OsMADS58 Stabilizes Gene Regulatory Circuits during Rice Stamen Development
Abstract
1. Introduction
2. Results
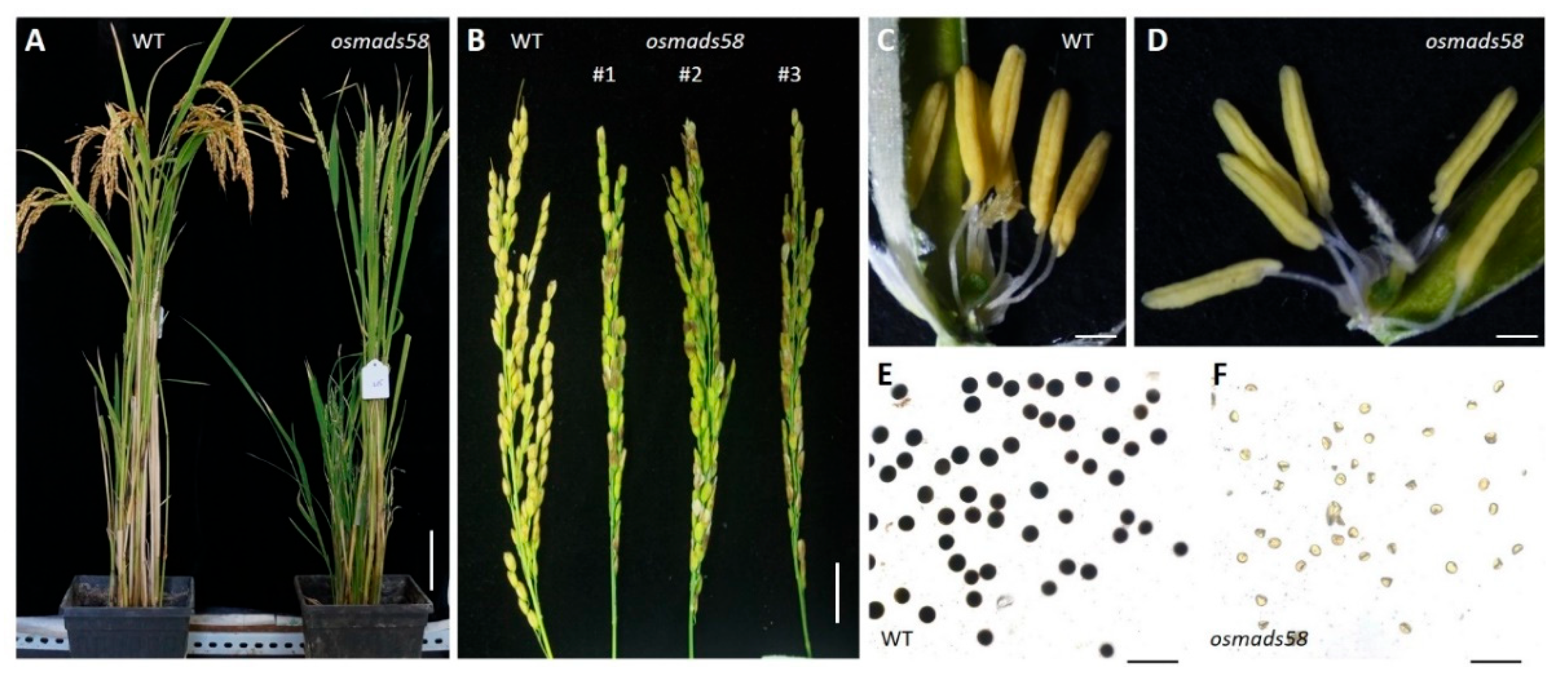
2.1. Osmads58 CRISPR Lines Exhibit Sterility
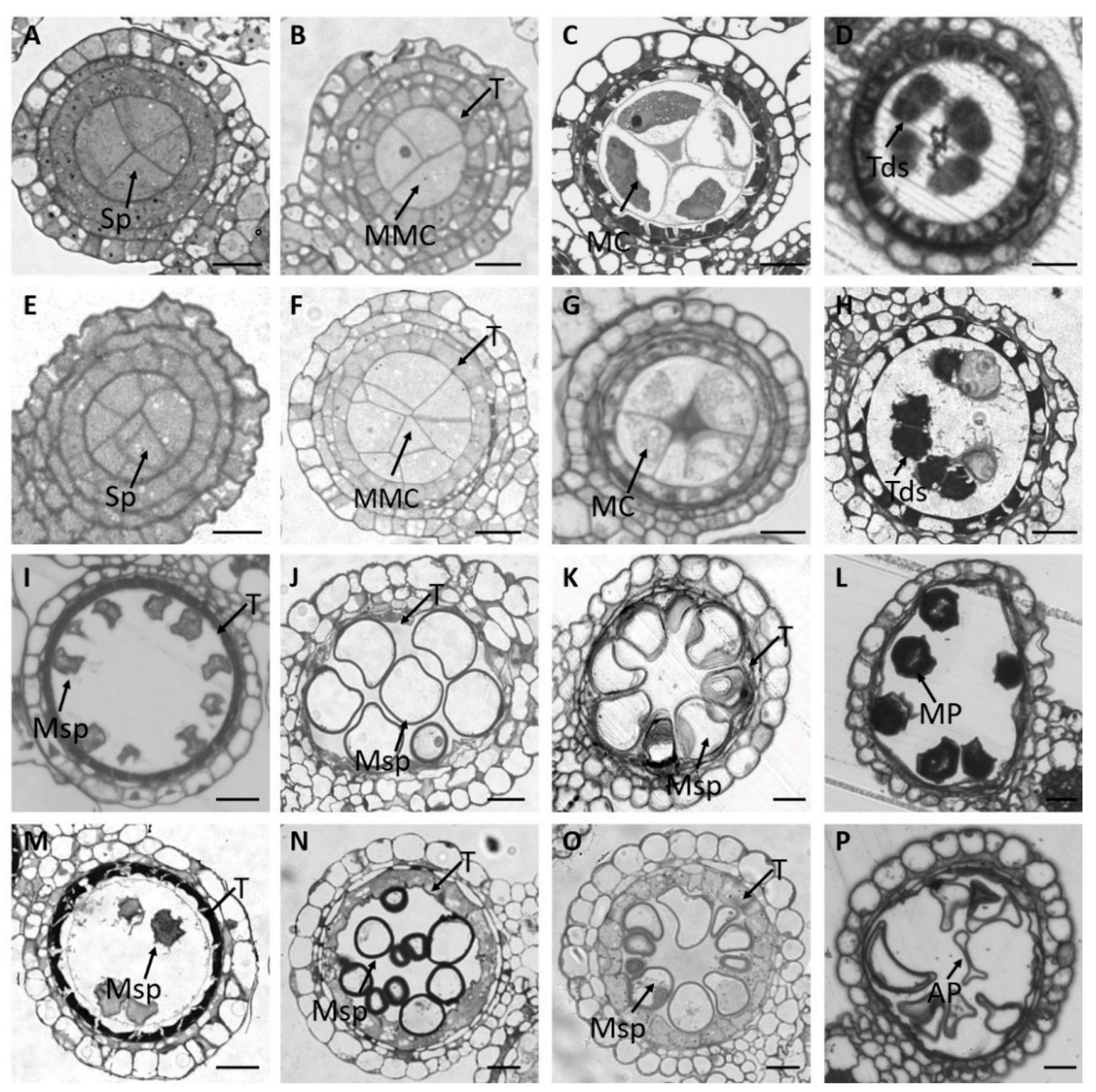
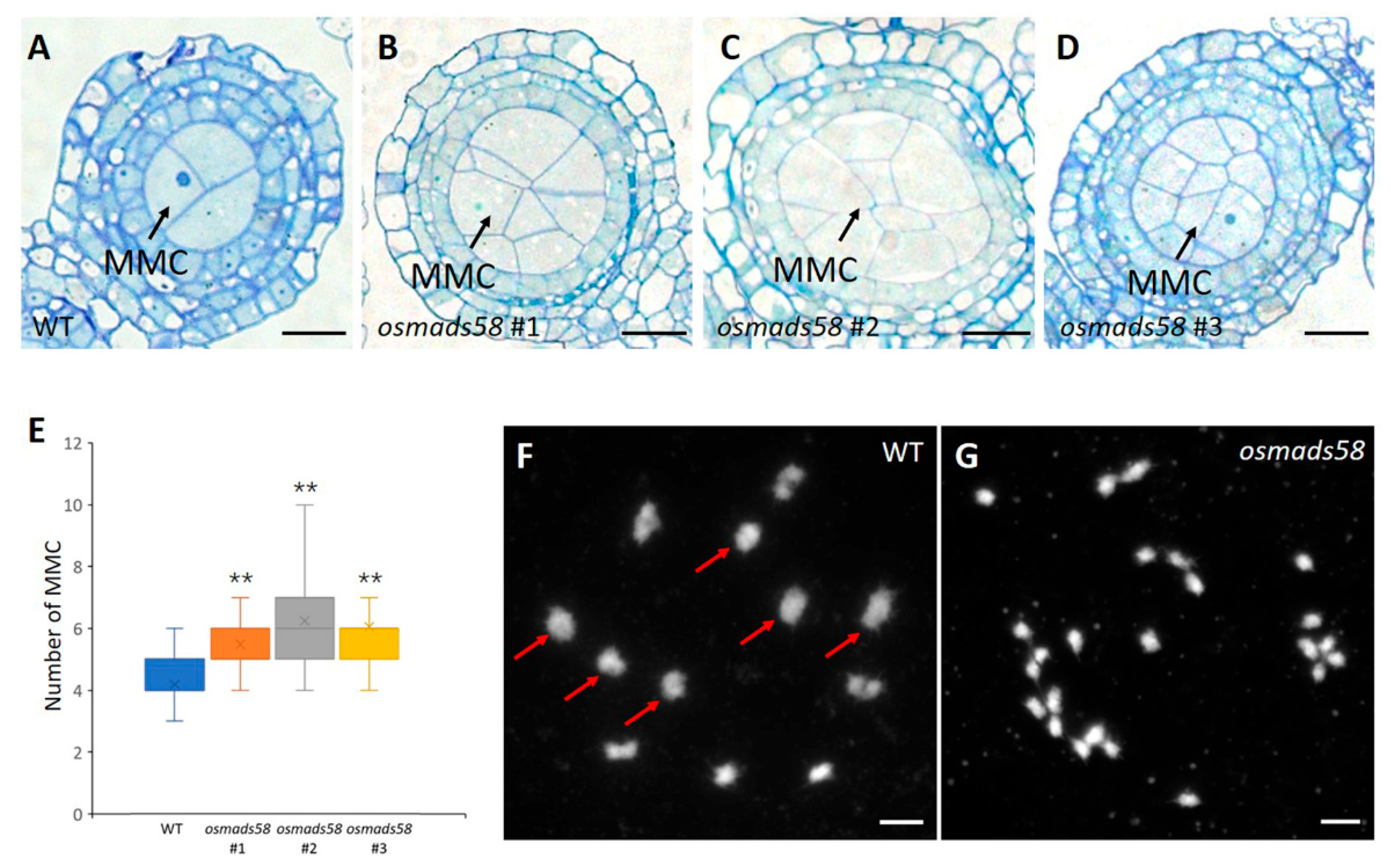
2.2. Both MMCs and Tapetum Are Aberrant in the osmads58 CRISPR Line
2.2.1. Disturbed Chromosome Behavior in Meiosis
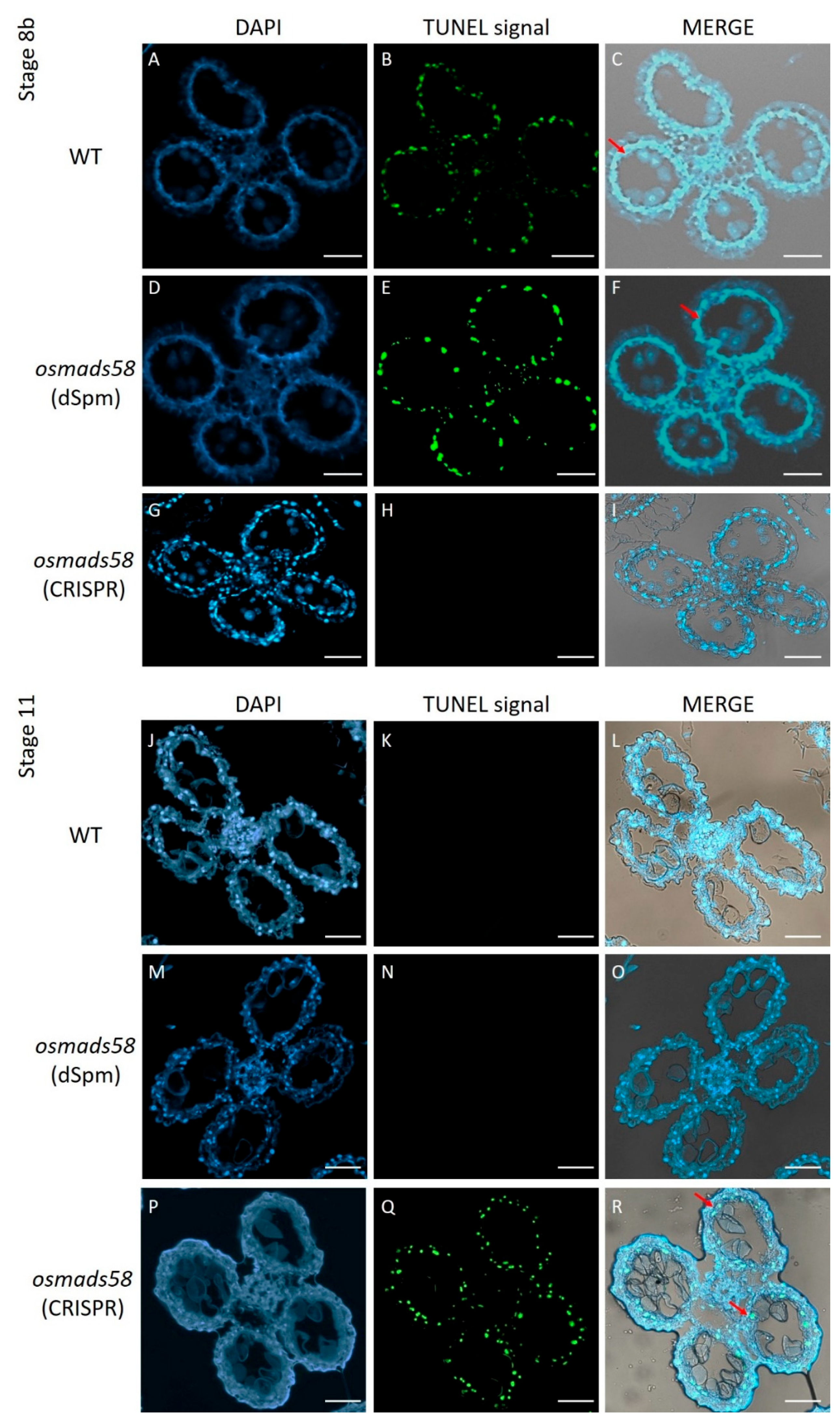
2.2.2. Delayed Tapetum Degeneration
2.3. Gene Expression Is Severely Disturbed in the osmads58 CRISPR Line
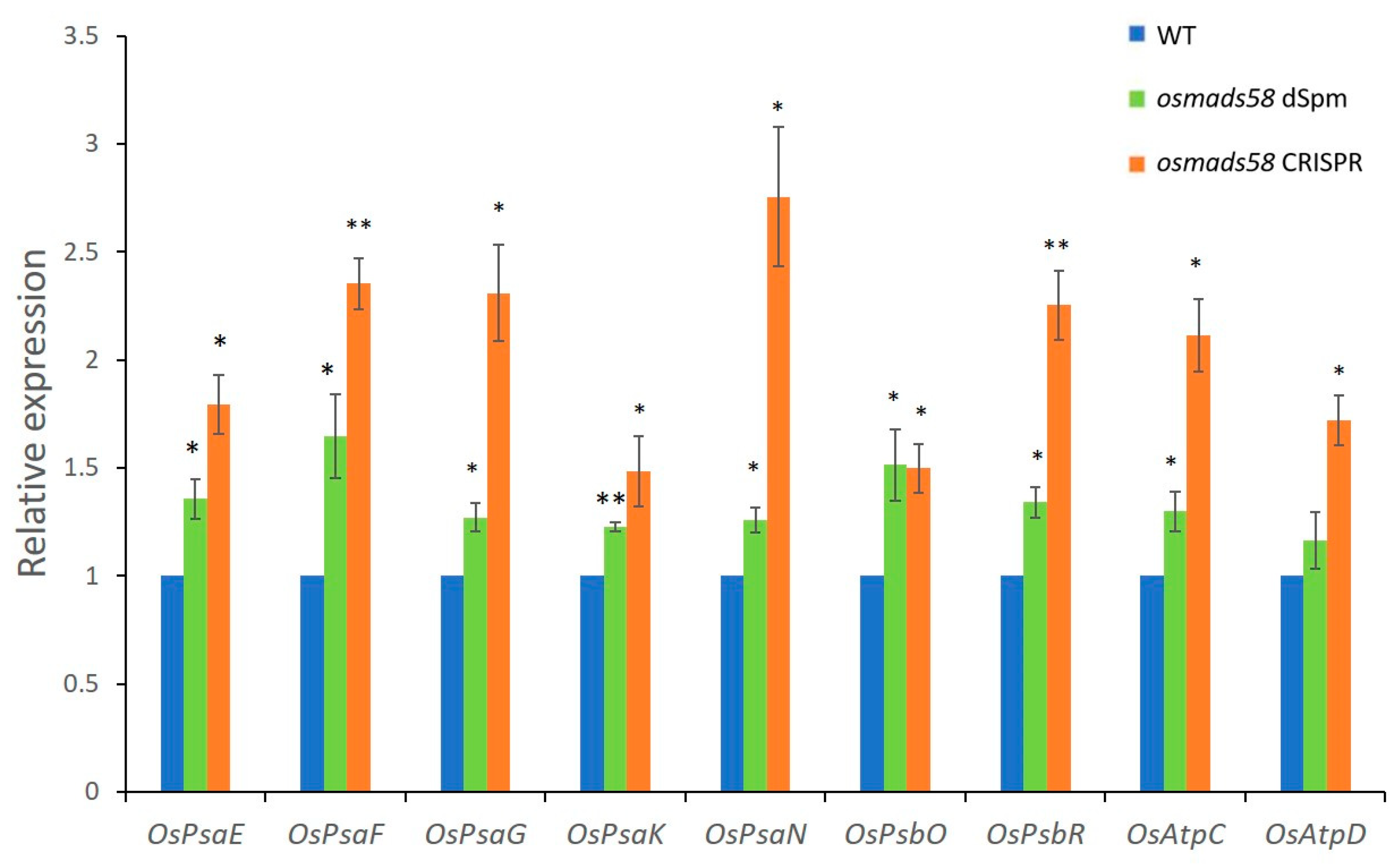
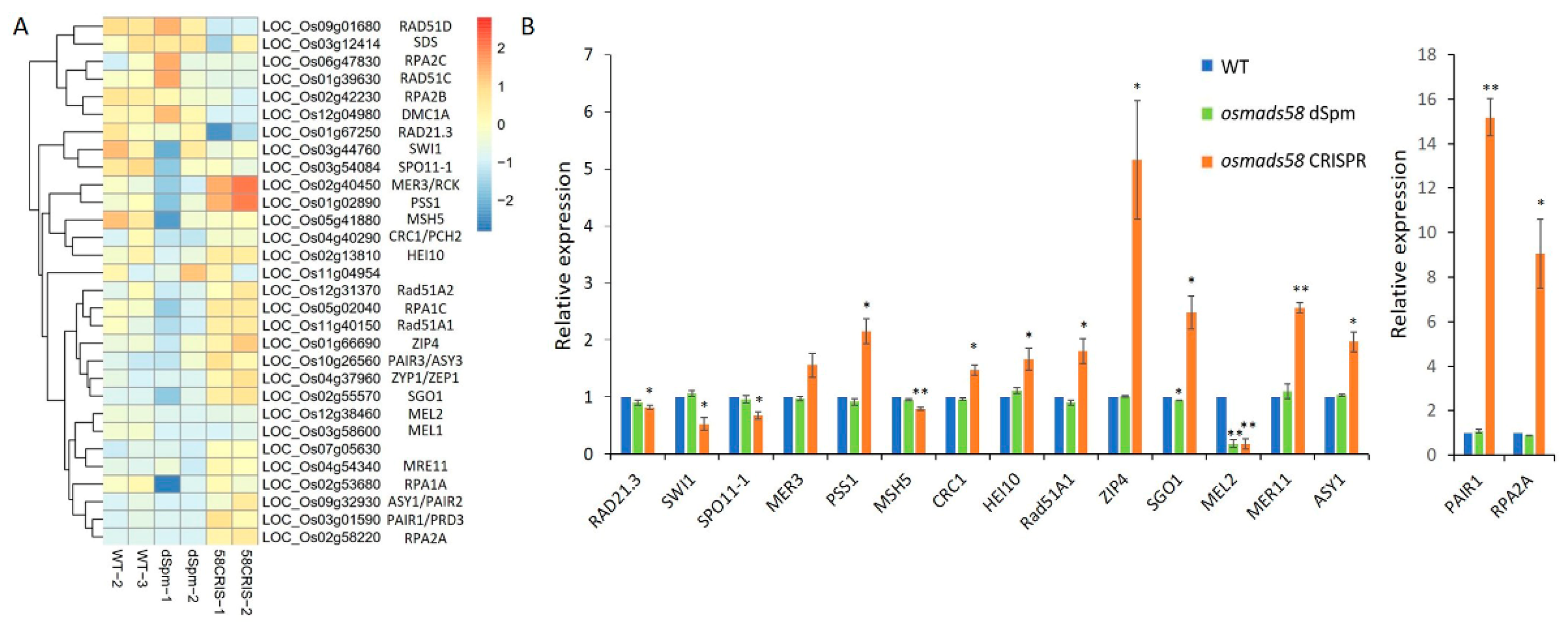
2.3.1. OsMADS58 Target Genes Have Higher Expression in the osmads58 CRISPR Line
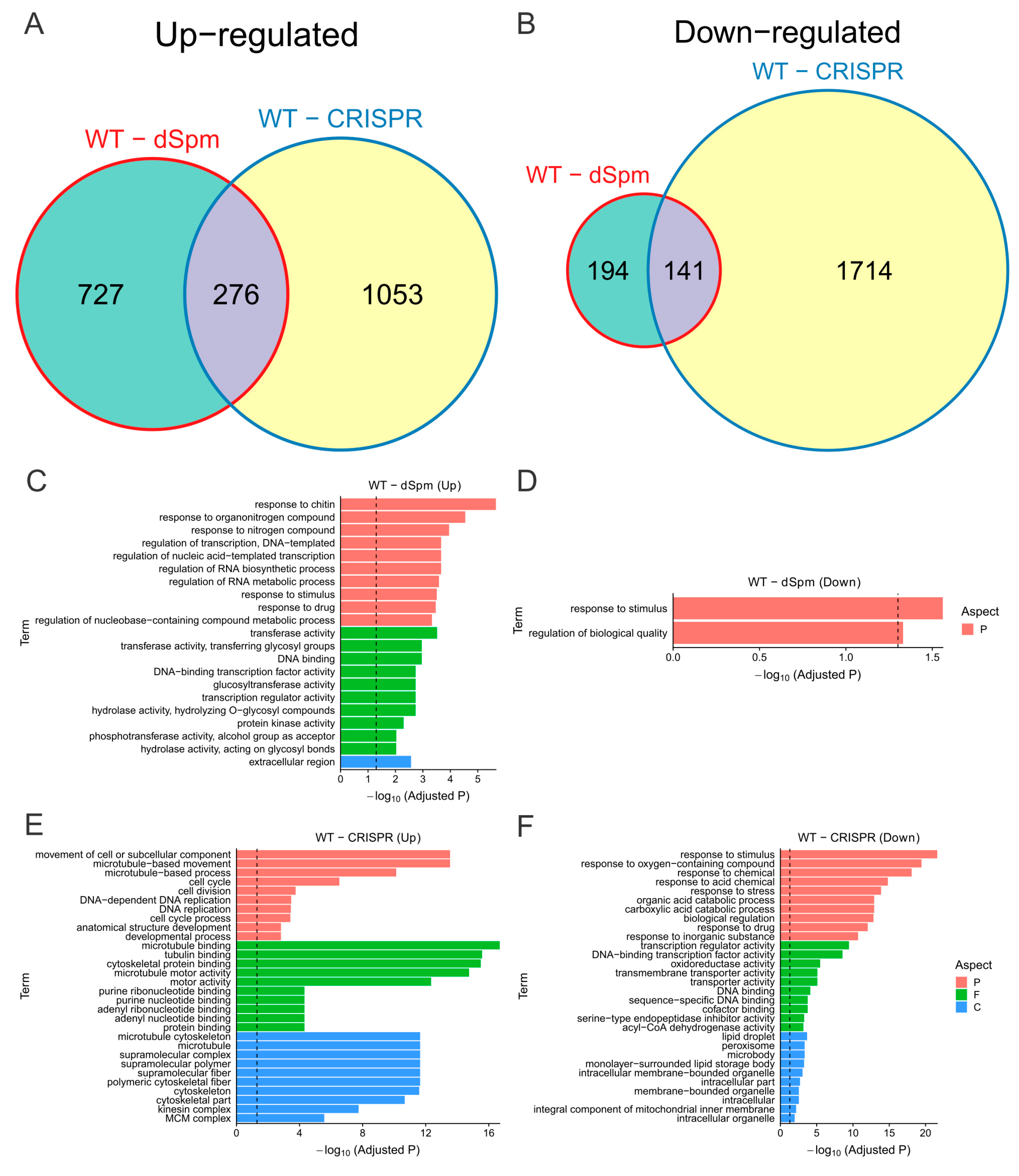
2.3.2. Null Mutation of OsMADS58 Causes Vast Variation in Gene Expression Profiles
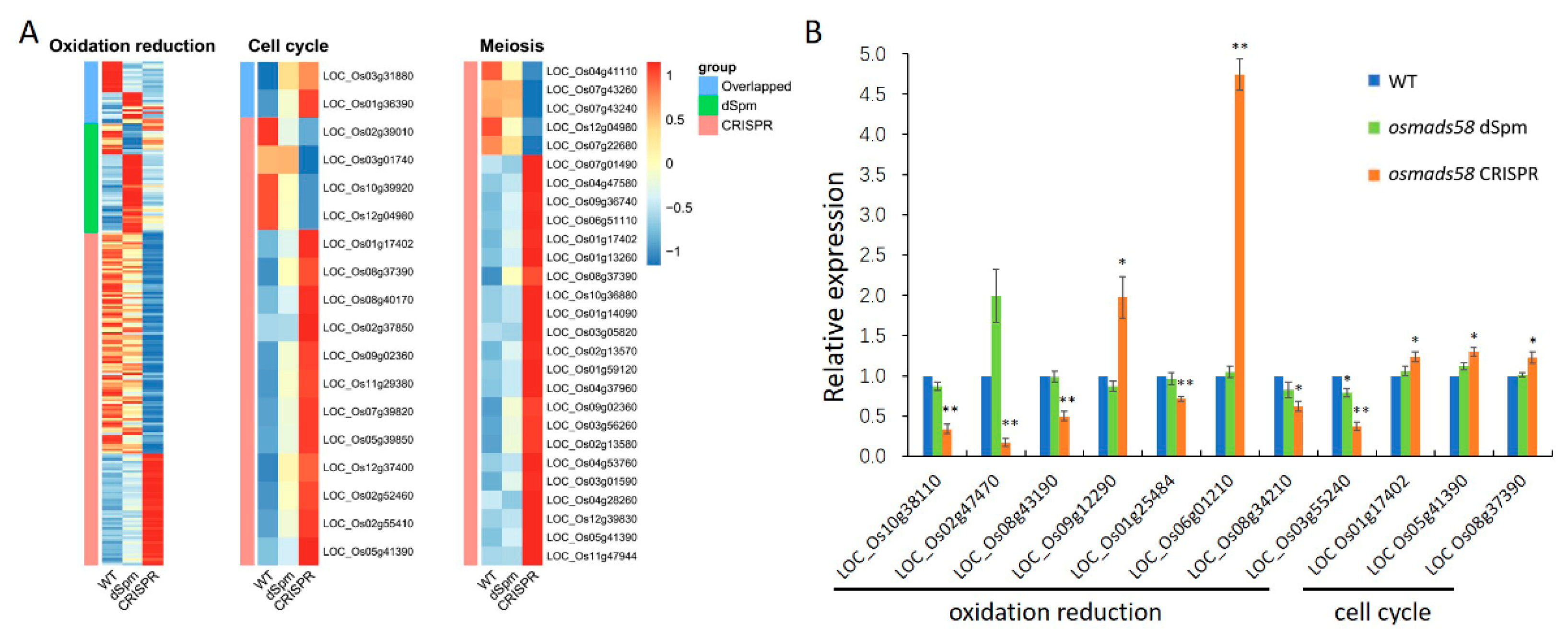
2.3.3. Null Mutation of OsMADS58 Markedly Alters the Expression of Genes in Key Functional Pathways in Stamen Development
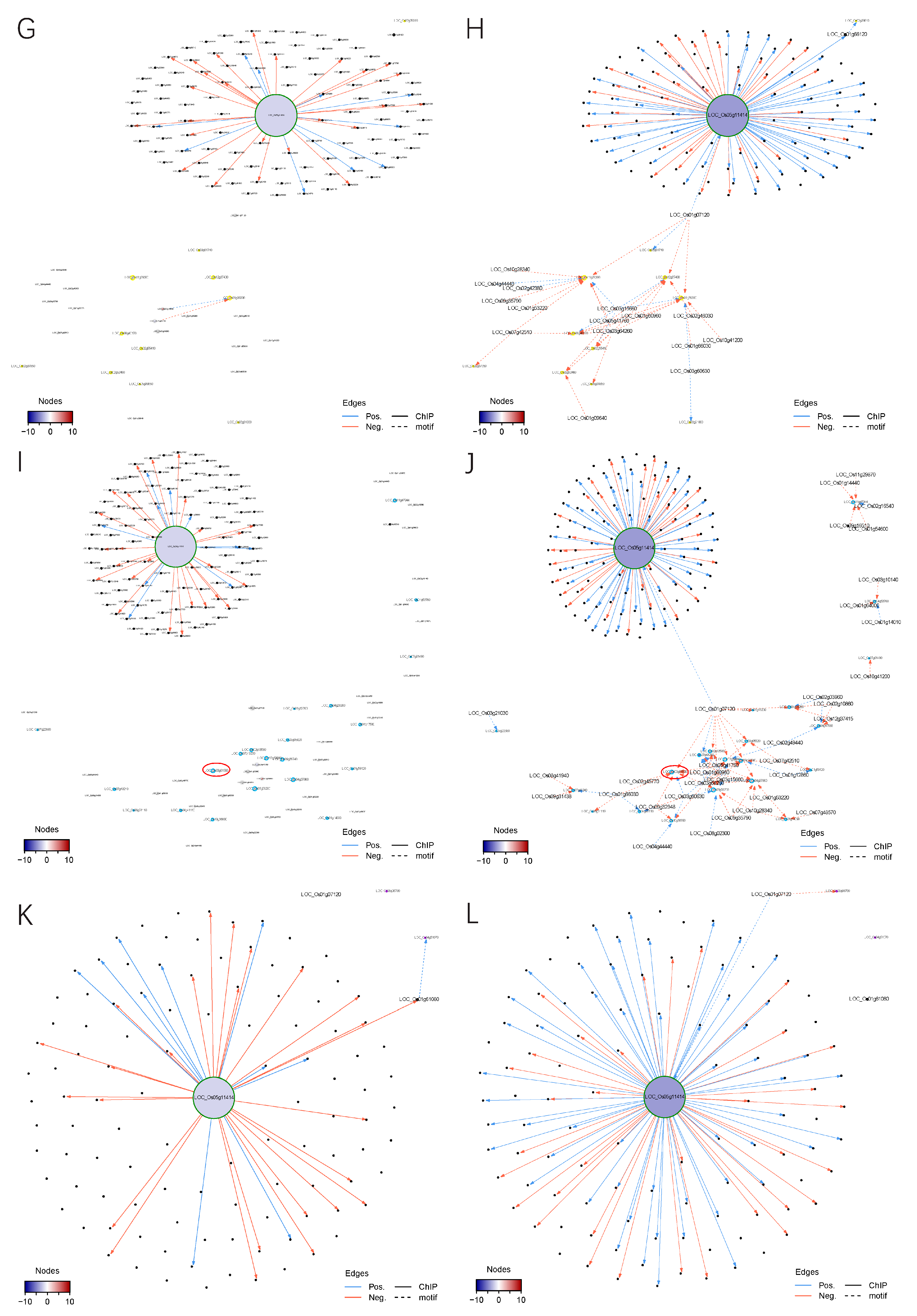
2.4. Correlations between Altered Developmental Events and Gene Expression Networks
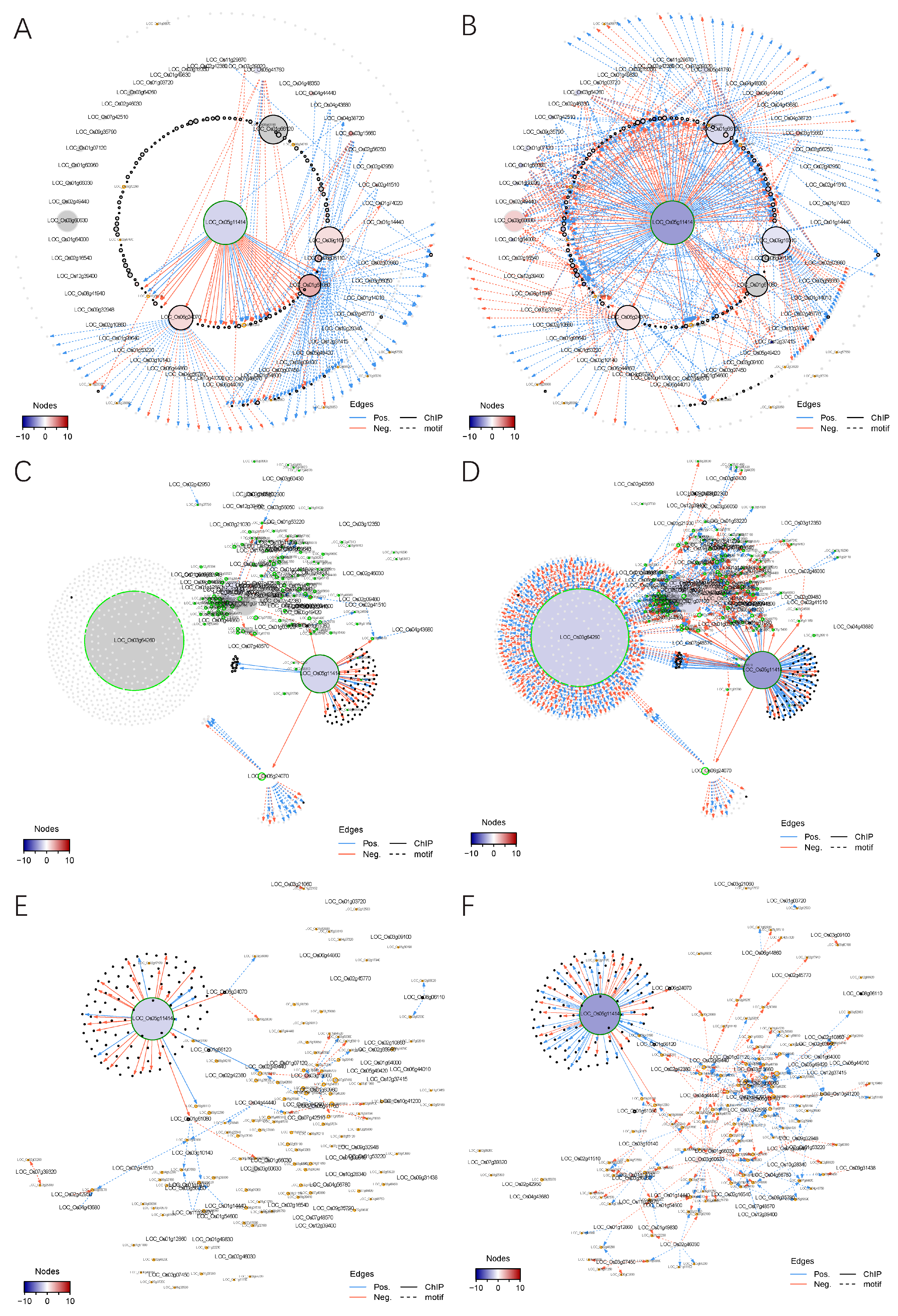
2.4.1. Construction of a Transcriptional Regulatory Network (TRN) Centered on OsMADS58
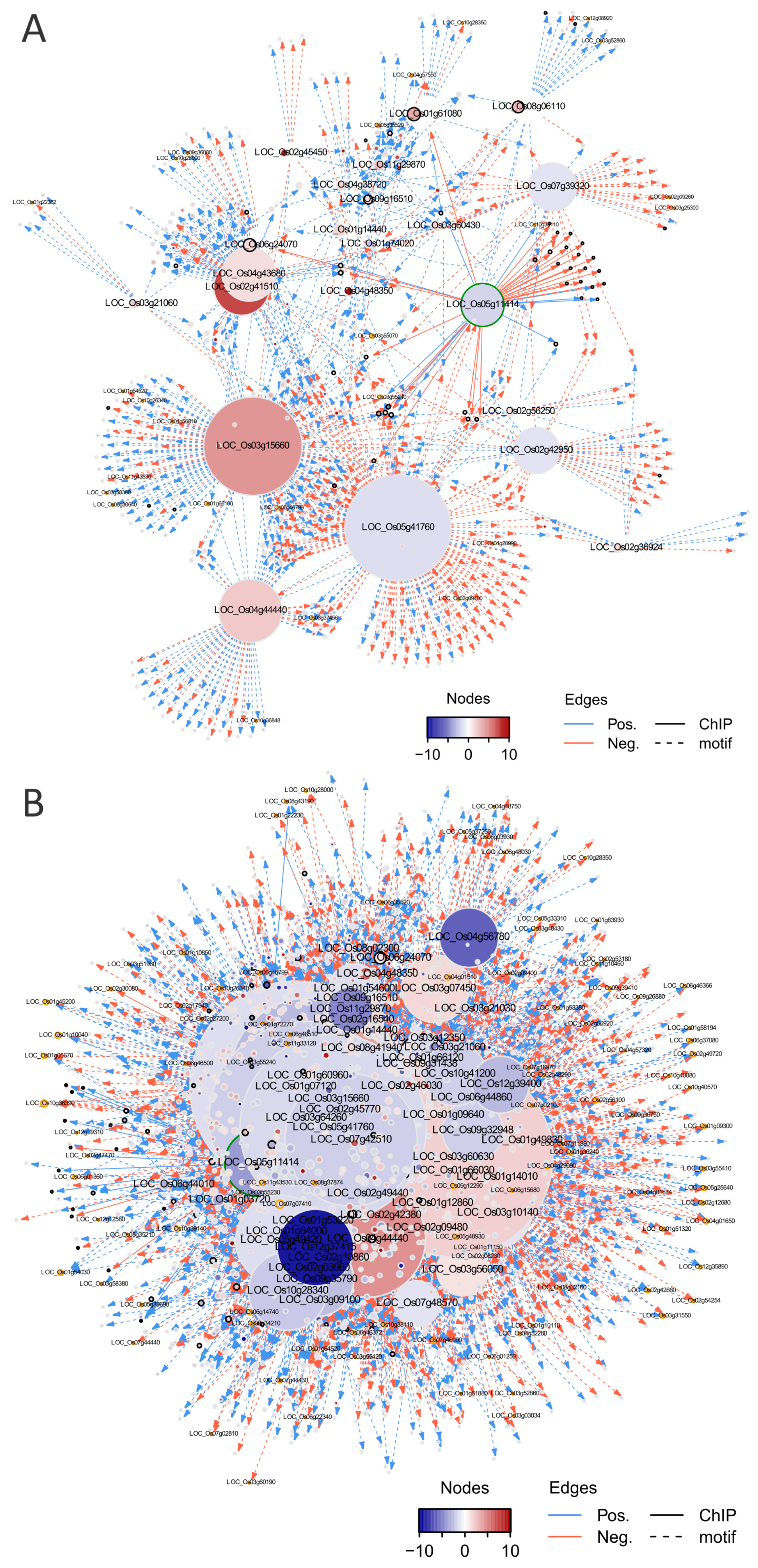
2.4.2. Alteration of TRNs during Stamen Development
2.4.3. Alteration of the Regulatory Circuits Underlying the Meiosis Aberrant
2.5. Network Topologies Explain the Phenotype Differences between dSpm and CRISPR Lines
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. In Vitro Translation of OsMADS58 and Mass Spectrometry Analysis
4.3. Genome Resequencing and SNP Calling of osmads58 CRISPR Lines
4.4. Characterization of Mutant Phenotypes
4.5. TUNEL Assay
4.6. Chromosome Spreads Staining
4.7. Real-Time PCR Expression Assay
4.8. RNA Extraction, cDNA Library Preparation, and Sequencing
4.9. Samples for RNA-Seq, Gene Expression Calculation and Differential Analysis
4.10. TRN Construction
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Nagato, Y. Flower development in rice. J. Exp. Bot. 2011, 62, 4719–4730. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L.; Smyth, D.R.; Meyerowitz, E.M. Genes directing flower development in Arabidopsis. Plant Cell 1989, 1, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L.; Smyth, D.R.; Meyerowitz, E.M. Genetic interactions among floral homeotic genes of Arabidopsis. Development 1991, 112, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Ma, H. Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 2005, 56, 393–434. [Google Scholar] [CrossRef]
- Yanofsky, M.F.; Ma, H.; Bowman, J.L.; Drews, G.N.; Feldmann, K.A.; Meyerowitz, E.M. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 1990, 346, 35–39. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Lee, D.Y.; Miyao, A.; Hirochika, H.; An, G.; Hirano, H.Y. Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa. Plant Cell 2006, 18, 15–28. [Google Scholar] [CrossRef]
- Dreni, L.; Pilatone, A.; Yun, D.; Erreni, S.; Pajoro, A.; Caporali, E.; Zhang, D.; Kater, M.M. Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy. Plant Cell 2011, 23, 2850–2863. [Google Scholar] [CrossRef]
- Yasui, Y.; Tanaka, W.; Sakamoto, T.; Kurata, T.; Hirano, H.Y. Genetic Enhancer Analysis Reveals that FLORAL ORGAN NUMBER2 and OsMADS3 Co-operatively Regulate Maintenance and Determinacy of the Flower Meristem in Rice. Plant Cell Physiol. 2017, 58, 893–903. [Google Scholar] [CrossRef]
- Hu, L.; Liang, W.; Yin, C.; Cui, X.; Zong, J.; Wang, X.; Hu, J.; Zhang, D. Rice MADS3 regulates ROS homeostasis during late anther development. Plant Cell 2011, 23, 515–533. [Google Scholar] [CrossRef]
- Sugiyama, S.H.; Yasui, Y.; Ohmori, S.; Tanaka, W.; Hirano, H.Y. Rice Flower Development Revisited: Regulation of Carpel Specification and Flower Meristem Determinacy. Plant Cell Physiol. 2019, 60, 1284–1295. [Google Scholar] [CrossRef]
- Chen, R.; Shen, L.P.; Wang, D.H.; Wang, F.G.; Zeng, H.Y.; Chen, Z.S.; Peng, Y.B.; Lin, Y.N.; Tang, X.; Deng, M.H.; et al. A Gene Expression Profiling of Early Rice Stamen Development that Reveals Inhibition of Photosynthetic Genes by OsMADS58. Mol. Plant 2015, 8, 1069–1089. [Google Scholar] [CrossRef]
- Schiefthaler, U.; Balasubramanian, S.; Sieber, P.; Chevalier, D.; Wisman, E.; Schneitz, K. Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1999, 96, 11664–11669. [Google Scholar] [CrossRef]
- Yang, W.C.; Ye, D.; Xu, J.; Sundaresan, V. The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 1999, 13, 2108–2117. [Google Scholar] [CrossRef]
- Ito, T.; Wellmer, F.; Yu, H.; Das, P.; Ito, N.; Alves-Ferreira, M.; Riechmann, J.L.; Meyerowitz, E.M. The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature 2004, 430, 356–360. [Google Scholar] [CrossRef]
- Ren, L.; Tang, D.; Zhao, T.; Zhang, F.; Liu, C.; Xue, Z.; Shi, W.; Du, G.; Shen, Y.; Li, Y.; et al. OsSPL regulates meiotic fate acquisition in rice. New Phytol. 2018, 218, 789–803. [Google Scholar] [CrossRef]
- Zhang, D.; Yang, L. Specification of tapetum and microsporocyte cells within the anther. Curr. Opin. Plant Biol. 2014, 17, 49–55. [Google Scholar] [CrossRef]
- Chen, Z.S.; Liu, X.F.; Wang, D.H.; Chen, R.; Zhang, X.L.; Xu, Z.H.; Bai, S.N. Transcription Factor OsTGA10 Is a Target of the MADS Protein OsMADS8 and Is Required for Tapetum Development. Plant Physiol. 2018, 176, 819–835. [Google Scholar] [CrossRef]
- Zhao, F.; Zheng, Y.F.; Zeng, T.; Sun, R.; Yang, J.Y.; Li, Y.; Ren, D.T.; Ma, H.; Xu, Z.H.; Bai, S.N. Phosphorylation of SPOROCYTELESS/NOZZLE by the MPK3/6 Kinase Is Required for Anther Development. Plant Physiol. 2017, 173, 2265–2277. [Google Scholar] [CrossRef]
- Hord, C.L.; Sun, Y.J.; Pillitteri, L.J.; Torii, K.U.; Wang, H.; Zhang, S.; Ma, H. Regulation of Arabidopsis early anther development by the mitogen-activated protein kinases, MPK3 and MPK6, and the ERECTA and related receptor-like kinases. Mol. Plant 2008, 1, 645–658. [Google Scholar] [CrossRef]
- Zhao, D.Z.; Wang, G.F.; Speal, B.; Ma, H. The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 2002, 16, 2021–2031. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wang, D.; Ye, S.; Chen, W.; Li, G.; Xu, Z.; Bai, S.; Zhao, F. Auxin guides germ-cell specification in Arabidopsis anthers. Proc. Natl. Acad. Sci. USA 2021, 118, e2101492118. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.; Liang, W.; Hu, J.; Zhang, D. MTR1 encodes a secretory fasciclin glycoprotein required for male reproductive development in rice. Dev. Cell 2012, 22, 1127–1137. [Google Scholar] [CrossRef] [PubMed]
- Miao, J.; Guo, D.; Zhang, J.; Huang, Q.; Qin, G.; Zhang, X.; Wan, J.; Gu, H.; Qu, L.J. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 2013, 23, 1233–1236. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.C.; Gong, H.Q.; Huang, M.L.; Bai, S.L.; He, Y.B.; Mao, X.; Geng, Z.; Li, S.G.; Wei, L.; Yuwen, J.S.; et al. Molecular analysis of early rice stamen development using organ-specific gene expression profiling. Plant Mol. Biol. 2006, 61, 845–861. [Google Scholar] [CrossRef]
- Wang, M.; Hoekstra, S.; van Bergen, S.; Lamers, G.E.; Oppedijk, B.J.; van der Heijden, M.W.; de Priester, W.; Schilperoort, R.A. Apoptosis in developing anthers and the role of ABA in this process during androgenesis in Hordeum vulgare L. Plant Mol. Biol. 1999, 39, 489–501. [Google Scholar] [CrossRef]
- Tian, F.; Yang, D.C.; Meng, Y.Q.; Jin, J.; Gao, G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2020, 48, D1104–D1113. [Google Scholar] [CrossRef]
- Ji, J.; Tang, D.; Shen, Y.; Xue, Z.; Wang, H.; Shi, W.; Zhang, C.; Du, G.; Li, Y.; Cheng, Z. P31comet, a member of the synaptonemal complex, participates in meiotic DSB formation in rice. Proc. Natl. Acad. Sci. USA 2016, 113, 10577–10582. [Google Scholar] [CrossRef]
- Miao, C.; Tang, D.; Zhang, H.; Wang, M.; Li, Y.; Tang, S.; Yu, H.; Gu, M.; Cheng, Z. Central region component1, a novel synaptonemal complex component, is essential for meiotic recombination initiation in rice. Plant Cell 2013, 25, 2998–3009. [Google Scholar] [CrossRef]
- Nonomura, K.; Nakano, M.; Eiguchi, M.; Suzuki, T.; Kurata, N. PAIR2 is essential for homologous chromosome synapsis in rice meiosis I. J. Cell Sci. 2006, 119, 217–225. [Google Scholar] [CrossRef]
- Wu, Z.; Ji, J.; Tang, D.; Wang, H.; Shen, Y.; Shi, W.; Li, Y.; Tan, X.; Cheng, Z.; Luo, Q. OsSDS is essential for DSB formation in rice meiosis. Front. Plant Sci. 2015, 6, 21. [Google Scholar] [CrossRef]
- Xue, Z.; Liu, C.; Shi, W.; Miao, Y.; Shen, Y.; Tang, D.; Li, Y.; You, A.; Xu, Y.; Chong, K.; et al. OsMTOPVIB is required for meiotic bipolar spindle assembly. Proc. Natl. Acad. Sci. USA 2019, 116, 15967–15972. [Google Scholar] [CrossRef]
- Yu, H.; Wang, M.; Tang, D.; Wang, K.; Chen, F.; Gong, Z.; Gu, M.; Cheng, Z. OsSPO11-1 is essential for both homologous chromosome pairing and crossover formation in rice. Chromosoma 2010, 119, 625–636. [Google Scholar] [CrossRef]
- Fang, Y.; Qin, X.; Liao, Q.; Du, R.; Luo, X.; Zhou, Q.; Li, Z.; Chen, H.; Jin, W.; Yuan, Y.; et al. The genome of homosporous maidenhair fern sheds light on the euphyllophyte evolution and defences. Nat. Plants 2021, 8, 1024–1037. [Google Scholar] [CrossRef]
- Bai, S.N.; Rao, G.Y.; Yang, J. Origins of the seed: The “golden-trio hypothesis”. Front Plant Sci. 2022, 13, 965000. [Google Scholar] [CrossRef] [PubMed]
- Musser, J.M.; Schippers, K.J.; Nickel, M.; Mizzon, G.; Kohn, A.B.; Pape, C.; Ronchi, P.; Papadopoulos, N.; Tarashansky, A.J.; Hammel, J.U.; et al. Profiling cellular diversity in sponges informs animal cell type and nervous system evolution. Science 2021, 374, 717–723. [Google Scholar] [CrossRef]
- Kelliher, T.; Walbot, V. Hypoxia triggers meiotic fate acquisition in maize. Science 2012, 337, 345–348. [Google Scholar] [CrossRef]
- Xie, H.T.; Wan, Z.Y.; Li, S.; Zhang, Y. Spatiotemporal Production of Reactive Oxygen Species by NADPH Oxidase Is Critical for Tapetal Programmed Cell Death and Pollen Development in Arabidopsis. Plant Cell 2014, 26, 2007–2023. [Google Scholar] [CrossRef]
- Yu, J.; Zhang, D. Molecular Control of Redox Homoeostasis in Specifying the Cell Identity of Tapetal and Microsporocyte Cells in Rice. Rice 2019, 12, 42. [Google Scholar] [CrossRef]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, L.; Tian, F.; Cheng, Z.; Zhao, Q.; Feng, Q.; Zhao, Y.; Han, B.; Fang, Y.; Lin, Y.; Chen, R.; et al. OsMADS58 Stabilizes Gene Regulatory Circuits during Rice Stamen Development. Plants 2022, 11, 2899. https://doi.org/10.3390/plants11212899
Shen L, Tian F, Cheng Z, Zhao Q, Feng Q, Zhao Y, Han B, Fang Y, Lin Y, Chen R, et al. OsMADS58 Stabilizes Gene Regulatory Circuits during Rice Stamen Development. Plants. 2022; 11(21):2899. https://doi.org/10.3390/plants11212899
Chicago/Turabian StyleShen, Liping, Feng Tian, Zhukuan Cheng, Qiang Zhao, Qi Feng, Yan Zhao, Bin Han, Yuhan Fang, Yanan Lin, Rui Chen, and et al. 2022. "OsMADS58 Stabilizes Gene Regulatory Circuits during Rice Stamen Development" Plants 11, no. 21: 2899. https://doi.org/10.3390/plants11212899
APA StyleShen, L., Tian, F., Cheng, Z., Zhao, Q., Feng, Q., Zhao, Y., Han, B., Fang, Y., Lin, Y., Chen, R., Wang, D., Sun, W., Sun, J., Zeng, H., Yao, N., Gao, G., Luo, J., Xu, Z., & Bai, S. (2022). OsMADS58 Stabilizes Gene Regulatory Circuits during Rice Stamen Development. Plants, 11(21), 2899. https://doi.org/10.3390/plants11212899









