmental DNA data for biomonitoring ecosystem change: Strengths and pitfalls. Mol. Eco. Resour. 2021, 21, 762–780. [Google Scholar] [CrossRef] [PubMed]
- Newman, M.E.J. Modularity and community structure in networks. Proc. Natl. Acad. Sci. USA 2006, 103, 8577–8582. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]
- Oliveros, J.C. Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 7 February 2022).
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012, 40, D109–D114. [Google Scholar] [CrossRef]
- Thijs, S.; Sillen, W.; Rineau, F.; Weyens, N.; Vangronsveld, J. Towards an enhanced understanding of plant-microbiome interactions to improve phytoremediation: Engineering the Metaorganism. Front. Microbiol. 2016, 7, 341. [Google Scholar] [CrossRef]
- Koskella, B. The phyllosphere. Curr. Biol. 2020, 30, R1143–R1146. [Google Scholar] [CrossRef]
- Knief, C. Analysis of plant microbe interactions in the era of next generation sequencing technologies. Front. Plant Sci. 2014, 5, 216. [Google Scholar] [CrossRef]
- Esser, D.S.; Leveau, J.H.; Meyer, K.; Wiegand, K. Spatial scales of interactions among bacteria and between bacteria and the leaf surface. FEMS Microbiol. Ecol. 2015, 91, fiu034. [Google Scholar] [CrossRef]
- Chan, C.; Panzeri, D.; Okuma, E.; Tõldsepp, K.; Wang, Y.Y.; Louh, G.Y.; Chin, T.C.; Yeh, Y.H.; Yekondi, S.; Huang, Y.H.; et al. Stress induced factor 2 regulates Arabidopsis stomatal immunity through phosphorylation of the anion channel SLAC1. Plant Cell 2020, 32, 2216–2236. [Google Scholar] [CrossRef]
- Yadav, R.K.P.; Bosabalidis, A.M.; Vokou, D. Leaf structural features of Mediterranean perennial species: Plasticity and life form specificity. J. Biol. 2004, 2, 21–34. [Google Scholar]
- Vokou, D.; Vareli, K.; Zarali, E.; Karamanoli, K.; Constantinidou, H.I.; Monokrousos, N.; Halley, J.M.; Sainis, I. Exploring biodiversity in the bacterial community of the Mediterranean phyllosphere and its relationship with airborne bacteria. Microb. Ecol. 2012, 64, 714–724. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Qiu, K.; Alahmad, A.; Pott, R. Particulate matter capturing capacity of roadside evergreen vegetation during the winter season. Urban For. Urban Green. 2020, 48, 12651. [Google Scholar] [CrossRef]
- Redondo-Bermúdez, M.D.C.; Gulenc, I.T.; Cameron, R.W.; Inkson, B.J. “Green barriers” for air pollutant capture: Leaf micromorphology as a mechanism to explain plants capacity to capture particulate matter. Environ. Pollut. 2021, 288, 117809. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Chen, C.; Lu, T.; Zhang, J.; Chen, J. Season impacts on estimating plant’s particulate retention: Field experiments and meta-analysis. Chemosphere 2022, 288, 132570. [Google Scholar] [CrossRef] [PubMed]
- Gomes, T.; Pereira, J.A.; Benhadi, J.; Lino-Neto, T.; Baptista, P. Endophytic and epiphytic phyllosphere fungal communities are shaped by different environmental factors in a Mediterranean ecosystem. Microb. Ecol. 2018, 76, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Ogaya, R.; Peñuelas, J. Tree growth, mortality, and above-ground biomass accumulation in a holm oak forest under a five-year experimental field drought. Plant Ecol. 2007, 189, 291–299. [Google Scholar] [CrossRef]
- Peñuelas, J.; Rico, L.; Ogaya, R.; Jump, A.S.; Terradas, J. Summer season and long-term drought increase the richness of bacteria and fungi in the foliar phyllosphere of Quercus ilex in a mixed Mediterranean forest. Plant Biol. 2012, 14, 565–575. [Google Scholar] [CrossRef]
- Kaneko, R.; Kaneko, S. The effect of bagging branches on levels of endophytic fungal infection in Japanese beech leaves. For. Pathol. 2004, 34, 65–78. [Google Scholar] [CrossRef]
- Osono, T. Endophytic and epiphytic phyllosphere fungi of Camellia japonica: Seasonal and leaf age-dependent variations. Mycologia 2008, 100, 387–391. [Google Scholar] [CrossRef]
- Jumpponen, A.; Jones, K.L. Seasonally dynamic fungal communities in the Quercus macrocarpa phyllosphere differ between urban and nonurban environments. New Phytol. 2010, 186, 496–513. [Google Scholar] [CrossRef] [PubMed]
- Stone, B.; Jackson, C.R. Seasonal patterns contribute more towards phyllosphere bacterial community structure than short-term perturbations. Microb. Ecol. 2021, 81, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Bowsher, A.W.; Gmn, B.; Bonito, G.; Shade, A. Seasonal dynamics of core fungi in the switchgrass phyllosphere, and co-occurrence with leaf bacteria. Phytobiomes J. 2021, 5, 60–68. [Google Scholar] [CrossRef]
- Grube, M.; Berg, G. Microbial consortia of bacteria and fungi with focus on the lichen symbiosis. Fungal Biol. 2009, 23, 72–85. [Google Scholar] [CrossRef]
- Spring, S.; Bunk, B.; Spröer, C.; Schumann, P.; Rohde, M.; Tindall, B.J.; Klenk, H.P. Characterization of the first cultured representative of Verrucomicrobia subdivision 5 indicates the proposal of a novel phylum. ISME J. 2016, 10, 2801–2816. [Google Scholar] [CrossRef]
- Sackett, J.D.; Kruger, B.R.; Becraft, E.D.; Jarett, J.K.; Stepanauskas, R.; Woyke, T.; Duane, P.M. Four draft single-cell genome sequences of novel, nearly identical Kiritimatiellaeota strains isolated from the continental deep subsurface. Microbiol. Resour. Announc. 2019, 8, 1218–1249. [Google Scholar] [CrossRef]
- Vogel, M.A.; Mason, O.U.; Miller, T.E. Environmental stressors alter the composition of seagrass phyllosphere microbial communities. Clim. Chang. Ecol. 2021, 2, 100042. [Google Scholar] [CrossRef]
- Köberl, M.; Müller, H.; Ramadan, E.M.; Berg, G. Desert farming benefits from microbial potential in arid soils and promotes diversity and plant health. PLoS ONE 2011, 6, e24452. [Google Scholar] [CrossRef]
- Ortega, R.A.; Mahnert, A.; Berg, C.; Müller, H.; Berg, G. The plant is crucial: Specific composition and function of the phyllosphere microbiome of indoor ornamentals. FEMS Microbiol. Ecol. 2016, 92, fiw173. [Google Scholar] [CrossRef]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; le Van, A.; Dufresne, A. The importance of the microbiome of the plant holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef]
- Zuzolo, D.; Sciarrillo, R.; Postiglione, A.; Guarino, C. The remediation potential for PAHs of Verbascum sinuatum L. combined with an enhanced rhizosphere landscape: A full-scale mesocosm experiment. Biotechnol. Rep. 2021, 31, e00657. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, C.; Song, G.; Liu, H.; Sheng, G.; Ding, Z.; Wang, Z.; Sun, Y.; Xu, Y.; Chen, J.; et al. Characterization of a protocatechuate catabolic gene cluster in Rhodococcus ruber OA1 involved in naphthalene degradation. Ann. Microbiol. 2016, 66, 469–478. [Google Scholar] [CrossRef]
- Sazonova, O.I.; Sokolov, S.L.; Prisyazhnaya, N.V.; Izmalkova, T.Y.; Kosheleva, I.A.; Boronin, A.M. Epiphytic microorganisms degrading aromatic hydrocarbons from the phyllosphere of urban woody plants. Microbiology 2017, 86, 82–88. [Google Scholar] [CrossRef]
- Cilentoediano 2022. Available online: http://www.cilentoediano.it (accessed on 1 November 2022).
- ARPAC. Available online: www.arpacampania.it (accessed on 1 November 2022).
- Bacaloni, A.; Cafaro, C.; de Giorgi, L.; Ruocco, R.; Zoccolillo, L. Improved analysis of Polycyclic Aromatic Hydrocarbons in atmospheric particulate matter by HPLC-fluorescence. Ann. Chim. 2004, 94, 751–759. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Chen, J. A comprehensive evaluation of microbial differential abundance analysis methods: Current status and potential solutions. Microbiome 2022, 10, 1–23. [Google Scholar] [CrossRef]
- Revelle, W. Psych: Procedures for Psychological, Psychometric, and Personality Research. Available online: https://CRAN.R-project.org/package=psych (accessed on 1 October 2022).
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An open source software for exploring and manipulating networks. In Proceedings of the International AAAI Conference on Weblogs and Social Media, San Jose, CA, USA, 19 March 2009; pp. 361–362. [Google Scholar] [CrossRef]
- Yin, Y.; Zhu, D.; Yang, G.; Su, J.; Duan, G. Diverse antibiotic resistance genes and potential pathogens inhabit in the phyllosphere of fresh vegetables. Sci. Total Environ. 2022, 815, 152851. [Google Scholar] [CrossRef]
- Fruchterman, T.M.J.; Reingold, E.M. Graph drawing by force-directed placement. Softw. Pract. Exp. 1991, 21, 1129–1164. [Google Scholar] [CrossRef]
- Barberán, A.; Bates, S.; Casamayor, E.O.; Fierer, N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012, 6, 343–351. [Google Scholar] [CrossRef]
- Xing, M.; Wang, Q.; Li, X.; Li, Y.; Zhou, X. Selection of keystone species based on stable carbon and nitrogen isotopes to construct a typical food web on the shore of Xingkai Lake, China. Ecol. Indic. 2021, 132, 108263. [Google Scholar] [CrossRef]
- Zaura, E.; Bjf, K.; Huse, S.M.; Crielaard, W. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 2009, 9, 259. [Google Scholar] [CrossRef]

| PM10 (ng cm−2) | Benzo(a)pyrene (ng cm−2) | Benzo(a)anthracene (ng cm−2) | Benzo(b)fluoranthene (ng cm−2) | Benzo(j)fluoranthene (ng cm−2) | Benzo[k]fluoranthene (ng cm−2) | Indeno[1,2,3-cd]pyrene (ng cm−2) | Dibenz[a,h]anthracene (ng cm−2) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H2O | ClCH3 | H2O | ClCH3 | H2O | ClCH3 | H2O | ClCH3 | H2O | ClCH3 | H2O | ClCH3 | H2O | ClCH3 | H2O | ClCH3 | |
| UA Winter | 91,628.54 ± 959.47 a | 38,926.46 ± 755.81 abc | 0.47 ± 0.12 | 0.1 ± 0.05 a | nd | nd a | 0.09 ± 0.04 a | nd a | 0.09 ± 0.05 | nd | nd | nd | nd | nd a | nd | nd |
| NA Winter | 37,789.85 ± 4549.16 b | 21,022.88 ± 1443.39 a | 0.42 ± 0.19 | 0.36 ± 0.16 ab | nd | 0.12 ± 0.05 b | nd b | nd a | nd | nd | nd | nd | nd | nd a | nd | nd |
| UA Summer | 91,375.23 ± 4978.18 a | 26,908.81 ± 1274.22 b | 0.49 ± 0.05 | 0.1 ± 0.05 a | nd | nd a | nd b | nd a | nd | nd | nd | nd | nd | nd a | nd | nd |
| NA Summer | 75571.41 ± 2889.44 ab | 27,465.91 ± 6274.23 c | 0.55 ± 0.25 | 0.72 ± 0.05 b | nd | 0.15 ± 0.07 a | 0.19 ± 0.08 b | 0.17 ± 0.08 a | nd | nd | nd | nd | nd | 0.28 ± 0.13 a | nd | nd |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Postiglione, A.; Prigioniero, A.; Zuzolo, D.; Tartaglia, M.; Scarano, P.; Maisto, M.; Ranauda, M.A.; Sciarrillo, R.; Thijs, S.; Vangronsveld, J.; et al. Quercus ilex Phyllosphere Microbiome Environmental-Driven Structure and Composition Shifts in a Mediterranean Contex. Plants 2022, 11, 3528. https://doi.org/10.3390/plants11243528
Postiglione A, Prigioniero A, Zuzolo D, Tartaglia M, Scarano P, Maisto M, Ranauda MA, Sciarrillo R, Thijs S, Vangronsveld J, et al. Quercus ilex Phyllosphere Microbiome Environmental-Driven Structure and Composition Shifts in a Mediterranean Contex. Plants. 2022; 11(24):3528. https://doi.org/10.3390/plants11243528
Chicago/Turabian StylePostiglione, Alessia, Antonello Prigioniero, Daniela Zuzolo, Maria Tartaglia, Pierpaolo Scarano, Maria Maisto, Maria Antonietta Ranauda, Rosaria Sciarrillo, Sofie Thijs, Jaco Vangronsveld, and et al. 2022. "Quercus ilex Phyllosphere Microbiome Environmental-Driven Structure and Composition Shifts in a Mediterranean Contex" Plants 11, no. 24: 3528. https://doi.org/10.3390/plants11243528
APA StylePostiglione, A., Prigioniero, A., Zuzolo, D., Tartaglia, M., Scarano, P., Maisto, M., Ranauda, M. A., Sciarrillo, R., Thijs, S., Vangronsveld, J., & Guarino, C. (2022). Quercus ilex Phyllosphere Microbiome Environmental-Driven Structure and Composition Shifts in a Mediterranean Contex. Plants, 11(24), 3528. https://doi.org/10.3390/plants11243528












