Reduced Expression of PRX2/ATPRX1, PRX8, PRX35, and PRX73 Affects Cell Elongation, Vegetative Growth, and Vasculature Structures in Arabidopsis thaliana
Abstract
1. Introduction
2. Results
2.1. Identification of Four PRX Genes Belonging to Class III Peroxidase
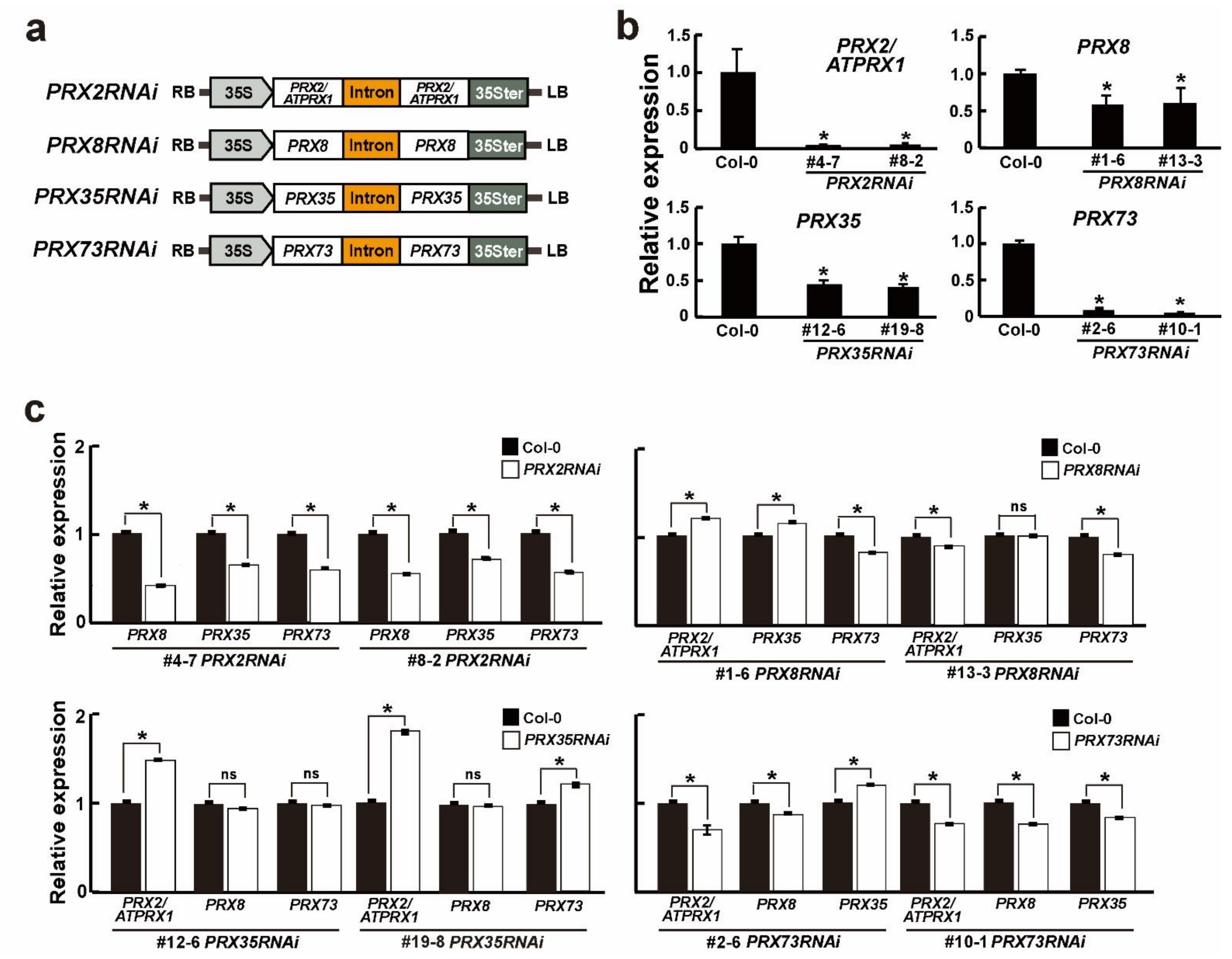
2.2. The Effect of RNAi-Mediated Silencing on PRX2/ATPRX1, PRX8, PRX35, and PRX73 Expression
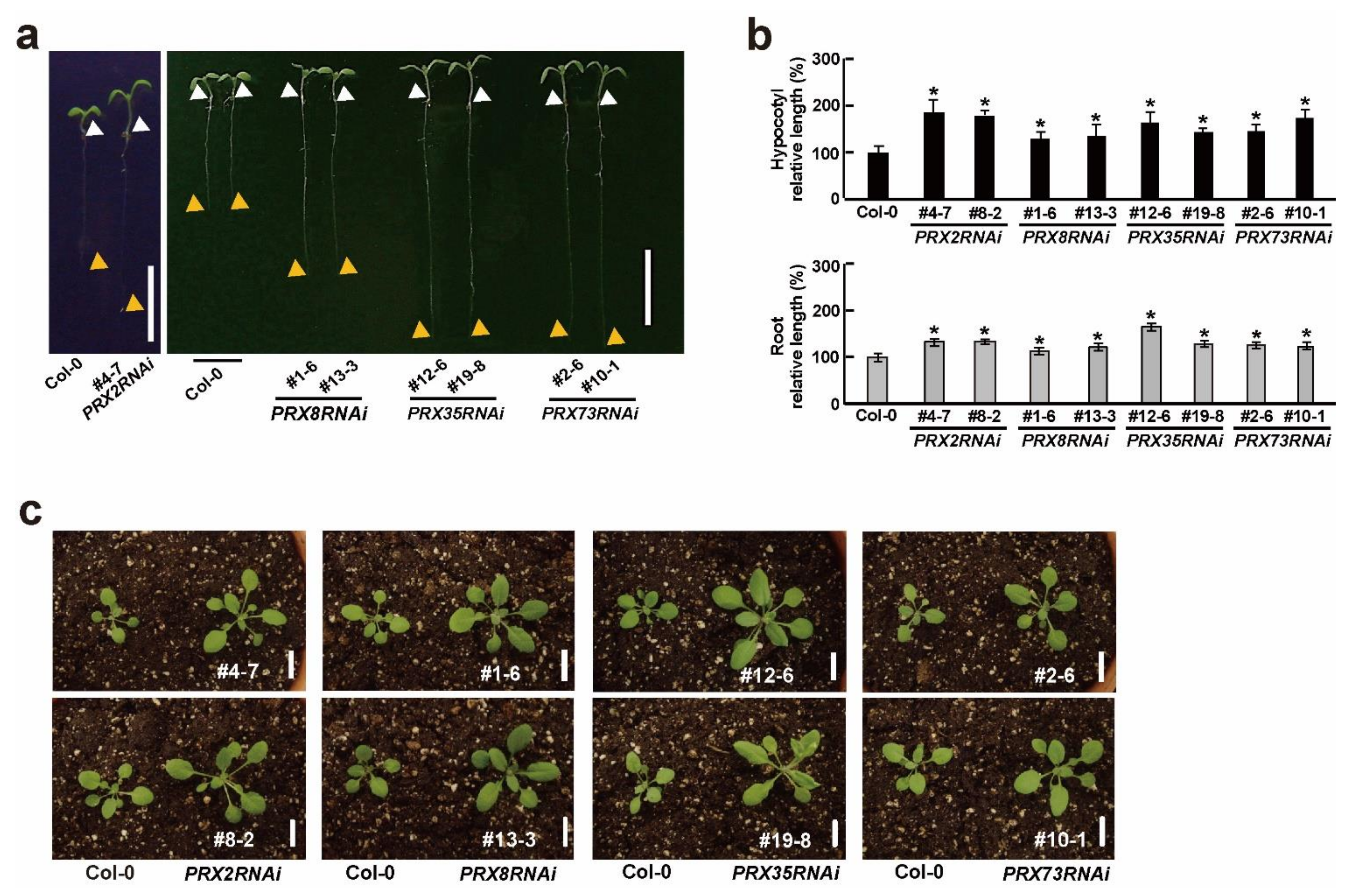
2.3. Growth and Developmental Changes in Four PRXsRNAi Plants
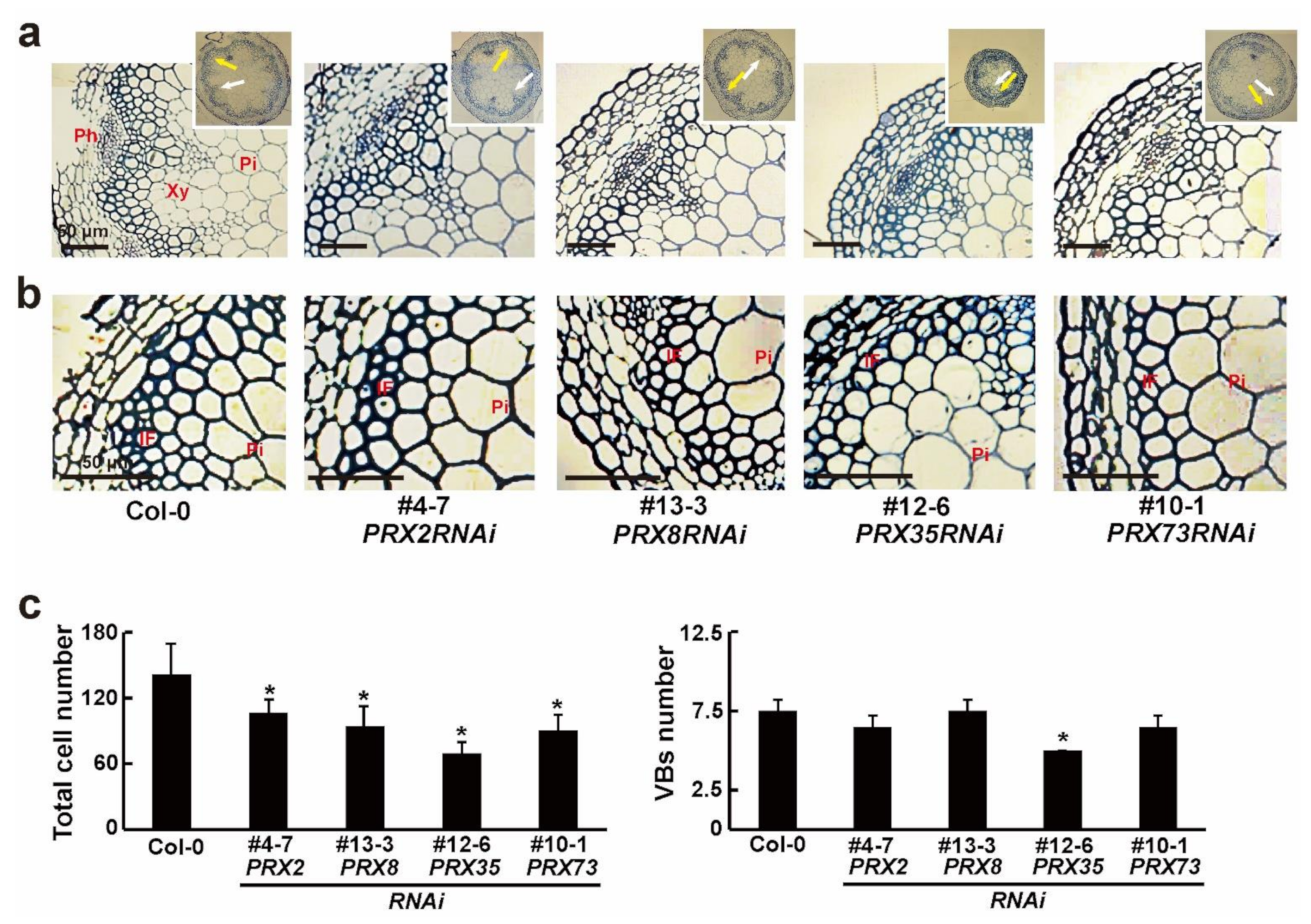
2.4. Phenotypic Changes of Vascular Patterns in Stems of Four PRXRNAi Plants
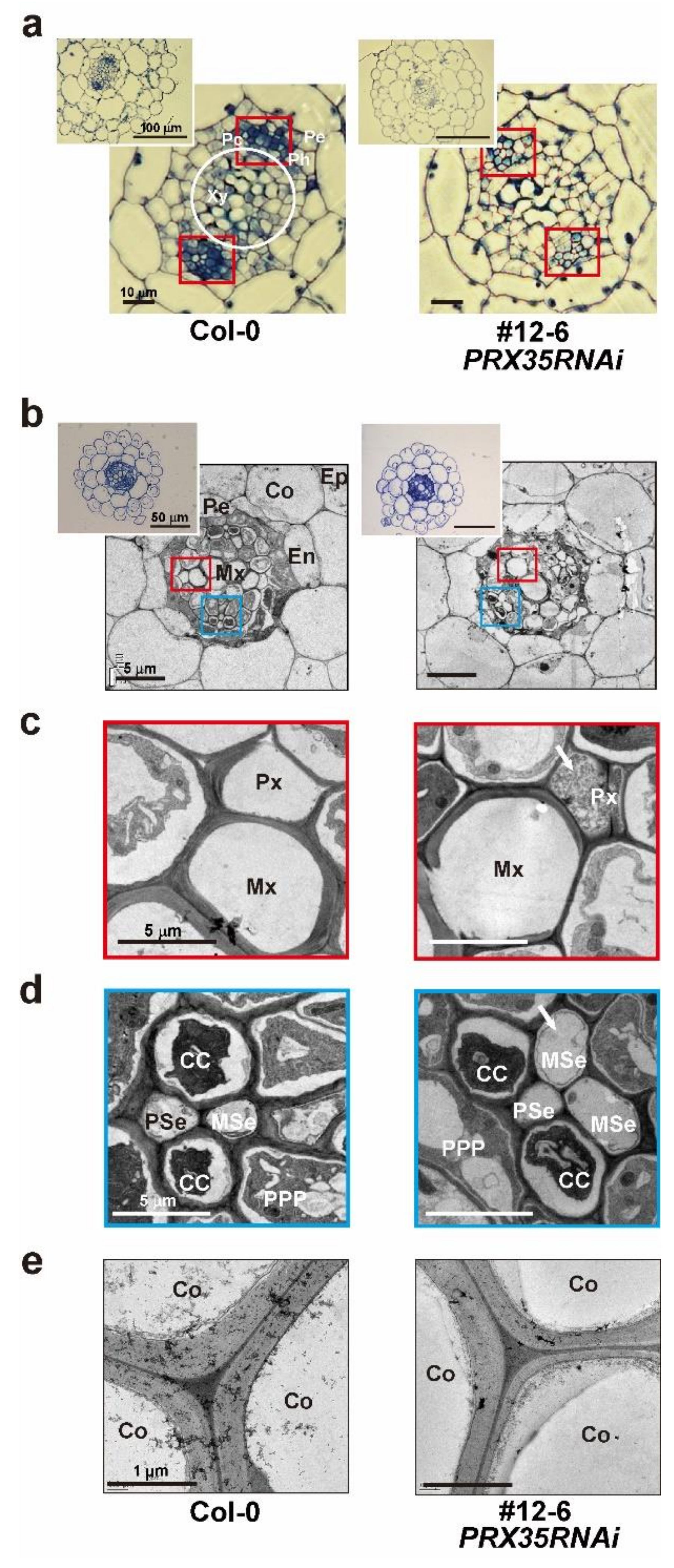
2.5. Phenotypic Changes in Hypocotyl and Root Vasculature Structures in PRX35RNAi Plants
2.6. Altered Vasculature Structures and Decreased mRNA Levels of Lignin Synthesis and Synthesis-Regulated Genes in Stems of PRX35RNAi Plants
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Generation of Transgenic Arabidopsis Plants
4.3. RNA Isolation and Gene Expression Analysis
4.4. Measurements of Chlorophyll and Anthocyanin Contents
4.5. Histochemical Staining
4.6. Transmission Electron Microscopy (TEM)
4.7. Quantitative Vascular Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shigeto, J.; Tsutsumi, Y. Diverse functions and reactions of class III peroxidases. New Phytol. 2016, 209, 1395–1402. [Google Scholar] [CrossRef]
- Marjamaa, K.; Kukkola, E.M.; Fagerstedt, K.V. The role of xylem class III peroxidases in lignification. J. Exp. Bot. 2009, 60, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Cosio, C.; Dunand, C. Specific functions of individual class III peroxidase genes. J. Exp. Bot. 2009, 60, 391–408. [Google Scholar] [CrossRef]
- Luthje, S.; Martinez-Cortes, T. Membrane-Bound Class III Peroxidases: Unexpected Enzymes with Exciting Functions. Int. J. Mol. Sci. 2018, 19, 2876. [Google Scholar] [CrossRef]
- Luthje, S.; Meisrimler, C.N.; Hopff, D.; Moller, B. Phylogeny, topology, structure and functions of membrane-bound class III peroxidases in vascular plants. Phytochemistry 2011, 72, 1124–1135. [Google Scholar] [CrossRef] [PubMed]
- Shigeto, J.; Itoh, Y.; Hirao, S.; Ohira, K.; Fujita, K.; Tsutsumi, Y. Simultaneously disrupting AtPrx2, AtPrx25 and AtPrx71 alters lignin content and structure in Arabidopsis stem. J. Integr. Plant Biol. 2015, 57, 349–356. [Google Scholar] [CrossRef]
- Kunieda, T.; Shimada, T.; Kondo, M.; Nishimura, M.; Nishitani, K.; Hara-Nishimura, I. Spatiotemporal secretion of PEROXIDASE36 is required for seed coat mucilage extrusion in Arabidopsis. Plant Cell 2013, 25, 1355–1367. [Google Scholar] [CrossRef]
- Liszkay, A.; Kenk, B.; Schopfer, P. Evidence for the involvement of cell wall peroxidase in the generation of hydroxyl radicals mediating extension growth. Planta 2003, 217, 658–667. [Google Scholar] [CrossRef] [PubMed]
- Raggi, S.; Ferrarini, A.; Delledonne, M.; Dunand, C.; Ranocha, P.; De Lorenzo, G.; Cervone, F.; Ferrari, S. The Arabidopsis Class III Peroxidase AtPRX71 Negatively Regulates Growth under Physiological Conditions and in Response to Cell Wall Damage. Plant Physiol. 2015, 169, 2513–2525. [Google Scholar] [CrossRef]
- Jemmat, A.M.; Ranocha, P.; Le Ru, A.; Neel, M.; Jauneau, A.; Raggi, S.; Ferrari, S.; Burlat, V.; Dunand, C. Coordination of five class III peroxidase-encoding genes for early germination events of Arabidopsis thaliana. Plant Sci. 2020, 298, 110565. [Google Scholar] [CrossRef]
- Francoz, E.; Ranocha, P.; Nguyen-Kim, H.; Jamet, E.; Burlat, V.; Dunand, C. Roles of cell wall peroxidases in plant development. Phytochemistry 2015, 112, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, N.; Benske, A.; Betz, H.; Schuetz, M.; Samuels, A.L. Laccases and Peroxidases Co-Localize in Lignified Secondary Cell Walls throughout Stem Development. Plant Physiol. 2020, 184, 806–822. [Google Scholar] [CrossRef] [PubMed]
- Keren-Keiserman, A.; Tanami, Z.; Shoseyov, O.; Ginzberg, I. Peroxidase activity associated with suberization processes of the muskmelon (Cucumis melo) rind. Physiol. Plant 2004, 121, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Zipor, G.; Duarte, P.; Carqueijeiro, I.; Shahar, L.; Ovadia, R.; Teper-Bamnolker, P.; Eshel, D.; Levin, Y.; Doron-Faigenboim, A.; Sottomayor, M.; et al. In planta anthocyanin degradation by a vacuolar class III peroxidase in Brunfelsia calycina flowers. New Phytol. 2015, 205, 653–665. [Google Scholar] [CrossRef]
- Gazaryan, I.G.; Lagrimini, L.M.; Ashby, G.A.; Thorneley, R.N. Mechanism of indole-3-acetic acid oxidation by plant peroxidases: Anaerobic stopped-flow spectrophotometric studies on horseradish and tobacco peroxidases. Biochem. J. 1996, 313, 841–847. [Google Scholar] [CrossRef]
- Pacheco, J.M.; Ranocha, P.; Kasulin, L.; Fusari, C.M.; Servi, L.; Aptekmann, A.A.; Gabarain, V.B.; Peralta, J.M.; Borassi, C.; Marzol, E.; et al. Apoplastic class III peroxidases PRX62 and PRX69 promote Arabidopsis root hair growth at low temperature. Nat. Commun. 2022, 13, 1310. [Google Scholar] [CrossRef]
- Passardi, F.; Theiler, G.; Zamocky, M.; Cosio, C.; Rouhier, N.; Teixera, F.; Margis-Pinheiro, M.; Ioannidis, V.; Penel, C.; Falquet, L.; et al. PeroxiBase: The peroxidase database. Phytochemistry 2007, 68, 1605–1611. [Google Scholar] [CrossRef]
- Severino, F.E.; Brandalise, M.; Costa, C.S.; Wilcken, S.R.; Maluf, M.P.; Goncalves, W.; Maia, I.G. CaPrx, a Coffea arabica gene encoding a putative class III peroxidase induced by root-knot nematode infection. Plant Sci. 2012, 191–192, 35–42. [Google Scholar] [CrossRef]
- Van Loon, L.C.; Rep, M.; Pieterse, C.M. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef]
- Wu, Y.; Yang, Z.; How, J.; Xu, H.; Chen, L.; Li, K. Overexpression of a peroxidase gene (AtPrx64) of Arabidopsis thaliana in tobacco improves plant’s tolerance to aluminum stress. Plant Mol. Biol. 2017, 95, 157–168. [Google Scholar] [CrossRef]
- Shigeto, J.; Nagano, M.; Fujita, K.; Tsutsumi, Y. Catalytic profile of Arabidopsis peroxidases, AtPrx-2, 25 and 71, contributing to stem lignification. PLoS ONE 2014, 9, e105332. [Google Scholar] [CrossRef]
- Blee, K.A.; Choi, J.W.; O’Connell, A.P.; Schuch, W.; Lewis, N.G.; Bolwell, G.P. A lignin-specific peroxidase in tobacco whose antisense suppression leads to vascular tissue modification. Phytochemistry 2003, 64, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Marjamaa, K.; Kukkola, E.; Lundell, T.; Karhunen, P.; Saranpaa, P.; Fagerstedt, K.V. Monolignol oxidation by xylem peroxidase isoforms of Norway spruce (Picea abies) and silver birch (Betula pendula). Tree Physiol. 2006, 26, 605–611. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, Y.; Kajita, S.; Kawai, S.; Katayama, Y.; Morohoshi, N. Down-regulation of an anionic peroxidase in transgenic aspen and its effect on lignin characteristics. J. Plant Res. 2003, 116, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, S.; Nishida, T.; Tsutsumi, Y.; Kondo, R. Lignin dehydrogenative polymerization mechanism: A poplar cell wall peroxidase directly oxidizes polymer lignin and produces in vitro dehydrogenative polymer rich in beta-O-4 linkage. FEBS Lett. 2004, 562, 197–201. [Google Scholar] [CrossRef]
- Herrero, J.; Fernandez-Perez, F.; Yebra, T.; Novo-Uzal, E.; Pomar, F.; Pedreno, M.A.; Cuello, J.; Guera, A.; Esteban-Carrasco, A.; Zapata, J.M. Bioinformatic and functional characterization of the basic peroxidase 72 from Arabidopsis thaliana involved in lignin biosynthesis. Planta 2013, 237, 1599–1612. [Google Scholar] [CrossRef]
- Fernandez-Perez, F.; Pomar, F.; Pedreno, M.A.; Novo-Uzal, E. The suppression of AtPrx52 affects fibers but not xylem lignification in Arabidopsis by altering the proportion of syringyl units. Physiol. Plant 2015, 154, 395–406. [Google Scholar] [CrossRef]
- Lee, Y.; Rubio, M.C.; Alassimone, J.; Geldner, N. A mechanism for localized lignin deposition in the endodermis. Cell 2013, 153, 402–412. [Google Scholar] [CrossRef]
- Taha, M.; Foda, M.; Shahsavari, E.; Aburto-Medina, A.; Adetutu, E.; Ball, A. Commercial feasibility of lignocellulose biodegradation: Possibilities and challenges. Curr. Opin. Biotechnol. 2016, 38, 190–197. [Google Scholar] [CrossRef]
- Yuan, Y.; Jiang, B.; Chen, H.; Wu, W.; Wu, S.; Jin, Y.; Xiao, H. Recent advances in understanding the effects of lignin structural characteristics on enzymatic hydrolysis. Biotechnol. Biofuels 2021, 14, 205. [Google Scholar] [CrossRef]
- Liu, X.; Bouxin, F.P.; Fan, J.; Budarin, V.L.; Hu, C.; Clark, J.H. Recent Advances in the Catalytic Depolymerization of Lignin towards Phenolic Chemicals: A Review. ChemSusChem 2020, 13, 4296–4317. [Google Scholar] [CrossRef]
- Luccioni, L.G.; Oliverio, K.A.; Yanovsky, M.J.; Boccalandro, H.E.; Casal, J.J. Brassinosteroid mutants uncover fine tuning of phytochrome signaling. Plant Physiol. 2002, 128, 173–181. [Google Scholar] [CrossRef]
- Oh, E.; Zhu, J.Y.; Wang, Z.Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 2012, 14, 802–809. [Google Scholar] [CrossRef]
- Kim, B.; Jeong, Y.J.; Corvalan, C.; Fujioka, S.; Cho, S.; Park, T.; Choe, S. Darkness and gulliver2/phyB mutation decrease the abundance of phosphorylated BZR1 to activate brassinosteroid signaling in Arabidopsis. Plant J. 2014, 77, 737–747. [Google Scholar] [CrossRef]
- Passardi, F.; Tognolli, M.; De Meyer, M.; Penel, C.; Dunand, C. Two cell wall associated peroxidases from Arabidopsis influence root elongation. Planta 2006, 223, 965–974. [Google Scholar] [CrossRef]
- Jin, J.; Hewezi, T.; Baum, T.J. Arabidopsis peroxidase AtPRX53 influences cell elongation and susceptibility to Heterodera schachtii. Plant Signal Behav. 2011, 6, 1778–1786. [Google Scholar] [CrossRef] [PubMed]
- Mei, W.; Qin, Y.; Song, W.; Li, J.; Zhu, Y. Cotton GhPOX1 encoding plant class III peroxidase may be responsible for the high level of reactive oxygen species production that is related to cotton fiber elongation. J. Genet. Genomics. 2009, 36, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Nooden, L.D.; Penney, J.P. Correlative controls of senescence and plant death in Arabidopsis thaliana (Brassicaceae). J. Exp. Bot. 2001, 52, 2151–2159. [Google Scholar] [CrossRef] [PubMed]
- Kang, X.; Kirui, A.; Dickwella Widanage, M.C.; Mentink-Vigier, F.; Cosgrove, D.J.; Wang, T. Lignin-polysaccharide interactions in plant secondary cell walls revealed by solid-state NMR. Nat. Commun. 2019, 10, 347. [Google Scholar] [CrossRef]
- Yuan, Y.; Teng, Q.; Zhong, R.; Haghighat, M.; Richardson, E.A.; Ye, Z.H. Mutations of Arabidopsis TBL32 and TBL33 Affect Xylan Acetylation and Secondary Wall Deposition. PLoS ONE 2016, 11, e0146460. [Google Scholar] [CrossRef]
- Kubo, M.; Udagawa, M.; Nishikubo, N.; Horiguchi, G.; Yamaguchi, M.; Ito, J.; Mimura, T.; Fukuda, H.; Demura, T. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005, 19, 1855–1860. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.L.; Lee, C.H.; Zhong, R.Q.; Ye, Z.H. MYB58 and MYB63 Are Transcriptional Activators of the Lignin Biosynthetic Pathway during Secondary Cell Wall Formation in Arabidopsis. Plant Cell 2009, 21, 248–266. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Choi, H.; An, G. Roles of lignin biosynthesis and regulatory genes in plant development. J. Integr. Plant. Biol. 2015, 57, 902–912. [Google Scholar] [CrossRef] [PubMed]
- Brownleader, M.D.; Ahmed, N.; Trevan, M.; Chaplin, M.F.; Dey, P.M. Purification and Partial Characterization of Tomato Extensin Peroxidase. Plant Physiol. 1995, 109, 1115–1123. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Chaplin, M.; Trevan, M.; Dey, P.; Brownleader, M.D. Purification and partial characterization of ‘extensin peroxidase’. Biochem. Soc. Trans. 1995, 23, 154. [Google Scholar] [CrossRef]
- Fry, S.C. Cross-linking of matrix polymers in the growing cell-walls of angiosperms. Annu. Rev. Plant. Physiol. Plant Mol. Biol. 1986, 37, 165–186. [Google Scholar] [CrossRef]
- Macadam, J.W.; Nelson, C.J.; Sharp, R.E. Peroxidase activity in the leaf elongation zone of tall fescue: I. Spatial distribution of ionically bound peroxidase activity in genotypes differing in length of the elongation zone. Plant Physiol. 1992, 99, 872–878. [Google Scholar] [CrossRef]
- Warneck, H.M.; Haug, T.; Seitz, U. Activation of cell wall associated peroxidase isoenzymes in pea epicotyls by a xyloglucan-derived nonasaccharide. J. Exp. Bot. 1996, 47, 1897–1904. [Google Scholar] [CrossRef]
- Wallace, G.; Fry, S.C. Action of diverse peroxidases and laccases on six cell wall-related phenolic compounds. Phytochemistry 1999, 52, 769–773. [Google Scholar] [CrossRef]
- Lu, D.; Wang, T.; Persson, S.; Mueller-Roeber, B.; Schippers, J.H. Transcriptional control of ROS homeostasis by KUODA1 regulates cell expansion during leaf development. Nat. Commun. 2014, 5, 3767. [Google Scholar] [CrossRef]
- Cosio, C.; Ranocha, P.; Francoz, E.; Burlat, V.; Zheng, Y.; Perry, S.E.; Ripoll, J.J.; Yanofsky, M.; Dunand, C. The class III peroxidase PRX17 is a direct target of the MADS-box transcription factor AGAMOUS-LIKE15 (AGL15) and participates in lignified tissue formation. New Phytol. 2017, 213, 250–263. [Google Scholar] [CrossRef] [PubMed]
- Passardi, F.; Penel, C.; Dunand, C. Performing the paradoxical: How plant peroxidases modify the cell wall. Trends Plant Sci. 2005, 9, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Ripperger, A.; Ye, Z.H. Ectopic deposition of lignin in the pith of stems of two Arabidopsis mutants. Plant Physiol. 2000, 123, 59–70. [Google Scholar] [CrossRef]
- Blaschek, L.; Pesquet, E. Phenoloxidases in Plants-How Structural Diversity Enables Functional Specificity. Front Plant Sci. 2021, 12, 754601. [Google Scholar] [CrossRef]
- Valerio, L.; De Meyer, M.; Penel, C.; Dunand, C. Expression analysis of the Arabidopsis peroxidase multigenic family. Phytochemistry 2004, 65, 1331–1342. [Google Scholar] [CrossRef]
- Huang, J.; Gu, M.; Lai, Z.; Fan, B.; Shi, K.; Zhou, Y.H.; Yu, J.Q.; Chen, Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef]
- Schilmiller, A.L.; Stout, J.; Weng, J.K.; Humphreys, J.; Ruegger, M.O.; Chapple, C. Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J. 2009, 60, 771–782. [Google Scholar] [CrossRef]
- Sibout, R.; Eudes, A.; Mouille, G.; Pollet, B.; Lapierre, C.; Jouanin, L.; Seguin, A. CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 2005, 17, 2059–2076. [Google Scholar] [CrossRef]
- Franke, R.; Hemm, M.R.; Denault, J.W.; Ruegger, M.O.; Humphreys, J.M.; Chapple, C. Changes in secondary metabolism and deposition of an unusual lignin in the ref8 mutant of Arabidopsis. Plant J. 2002, 30, 47–59. [Google Scholar] [CrossRef]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R.; et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar] [CrossRef]
- Jeong, Y.J.; Lee, J.S.; Kim, D.G. Characterization of BRASSINOSTEROID F-BOX Proteins BRFPs that Regulate BRASSINOSTEROID-INSENSITIVE 2 Kinase. J. Plant Biol. 2022, 65, 53–63. [Google Scholar] [CrossRef]
- Jeong, Y.J. Putative E3 ligases as candidates controlling BRASSINOSTEROID INSENSITIVE 2 (BIN2) kinase in Arabidopsis. Plant Biotechnol. Rep. 2020, 14, 703–712. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.; Corvalan, C.; Kwon, S.I.; Choe, S. Analysis of anti-BZR1 antibody reveals the roles BES1 in maintaining the BZR1 levels in Arabidopsis. J. Plant Biol. 2015, 58, 87–95. [Google Scholar] [CrossRef]
- Jeong, Y.J.; Park, S.; Suh, S.J.; Kwon, S.I.; Cha, R.; Kim, Y.E.; Choe, S. Overexpression of the 3’ half of the PHYB partially suppresses dwarfism in the brassinosteroid-insensitive bri1-5 mutant. J. Plant Biol. 2016, 59, 83–91. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Chlorophylls and Carotenoids—Pigments of Photosynthetic Biomembranes. Methods Enzymol. 1987, 148, 350–382. [Google Scholar]
- Mita, S.; Murano, N.; Akaike, M.; Nakamura, K. Mutants of Arabidopsis thaliana with pleiotropic effects on the expression of the gene for beta-amylase and on the accumulation of anthocyanin that are inducible by sugars. Plant J. 1997, 11, 841–851. [Google Scholar] [CrossRef]
- Spurr, A.R. A Low-Viscosity Epoxy Resin Embedding Medium for Electron Microscopy. J. Ultrastruct. Res. 1969, 26, 31–43. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, Y.J.; Kim, Y.-C.; Lee, J.S.; Kim, D.-G.; Lee, J.H. Reduced Expression of PRX2/ATPRX1, PRX8, PRX35, and PRX73 Affects Cell Elongation, Vegetative Growth, and Vasculature Structures in Arabidopsis thaliana. Plants 2022, 11, 3353. https://doi.org/10.3390/plants11233353
Jeong YJ, Kim Y-C, Lee JS, Kim D-G, Lee JH. Reduced Expression of PRX2/ATPRX1, PRX8, PRX35, and PRX73 Affects Cell Elongation, Vegetative Growth, and Vasculature Structures in Arabidopsis thaliana. Plants. 2022; 11(23):3353. https://doi.org/10.3390/plants11233353
Chicago/Turabian StyleJeong, Yu Jeong, Young-Cheon Kim, June Seung Lee, Dong-Gwan Kim, and Jeong Hwan Lee. 2022. "Reduced Expression of PRX2/ATPRX1, PRX8, PRX35, and PRX73 Affects Cell Elongation, Vegetative Growth, and Vasculature Structures in Arabidopsis thaliana" Plants 11, no. 23: 3353. https://doi.org/10.3390/plants11233353
APA StyleJeong, Y. J., Kim, Y.-C., Lee, J. S., Kim, D.-G., & Lee, J. H. (2022). Reduced Expression of PRX2/ATPRX1, PRX8, PRX35, and PRX73 Affects Cell Elongation, Vegetative Growth, and Vasculature Structures in Arabidopsis thaliana. Plants, 11(23), 3353. https://doi.org/10.3390/plants11233353










