Genome-Wide Characterization and Expression Profiling of HD-Zip Genes in ABA-Mediated Processes in Fragaria vesca
Abstract
1. Introduction
2. Results
2.1. Identification of HD-Zip Family Members in F. vesca
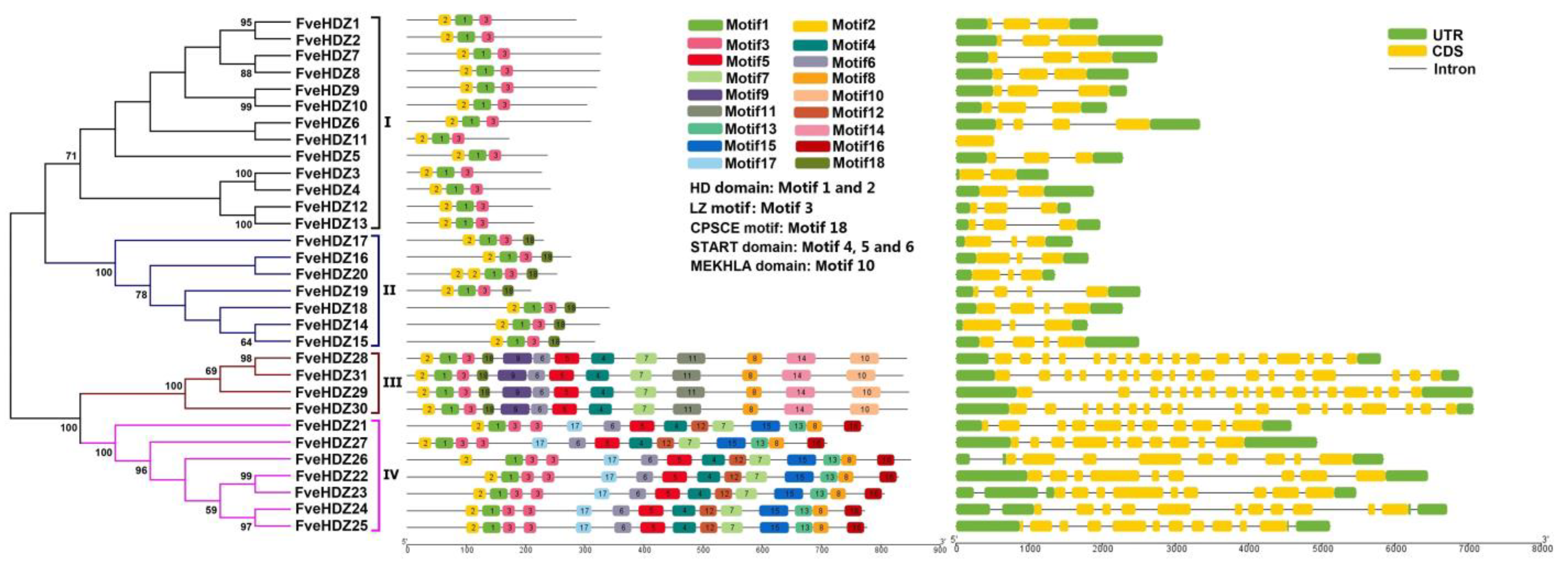
2.2. Phylogenetic and Motif Analyses of FveHDZ Proteins
2.3. Chromosomal Location, Gene Structure, and Synteny Analyses of FveHDZs
2.4. Promoter Analysis of FveHDZs
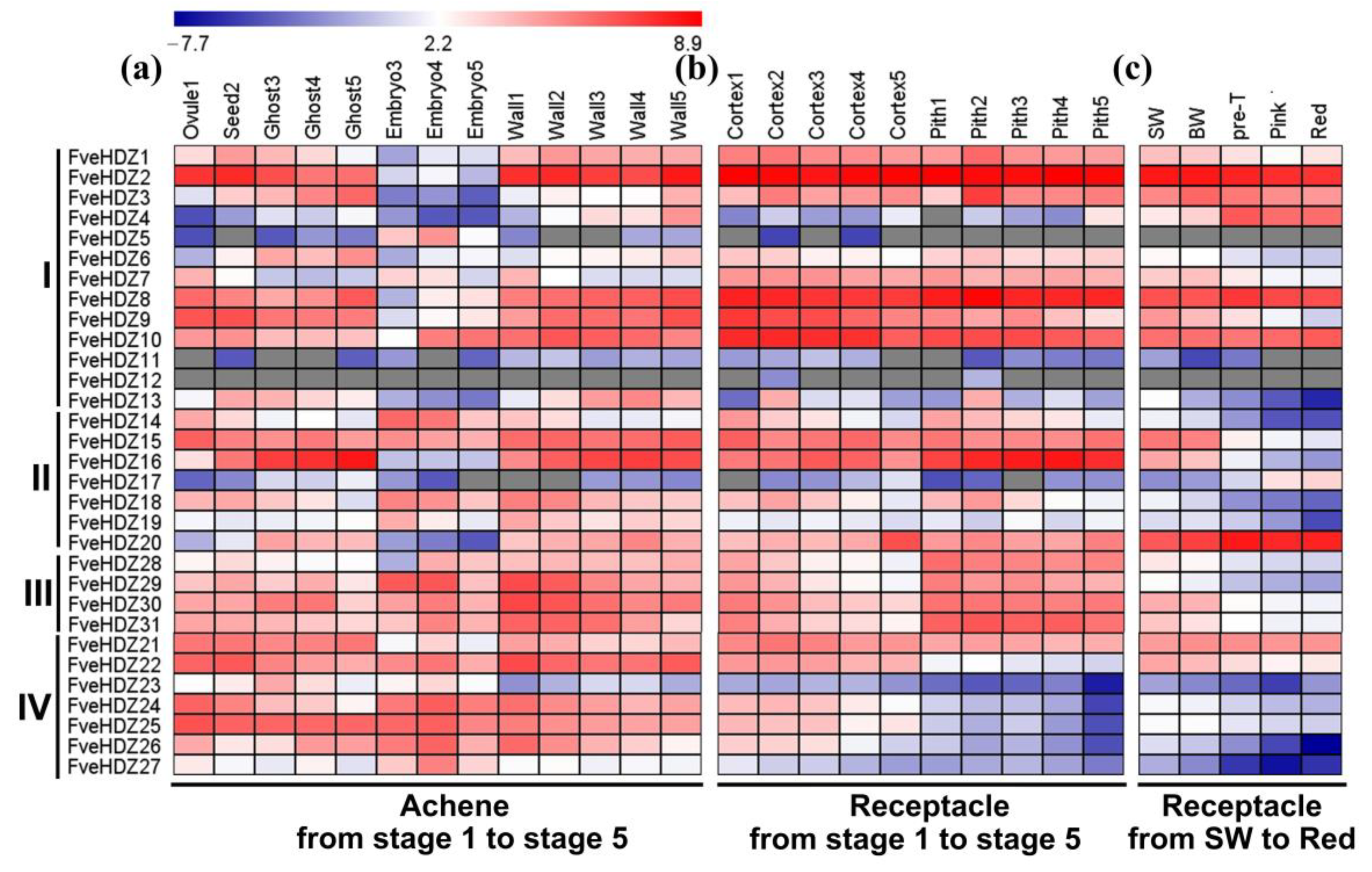
2.5. Expression Analysis of FveHDZs during Fruit Development and Ripening
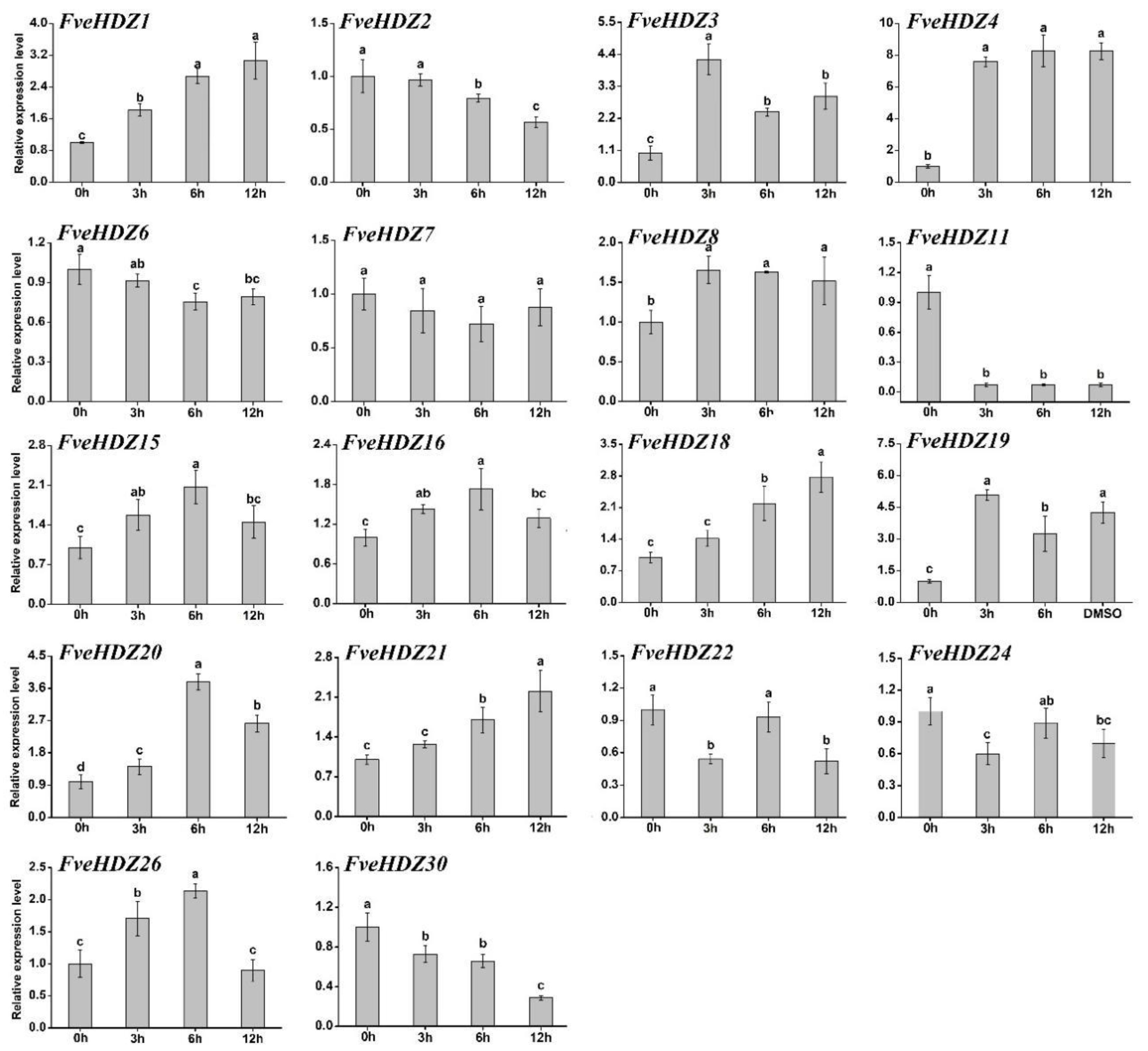
2.6. Expression Analysis of FveHDZs in Response to ABA
2.7. Expression Analysis of FveHDZs in ABA-Induced Leaf Senescence
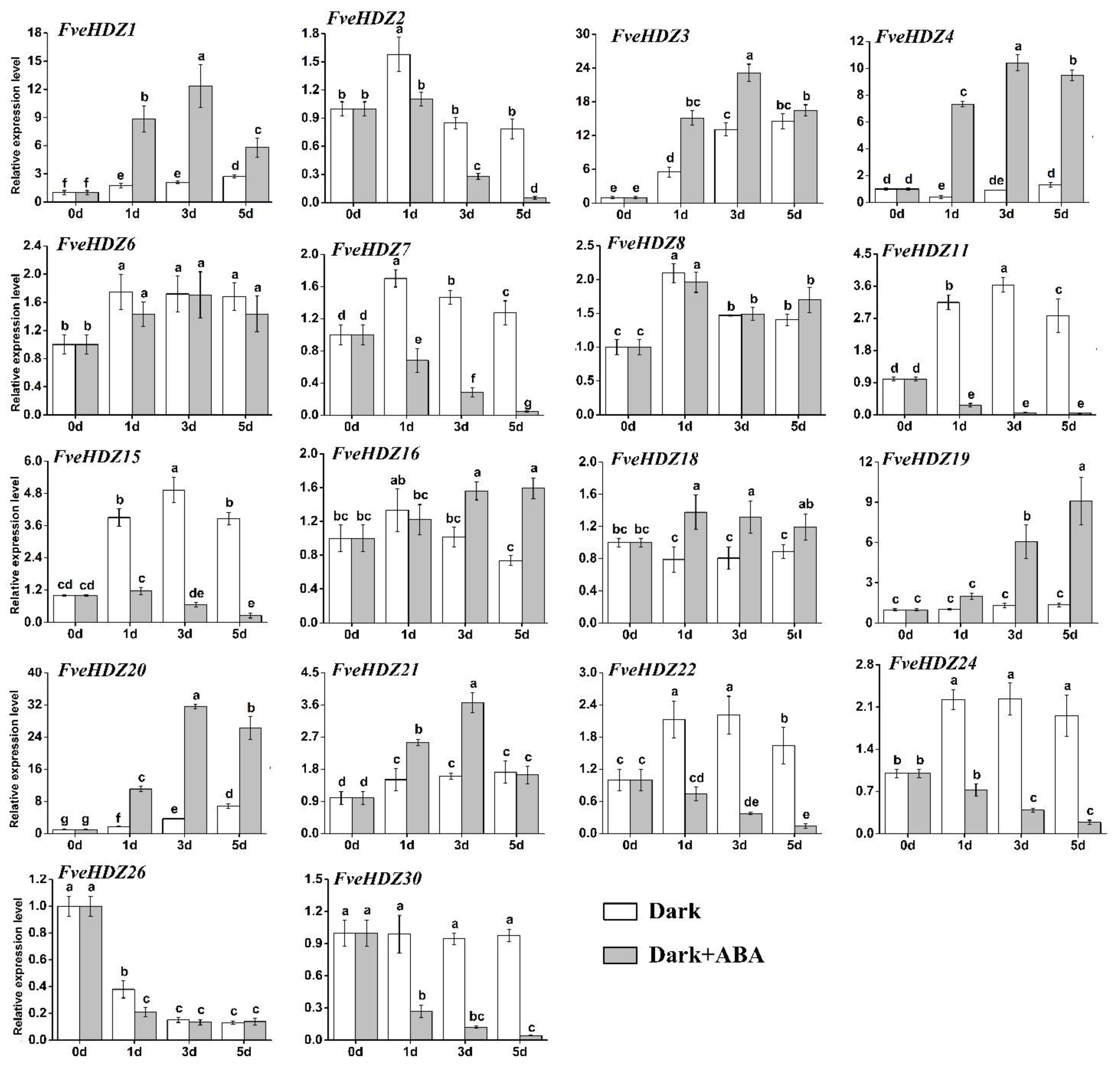
2.8. Expression Analysis of FveHDZs in Response to Dehydration
2.9. Binding of FveHDZs to the Promoter Fragments of Protein Phosphatase 2C Genes (PP2Cs) in Y1H Assays
3. Discussion
4. Materials and Methods
4.1. Identification and Analysis of HD-Zip Family Proteins in F. vesca
4.2. Phylogenetic Analysis and Gene Nomenclature
4.3. Gene Structure, Chromosomal Location, and Gene Duplication
4.4. Identification of Putative Promoter Cis-Acting Elements
4.5. Plant Materials and Stress Treatments
4.6. Transcriptome Retrieve
4.7. Yeast One-Hybrid Assay
4.8. RNA Extraction, RT-qPCR Analysis, and the Selection of FveHDZs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Riano-Pachon, D.M.; Ruzicic, S.; Dreyer, I.; Mueller-Roeber, B. PlnTFDB: An integrative plant transcription factor database. BMC Bioinform. 2007, 8, 42. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Z. Overexpression analysis of plant transcription factors. Curr. Opin. Plant Biol. 2003, 6, 430–440. [Google Scholar] [CrossRef]
- Bharathan, G.; Janssen, B.J.; Kellogg, E.A.; Sinha, N. Did homeodomain proteins duplicate before the origin of angiosperms, fungi, and metazoa? Proc. Natl. Acad. Sci. USA 1997, 94, 13749–13753. [Google Scholar] [CrossRef] [PubMed]
- Ariel, F.D.; Manavella, P.A.; Dezar, C.A.; Chan, R.L. The true story of the HD-Zip family. Trends Plant Sci. 2007, 12, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Johannesson, H.; Wang, Y.; Engstrom, P. DNA-binding and dimerization preferences of Arabidopsis homeodomain-leucine zipper transcription factors in vitro. Plant Mol. Biol. 2001, 45, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Chi, X.; Chai, G.; Kong, Y.; He, G.; Wang, X.; Shi, D.; Zhang, D.; Zhou, G. Genome-wide identification, evolutionary expansion, and expression profile of homeodomain-leucine zipper gene family in poplar (Populus trichocarpa). PLoS ONE 2012, 7, e31149. [Google Scholar] [CrossRef]
- Agalou, A.; Purwantomo, S.; Overnaes, E.; Johannesson, H.; Zhu, X.; Estiati, A.; de Kam, R.J.; Engstroem, P.; Slamet-Loedin, I.H.; Zhu, Z.; et al. A genome-wide survey of HD-Zip genes in rice and analysis of drought-responsive family members. Plant Mol. Biol. 2008, 66, 87–103. [Google Scholar] [CrossRef]
- Elhiti, M.; Stasolla, C. Structure and function of homodomain-leucine zipper (HD-Zip) proteins. Plant Signal. Behav. 2009, 4, 86–88. [Google Scholar] [CrossRef]
- Aoyama, T.; Dong, C.H.; Wu, Y.; Carabelli, M.; Sessa, G.; Ruberti, I.; Morelli, G.; Chua, N.H. Ectopic expression of the Arabidopsis transcriptional tctivator Athb-1 alters leaf cell fate in tobacco. Plant Cell 1995, 7, 1773–1785. [Google Scholar] [CrossRef]
- Sharif, R.; Raza, A.; Chen, P.; Li, Y.; El-Ballat, E.M.; Rauf, A.; Hano, C.; El-Esawi, M.A. HD-ZIP Gene Family: Potential roles in improving plant growth and regulating stress-responsive mechanisms in plants. Genes 2021, 12, 1256. [Google Scholar] [CrossRef]
- Gong, S.; Ding, Y.; Hu, S.; Ding, L.; Chen, Z.; Zhu, C. The role of HD-Zip class I transcription factors in plant response to abiotic stresses. Physiol. Plant. 2019, 167, 516–525. [Google Scholar] [CrossRef] [PubMed]
- Ré, D.A.; Capella, M.; Bonaventure, G.; Chan, R.L. Arabidopsis AtHB7 and AtHB12 evolved divergently to fine tune processes associated with growth and responses to water stress. BMC Plant Biol. 2014, 14, 150. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; Sun, Q.; Lu, S.; Chai, L.; Ye, J.; Deng, X. Citrus transcription factor CsHB5 regulates abscisic acid biosynthetic genes and promotes senescence. Plant J. 2021, 108, 151–168. [Google Scholar] [CrossRef]
- Ma, Y.J.; Li, P.T.; Sun, L.M.; Zhou, H.; Zeng, R.F.; Ai, X.Y.; Zhang, J.Z.; Hu, C.G. HD-ZIP I Transcription Factor (PtHB13) Negatively Regulates Citrus Flowering through Binding to FLOWERING LOCUS C Promoter. Plants 2020, 9, 114. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.F.; Hong, Y.G.; Yin, M.G.; Li, C.Y.; Zhang, K.; Grierson, D. A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J. 2008, 55, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Guo, Z.H.; Cheng, H.Y.; Zhou, Y.H.; Qi, K.J.; Wang, G.M.; Zhang, S.L. A HD-ZIP II HOMEBOX transcription factor, PpHB.G7, mediates ethylene biosynthesis during fruit ripening in peach. Plant Sci. 2019, 278, 12–19. [Google Scholar] [CrossRef]
- Rueda, E.C.; Dezar, C.A.; Gonzalez, D.H.; Chan, R.L. Hahb-10, a sunflower homeobox-leucine zipper gene, is regulated by light quality and quantity, and promotes early flowering when expressed in Arabidopsis. Plant Cell Physiol. 2005, 46, 1954–1963. [Google Scholar] [CrossRef]
- He, G.; Liu, P.; Zhao, H.; Sun, J. The HD-ZIP II transcription factors regulate plant architecture through the auxin pathway. Int. J. Mol. Sci. 2020, 21, 3250. [Google Scholar] [CrossRef]
- Sessa, G.; Carabelli, M.; Sassi, M.; Ciolfi, A.; Possenti, M.; Mittempergher, F.; Becker, J.; Morelli, G.; Ruberti, I. A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev. 2005, 19, 2811–2815. [Google Scholar] [CrossRef] [PubMed]
- Prigge, M.J.; Otsuga, D.; Alonso, J.M.; Ecker, J.R.; Drews, G.N.; Clark, S.E. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 2005, 17, 61–76. [Google Scholar] [CrossRef]
- Baima, S.; Possenti, M.; Matteucci, A.; Wisman, E.; Altamura, M.M.; Ruberti, I.; Morelli, G. The Arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol. 2001, 126, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Williams, L.; Grigg, S.P.; Xie, M.T.; Christensen, S.; Fletcher, J.C. Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development 2005, 132, 3657–3668. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Chen, J.; Feng, L.; Wang, Q.; Li, S.; Tan, X.; Yang, F.; Yang, W. HD-Zip III Gene Family: Identification and Expression Profiles during Leaf Vein Development in Soybean. Plants 2022, 11, 1728. [Google Scholar] [CrossRef]
- Xie, Q.; Gao, Y.; Li, J.; Yang, Q.; Qu, X.; Li, H.; Zhang, J.; Wang, T.; Ye, Z.; Yang, C. The HD-Zip IV transcription factor SlHDZIV8 controls multicellular trichome morphology by regulating the expression of Hairless-2. J. Exp. Bot. 2020, 71, 7132–7145. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Bartholomew, E.S.; Dong, M.; Zhai, X.; Yin, S.; Zhang, Y.; Feng, Z.; Wu, L.; Liu, W.; Shan, N.; et al. The HD-ZIP IV transcription factor GL2-LIKE regulates male flowering time and fertility in cucumber. J. Exp. Bot. 2020, 71, 5425–5437. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Katsumata, H.; Abe, M.; Yabe, N.; Komeda, Y.; Yamamoto, K.T.; Takahashi, T. Characterization of the class IV homeodomain-leucine zipper gene family in Arabidopsis. Plant Physiol. 2006, 141, 1363–1375. [Google Scholar] [CrossRef]
- Zhao, Y.; Chan, Z.; Gao, J.; Xing, L.; Cao, M.; Yu, C.; Hu, Y.; You, J.; Shi, H.; Zhu, Y.; et al. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. USA 2016, 113, 1949–1954. [Google Scholar] [CrossRef]
- Ali, F.; Qanmber, G.; Li, F.; Wang, Z. Updated role of ABA in seed maturation, dormancy, and germination. J. Adv. Res. 2022, 35, 199–214. [Google Scholar] [CrossRef]
- Yang, X.; Jia, Z.; Pu, Q.; Tian, Y.; Zhu, F.; Liu, Y. ABA Mediates Plant Development and Abiotic Stress via Alternative Splicing. Int. J. Mol. Sci. 2022, 23, 3796. [Google Scholar] [CrossRef]
- Soderman, E.; Mattsson, J.; Engstrom, P. The Arabidopsis homeobox gene ATHB-7 is induced by water deficit and by abscisic acid. Plant J. 1996, 10, 375–381. [Google Scholar] [CrossRef]
- Himmelbach, A.; Hoffmann, T.; Leube, M.; Hohener, B.; Grill, E. Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. Embo. J. 2002, 21, 3029–3038. [Google Scholar] [CrossRef] [PubMed]
- Valdes, A.E.; Overnas, E.; Johansson, H.; Rada-Iglesias, A.; Engstrom, P. The homeodomain-leucine zipper (HD-Zip) class I transcription factors ATHB7 and ATHB12 modulate abscisic acid signalling by regulating protein phosphatase 2C and abscisic acid receptor gene activities. Plant Mol. Biol. 2012, 80, 405–418. [Google Scholar] [CrossRef] [PubMed]
- Olsson, A.; Engström, P.; Söderman, E. The homeobox genes ATHB12 and ATHB7encode potential regulators of growth in response to water deficit in Arabidopsis. Plant Mol. Biol. 2004, 55, 663–677. [Google Scholar] [CrossRef] [PubMed]
- Dezar, C.A.; Gago, G.M.; Gonzalez, D.H.; Chan, R.L. Hahb-4, a sunflower homeobox-leucine zipper gene, is a developmental regulator and confers drought tolerance to Arabidopsis thaliana plants. Transgenic Res. 2005, 14, 429–440. [Google Scholar] [CrossRef]
- Gago, G.M.; Almoguera, C.; Jordano, J.; Gonzalez, D.H.; Chan, R.L. Hahb-4, a homeobox-leucine zipper gene potentially involved in abscisic acid-dependent responses to water stress in sunflower. Plant Cell Environ. 2002, 25, 633–640. [Google Scholar] [CrossRef]
- Jia, H.F.; Chai, Y.M.; Li, C.L.; Lu, D.; Luo, J.J.; Qin, L.; Shen, Y.Y. Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol. 2011, 157, 188–199. [Google Scholar] [CrossRef]
- Symons, G.M.; Chua, Y.J.; Ross, J.J.; Quittenden, L.J.; Davies, N.W.; Reid, J.B. Hormonal changes during non-climacteric ripening in strawberry. J. Exp. Bot. 2012, 63, 4741–4750. [Google Scholar] [CrossRef]
- Slovin, J.P.; Schmitt, K.; Folta, K.M. An inbred line of the diploid strawberry Fragaria vesca f. semperflorens for genomic and molecular genetic studies in the Rosaceae. Plant Methods 2009, 5, 15. [Google Scholar] [CrossRef]
- Shulaev, V.; Sargent, D.J.; Crowhurst, R.N.; Mockler, T.C.; Folkerts, O.; Delcher, A.L.; Jaiswal, P.; Mockaitis, K.; Liston, A.; Mane, S.P.; et al. The genome of woodland strawberry (Fragaria vesca). Nat. Genet. 2011, 43, 109–116. [Google Scholar] [CrossRef]
- Hollender, C.A.; Geretz, A.C.; Slovin, J.P.; Liu, Z.C. Flower and early fruit development in a diploid strawberry, Fragaria vesca. Planta 2012, 235, 1123–1139. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef]
- Kang, C.; Darwish, O.; Geretz, A.; Shahan, R.; Alkharouf, N.; Liu, Z. Genome-Scale Transcriptomic Insights into Early-Stage Fruit Development in Woodland Strawberry Fragaria vesca. Plant Cell 2013, 25, 1960–1978. [Google Scholar] [CrossRef]
- Hollender, C.A.; Kang, C.; Darwish, O.; Geretz, A.; Matthews, B.F.; Slovin, J.; Alkharouf, N.; Liu, Z. Floral Transcriptomes in Woodland Strawberry Uncover Developing Receptacle and Anther Gene Networks. Plant Physiol. 2014, 165, 1062–1075. [Google Scholar] [CrossRef] [PubMed]
- Gu, T.T.; Jia, S.F.; Huang, X.R.; Wang, L.; Fu, W.M.; Huo, G.T.; Gan, L.J.; Ding, J.; Li, Y. Transcriptome and hormone analyses provide insights into hormonal regulation in strawberry ripening. Planta 2019, 250, 145–162. [Google Scholar] [CrossRef]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 2016, 7, 571. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ding, G.Q.; Gu, T.T.; Ding, J.; Li, Y. Bioinformatic and expression analyses on carotenoid dioxygenase genes in fruit development and abiotic stress responses in Fragaria vesca. Mol. Genet. Genom. 2017, 292, 895–907. [Google Scholar] [CrossRef]
- Zhang, C.H.; Ma, R.J.; Shen, Z.J.; Sun, X.; Korir, N.K.; Yu, M.L. Genome-wide analysis of the homeodomain-leucine zipper (HD-ZIP) gene family in peach (Prunus persica). Genet. Mol. Res. 2014, 13, 2654–2668. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhou, Y.Q.; Jiang, H.Y.; Li, X.Y.; Gan, D.F.; Peng, X.J.; Zhu, S.W.; Cheng, B.J. Systematic analysis of sequences and expression patterns of drought-responsive members of the HD-Zip gene family in maize. PLoS ONE 2011, 6, e28488. [Google Scholar] [CrossRef] [PubMed]
- Van de Peer, Y.; Mizrachi, E.; Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017, 18, 411–424. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, C.; Guo, Y.; Niu, W.; Wang, Y.; Xu, Y. Evolution and expression analysis reveal the potential role of the HD-Zip gene family in regulation of embryo abortion in grapes (Vitis vinifera L.). BMC Genom. 2017, 18, 744. [Google Scholar] [CrossRef]
- Lee, Y.H.; Oh, H.S.; Cheon, C.I.; Hwang, I.T.; Kim, Y.J.; Chun, J.Y. Structure and expression of the Arabidopsis thaliana homeobox gene Athb-12. Biochem. Biophys. Res. Commun. 2001, 284, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Jiao, P.; Jiang, Z.; Wei, X.; Liu, S.; Qu, J.; Guan, S.; Ma, Y. Overexpression of the homeobox-leucine zipper protein ATHB-6 improves the drought tolerance of maize (Zea mays L.). Plant Sci. 2022, 316, 111159. [Google Scholar] [CrossRef] [PubMed]
- Manavella, P.A.; Arce, A.L.; Dezar, C.A.; Bitton, F.; Renou, J.P.; Crespi, M.; Chan, R.L. Cross-talk between ethylene and drought signalling pathways is mediated by the sunflower Hahb-4 transcription factor. Plant J. 2006, 48, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Manavella, P.A.; Dezar, C.A.; Bonaventure, G.; Baldwin, I.T.; Chan, R.L. HAHB4, a sunflower HD-Zip protein, integrates signals from the jasmonic acid and ethylene pathways during wounding and biotic stress responses. Plant J. 2008, 56, 376–388. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Ruan, J.; Ho, T.H.; You, Y.; Yu, T.; Quatrano, R.S. Cis-regulatory element based targeted gene finding: Genome-wide identification of abscisic acid- and abiotic stress-responsive genes in Arabidopsis thaliana. Bioinformatics 2005, 21, 3074–3081. [Google Scholar] [CrossRef] [PubMed]
- Hauser, F.; Li, Z.X.; Waadt, R.; Schroeder, J.I. SnapShot: Abscisic Acid Signaling. Cell 2017, 171, 1708–1708.e0. [Google Scholar] [CrossRef]
- Bobb, A.J.; Chern, M.S.; Bustos, M.M. Conserved RY-repeats mediate transactivation of seed-specific promoters by the developmental regulator PvALF. Nucleic Acids Res. 1997, 25, 641–647. [Google Scholar] [CrossRef]
- Guerriero, G.; Martin, N.; Golovko, A.; Sundstrom, J.F.; Rask, L.; Ezcurra, I. The RY/Sph element mediates transcriptional repression of maturation genes from late maturation to early seedling growth. New Phytol. 2009, 184, 552–565. [Google Scholar] [CrossRef]
- Eddy, S.R. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009, 23, 205–211. [Google Scholar] [CrossRef]
- Bhattacharjee, A.; Ghangal, R.; Garg, R.; Jain, M. Genome-wide analysis of homeobox gene family in legumes: Identification, gene duplication and expression profiling. PLoS ONE 2015, 10, e0119198. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.D.; Zhu, Y.X.; Yao, J.; Zhang, S.L.; Wang, L.; Guo, C.L.; van Nocker, S.; Wang, X.P. Genome-wide identification and expression analyses of the homeobox transcription factor family during ovule development in seedless and seeded grapes. Sci. Rep. 2017, 7, 16067. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Jung, S.; Bassett, C.; Bielenberg, D.G.; Cheng, C.H.; Dardick, C.; Main, D.; Meisel, L.; Slovin, J.; Troggio, M.; Schaffer, R.J. A standard nomenclature for gene designation in the Rosaceae. Tree Genet. Genomes 2015, 11, 108. [Google Scholar] [CrossRef]
- Darwish, O.; Slovin, J.P.; Kang, C.Y.; Hollender, C.A.; Geretz, A.; Houston, S.; Liu, Z.C. Alkharouf NW: SGR: An online genomic resource for the woodland strawberry. BMC Plant Biol. 2013, 13, 223. [Google Scholar] [CrossRef]
- Yu, D.; Tang, H.; Zhang, Y.; Du, Z.; Yu, H.; Chen, Q. Comparison and improvement of different methods of RNA isolation from strawberry (Fragria× ananassa). J. Agric. Sci. 2012, 4, 51–56. [Google Scholar] [CrossRef]
- Liu, D.; Huang, X.; Lin, Y.; Wang, X.; Yan, Z.; Wang, Q.; Ding, J.; Gu, T.; Li, Y. Identification of reference genes for transcript normalization in various tissue types and seedlings subjected to different abiotic stresses of woodland strawberry Fragaria vesca. Sci. Hortic. 2020, 261, 108840. [Google Scholar] [CrossRef]



| Gene Name | Subfamily | Intron No. | Protein Features | Duplication Type | ||||
|---|---|---|---|---|---|---|---|---|
| Protein Length (aa 1) | Molecular Weight (kDa) | Isoelectric Point (PI) | GRAVY 2 | Subcellular localization | ||||
| FveHDZ1 | I | 2 | 286 | 32.78210 | 5.30 | −1.076 | Nucleus | WGD/segmental |
| FveHDZ2 | I | 2 | 329 | 37.20892 | 5.18 | −0.881 | Nucleus | WGD/segmental |
| FveHDZ3 | I | 1 | 228 | 26.55655 | 5.54 | −1.125 | Nucleus | WGD/segmental |
| FveHDZ4 | I | 1 | 243 | 27.91169 | 4.9 | −1.095 | Nucleus | WGD/segmental |
| FveHDZ5 | I | 2 | 237 | 27.45371 | 8.49 | −1.033 | Nucleus | dispersed |
| FveHDZ6 | I | 3 | 310 | 35.28906 | 4.84 | −0.815 | Nucleus | dispersed |
| FveHDZ7 | I | 2 | 327 | 37.14513 | 4.93 | −0.835 | Nucleus | WGD/segmental |
| FveHDZ8 | I | 2 | 326 | 36.83849 | 4.74 | −0.802 | Nucleus | WGD/segmental |
| FveHDZ9 | I | 2 | 320 | 35.90381 | 6.4 | −0.932 | Nucleus | WGD/segmental |
| FveHDZ10 | I | 2 | 304 | 34.48854 | 6.12 | −0.846 | Nucleus | WGD/segmental |
| FveHDZ11 | I | 0 | 172 | 19.76543 | 9.1 | −0.701 | Nucleus | dispersed |
| FveHDZ12 | I | 2 | 212 | 24.67798 | 9.2 | −0.924 | Nucleus | dispersed |
| FveHDZ13 | I | 2 | 215 | 24.78802 | 6.47 | −0.886 | Nucleus | dispersed |
| FveHDZ14 | II | 2 | 326 | 36.00138 | 6.24 | −0.525 | Nucleus | WGD/segmental |
| FveHDZ15 | II | 3 | 317 | 35.68198 | 8.48 | −0.874 | Nucleus | WGD/segmental |
| FveHDZ16 | II | 2 | 277 | 31.13987 | 8.43 | −0.851 | Nucleus | WGD/segmental |
| FveHDZ17 | II | 2 | 231 | 26.44407 | 8.84 | −0.848 | Nucleus | dispersed |
| FveHDZ18 | II | 3 | 342 | 38.01078 | 8.68 | −0.695 | Nucleus | dispersed |
| FveHDZ19 | II | 3 | 209 | 23.31446 | 8.94 | −0.732 | Nucleus | dispersed |
| FveHDZ20 | II | 2 | 253 | 28.40885 | 7.69 | −0.872 | Nucleus | WGD/segmental |
| FveHDZ21 | IV | 10 | 771 | 85.78192 | 5.73 | −0.498 | Nucleus | dispersed |
| FveHDZ22 | IV | 8 | 830 | 89.98074 | 5.94 | −0.344 | Nucleus | dispersed |
| FveHDZ23 | IV | 8 | 807 | 88.41103 | 5.41 | −0.306 | Nucleus | dispersed |
| FveHDZ24 | IV | 10 | 773 | 84.29501 | 5.55 | −0.36 | Nucleus | proximal |
| FveHDZ25 | IV | 10 | 777 | 85.43578 | 5.84 | −0.299 | Nucleus | WGD/segmental |
| FveHDZ26 | IV | 8 | 851 | 94.79136 | 5.68 | −0.38 | Nucleus | dispersed |
| FveHDZ27 | IV | 9 | 709 | 78.56529 | 6.43 | −0.322 | Nucleus | dispersed |
| FveHDZ28 | III | 17 | 844 | 92.98051 | 5.94 | −0.134 | Nucleus | dispersed |
| FveHDZ29 | III | 17 | 847 | 92.77479 | 6.07 | −0.138 | Nucleus | dispersed |
| FveHDZ30 | III | 17 | 845 | 92.88056 | 6.06 | −0.148 | Nucleus | dispersed |
| FveHDZ31 | III | 17 | 838 | 91.87144 | 6.03 | −0.086 | Nucleus | dispersed |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Fan, J.; Wu, X.; Guan, L.; Li, C.; Gu, T.; Li, Y.; Ding, J. Genome-Wide Characterization and Expression Profiling of HD-Zip Genes in ABA-Mediated Processes in Fragaria vesca. Plants 2022, 11, 3367. https://doi.org/10.3390/plants11233367
Wang Y, Fan J, Wu X, Guan L, Li C, Gu T, Li Y, Ding J. Genome-Wide Characterization and Expression Profiling of HD-Zip Genes in ABA-Mediated Processes in Fragaria vesca. Plants. 2022; 11(23):3367. https://doi.org/10.3390/plants11233367
Chicago/Turabian StyleWang, Yong, Junmiao Fan, Xinjie Wu, Ling Guan, Chun Li, Tingting Gu, Yi Li, and Jing Ding. 2022. "Genome-Wide Characterization and Expression Profiling of HD-Zip Genes in ABA-Mediated Processes in Fragaria vesca" Plants 11, no. 23: 3367. https://doi.org/10.3390/plants11233367
APA StyleWang, Y., Fan, J., Wu, X., Guan, L., Li, C., Gu, T., Li, Y., & Ding, J. (2022). Genome-Wide Characterization and Expression Profiling of HD-Zip Genes in ABA-Mediated Processes in Fragaria vesca. Plants, 11(23), 3367. https://doi.org/10.3390/plants11233367






