Genome Editing in Crop Plant Research—Alignment of Expectations and Current Developments
Abstract
:1. Introduction
2. Results
2.1. Expectations of Genome Editing in Crop Plant Development Are Diverse, of Abstract Nature, and Differ among Stakeholders
2.2. Plant Traits Linked to Sustainable Development Goals
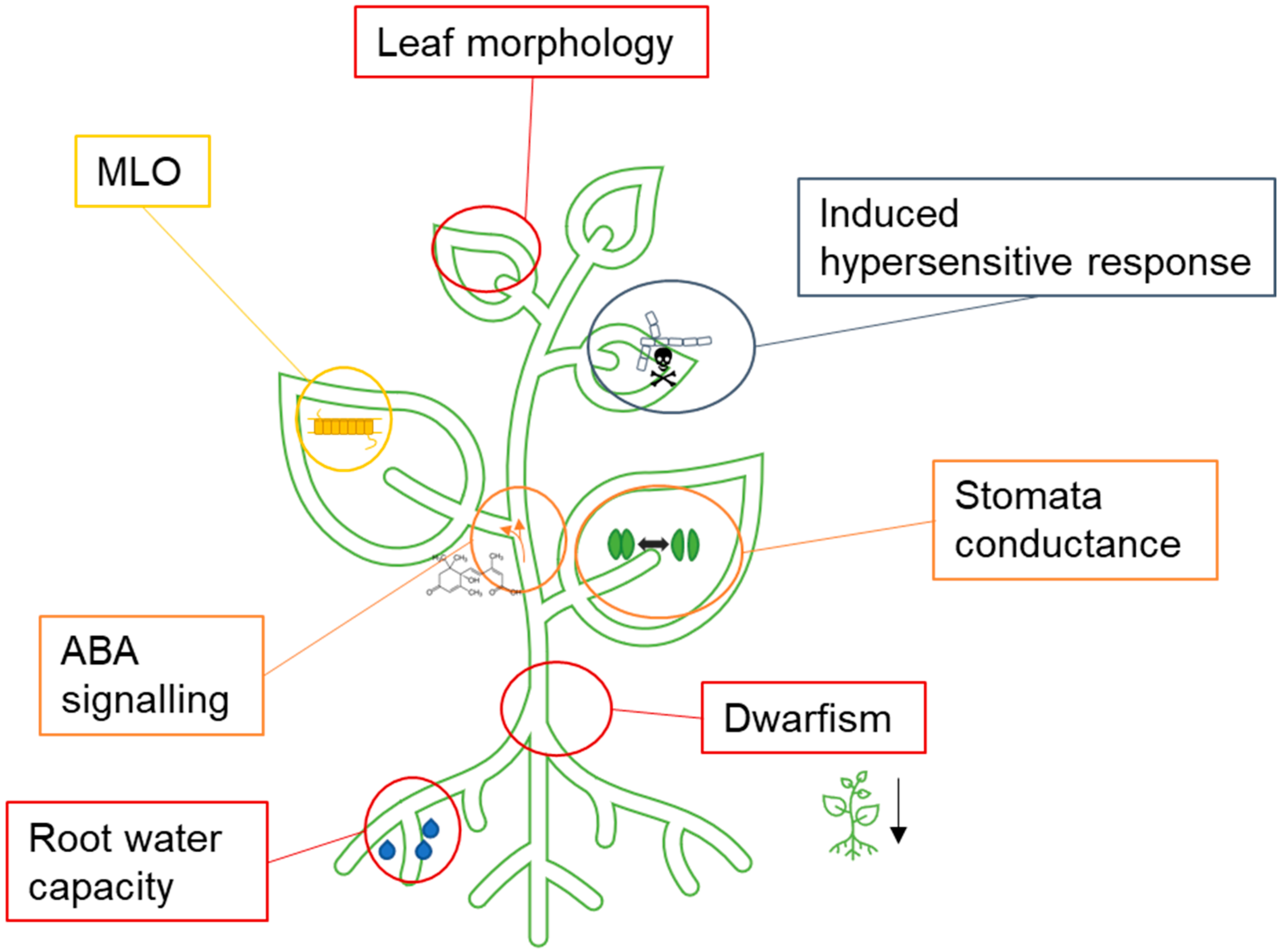
2.3. Overview on the Status of Plant Traits Addressed with Genome Editing
2.4. Crop Development and Research Status on Drought Tolerance
2.4.1. Possible Setscrews for Adapting to Immediate Drought Tolerance
2.4.2. Changes Altering Drought Tolerance Implemented with the Help of NGT
2.4.3. Future Options
2.5. Molecular Mechanisms of Plant Filamentous Pathogen Resistance
2.5.1. Different Classes of Resistance-Mediating Gene Loci Distinguished in the Literature
2.5.2. Changes Altering Pathogen Resistance Implemented with the Help of NGT
3. Discussion
3.1. Identifying Political, Societal, and Economic Expectations for Use of NGT Plants in Agriculture
3.1.1. Aligning Identified Expectations with Scientific Development
3.1.2. Resistance against Fungal Pathogens
3.1.3. Aiming at Drought Tolerance
4. Materials and Methods
Content Analysis of Political Expectations towards Plant Development
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lusser, M.; Parisi, C.; Plan, D.; Rodríguez-Cerezo, E. New Plant Breeding Techniques: State-of-the-Art and Prospects for Commercial Development; Publications Office of the European Union: Luxembourg, 2011. [Google Scholar] [CrossRef]
- Broothaerts, W.; Jacchia, S.; Angers, A.; Petrillo, M.; Querci, M.; Savini, C.; Van den Eede, G.; Emons, H. New Genomic Techniques: State-of-the-Art Review; EUR 30430 EN; Publications Office of the European Union: Luxembourg, 2021; ISBN 978-92-76-24696-1. JRC121847. [Google Scholar] [CrossRef]
- EFSA Panel on Genetically Modified Organisms (GMO). Scientific opinion addressing the safety assessment of plants developed using Zinc Finger Nuclease 3 and other Site-Directed Nucleases with similar function. EFSA J. 2012, 10, 2193. [Google Scholar] [CrossRef]
- Li, S.; Li, J.; He, Y.; Xu, M.; Zhang, J.; Du, W.; Zhao, Y.; Xia, L. Precise gene replacement in rice by RNA transcript-templated homologous recombination. Nat. Biotechnol. 2019, 37, 445–450. [Google Scholar] [CrossRef]
- Eckerstorfer, M.F.; Heissenberger, A.; Reichenbecher, W.; Steinbrecher, R.A.; Waßmann, F. An EU perspective on biosafety considerations for plants developed by genome editing and other new genetic modification techniques (nGMs). Front. Bioeng. Biotechnol. 2019, 7, 31. [Google Scholar] [CrossRef] [Green Version]
- Massel, K.; Lam, Y.; Wong, A.C.S.; Hickey, L.T.; Borrell, A.K.; Godwin, I.D. Hotter, drier, CRISPR: The latest edit on climate change. Theor. Appl. Genet. 2021, 134, 1691–1709. [Google Scholar] [CrossRef]
- Huang, T.K.; Puchta, H. Novel CRISPR/Cas applications in plants: From prime editing to chromosome engineering. Transgenic Res. 2021, 30, 529–549. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Brinkman, E.K.; Chen, T.; de Haas, M.; Holland, H.A.; Akhtar, W.; van Steensel, B. Kinetics and Fidelity of the Repair of Cas9-Induced Double-Strand DNA Breaks. Mol. Cell 2018, 70, 801–813.e6. [Google Scholar] [CrossRef] [Green Version]
- Kawall, K. New possibilities on the horizon: Genome editing makes the whole genome accessible for changes. Front. Plant Sci. 2019, 10, 525. [Google Scholar] [CrossRef]
- Kawall, K. Genome-edited Camelina sativa with a unique fatty acid content and its potential impact on ecosystems. Environ. Sci. Eur. 2021, 33, 38. [Google Scholar] [CrossRef]
- Puchta, H. Applying CRISPR/Cas for genome engineering in plants: The best is yet to come. Curr. Opin. Plant Biol. 2017, 36, 1–8. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, H.; Zhu, H. CRISPR technology is revolutionizing the improvement of tomato and other fruit crops. Hortic. Res. 2019, 6, 77. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Li, C.; Gao, C. Applications of CRISPR–Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 2020, 21, 661–677. [Google Scholar] [CrossRef] [PubMed]
- Hahn, F.; Nekrasov, V. CRISPR/Cas precision: Do we need to worry about off-targeting in plants? Plant Cell Rep. 2019, 38, 437–441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Modrzejewski, D.; Hartung, F.; Lehnert, H.; Sprink, T.; Kohl, C.; Keilwagen, J.; Wilhelm, R. Which Factors Affect the Occurrence of Off-Target Effects Caused by the Use of CRISPR/Cas: A Systematic Review in Plants. Front. Plant Sci. 2020, 11, 1838. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhu, J.-K. Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system. Mol. Plant 2017, 10, 523–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alok, A.; Sandhya, D.; Jogam, P.; Rodrigues, V.; Bhati, K.K.; Sharma, H.; Kumar, J. The Rise of the CRISPR/Cpf1 System for Efficient Genome Editing in Plants. Front. Plant Sci. 2020, 11, 264. [Google Scholar] [CrossRef]
- Tian, S.; Jiang, L.; Cui, X.; Zhang, J.; Guo, S.; Li, M.; Zhang, H.; Ren, Y.; Gong, G.; Zong, M.; et al. Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Rep. 2018, 37, 1353–1356. [Google Scholar] [CrossRef]
- Springer, N.M.; Schmitz, R.J. Exploiting induced and natural epigenetic variation for crop improvement. Nat. Rev. Genet. 2017, 18, 563–575. [Google Scholar] [CrossRef]
- Begemann, M.B.; Gray, B.N.; January, E.; Gordon, G.C.; He, Y.; Liu, H.; Wu, X.; Brutnell, T.P.; Mockler, T.C.; Oufattole, M. Precise insertion and guided editing of higher plant genomes using Cpf1 CRISPR nucleases. Sci. Rep. 2017, 7, 11606. [Google Scholar] [CrossRef] [Green Version]
- Roca Paixão, J.F.; Gillet, F.X.; Ribeiro, T.P.; Bournaud, C.; Lourenço-Tessutti, I.T.; Noriega, D.D.; de Melo, B.P.; de Almeida-Engler, J.; Grossi-de-Sa, M.F. Improved drought stress tolerance in Arabidopsis by CRISPR/dCas9 fusion with a Histone AcetylTransferase. Sci. Rep. 2019, 9, 8080. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.; Wang, Y.; Zhang, R.; Zhang, H.; Gao, C. CRISPR/Cas Genome Editing and Precision Plant Breeding in Agriculture. Annu. Rev. Plant Biol. 2019, 70, 667–697. [Google Scholar] [CrossRef]
- Zhang, Y.; Malzahn, A.A.; Sretenovic, S.; Qi, Y. The emerging and uncultivated potential of CRISPR technology in plant science. Nat. Plants 2019, 5, 778–794. [Google Scholar] [CrossRef] [PubMed]
- Ghoshal, B.; Picard, C.L.; Vong, B.; Feng, S.; Jacobsen, S.E. CRISPR-based targeting of DNA methylation in Arabidopsis thaliana by a bacterial CG-specific DNA methyltransferase. Proc. Natl. Acad. Sci. USA 2021, 118, e2125016118. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Gao, H.; Wang, H.; Lafitte, H.R.; Archibald, R.L.; Yang, M.; Hakimi, S.M.; Mo, H.; Habben, J.E. ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 2017, 15, 207–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Meng, X.; Zong, Y.; Chen, K.; Zhang, H.; Liu, J.; Li, J.; Gao, C. Gene replacements and insertions in rice by intron targeting using CRISPR-Cas9. Nat. Plants 2016, 2, 16139. [Google Scholar] [CrossRef]
- Zhou, J.; Xin, X.; He, Y.; Chen, H.; Li, Q.; Tang, X.; Zhong, Z.; Deng, K.; Zheng, X.; Akher, S.A.; et al. Multiplex QTL editing of grain-related genes improves yield in elite rice varieties. Plant Cell Rep. 2019, 38, 475–485. [Google Scholar] [CrossRef]
- Naeem, M.; Majeed, S.; Hoque, M.Z.; Ahmad, I. Latest Developed Strategies to Minimize the Off-Target Effects in CRISPR-Cas-Mediated Genome Editing. Cells 2020, 9, 1608. [Google Scholar] [CrossRef] [PubMed]
- Murugan, K.; Seetharam, A.S.; Severin, A.J.; Sashital, D.G. CRISPR-Cas12a has widespread off-target and dsDNA-nicking effects. J. Biol. Chem. 2020, 295, 5538–5553. [Google Scholar] [CrossRef] [Green Version]
- Höijer, I.; Johansson, J.; Gudmundsson, S.; Chin, C.S.; Bunikis, I.; Häggqvist, S.; Emmanouilidou, A.; Wilbe, M.; den Hoed, M.; Bondeson, M.L.; et al. Amplification-free long read sequencing reveals unforeseen CRISPR-Cas9 off-target activity. bioRxiv 2020, 1–19. [Google Scholar] [CrossRef]
- Mou, H.; Smith, J.L.; Peng, L.; Yin, H.; Moore, J.; Zhang, X.O.; Song, C.Q.; Sheel, A.; Wu, Q.; Ozata, D.M.; et al. CRISPR/Cas9-mediated genome editing induces exon skipping by alternative splicing or exon deletion. Genome Biol. 2017, 18, 4–11. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Liu, G.; Zhou, J.; Ren, Q.; You, Q.; Tian, L.; Xin, X.; Zhong, Z.; Liu, B.; Zheng, X.; et al. A large-scale whole-genome sequencing analysis reveals highly specific genome editing by both Cas9 and Cpf1 (Cas12a) nucleases in rice. Genome Biol. 2018, 19, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.; Ma, B.L. Assessment of canola crop lodging under elevated temperatures for adaptation to climate change. Agric. For. Meteorol. 2018, 248, 329–338. [Google Scholar] [CrossRef]
- Li, C.; Hao, M.; Wang, W.; Wang, H.; Chen, F.; Chu, W.; Zhang, B.; Mei, D.; Cheng, H.; Hu, Q. An Efficient CRISPR/Cas9 Platform for Rapidly Generating Simultaneous Mutagenesis of Multiple Gene Homoeologs in Allotetraploid Oilseed Rape. Front. Plant Sci. 2018, 9, 442. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Yang, X.; Yu, Y.; Si, X.; Zhai, X.; Zhang, H.; Dong, W.; Gao, C.; Xu, C. Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 2018, 36, 1160–1163. [Google Scholar] [CrossRef]
- Yang, Y.; Zhu, K.; Li, H.; Han, S.; Meng, Q.; Khan, S.U.; Fan, C.; Xie, K.; Zhou, Y. Precise editing of CLAVATA genes in Brassica napus L. regulates multilocular silique development. Plant Biotechnol. J. 2018, 16, 1322–1335. [Google Scholar] [CrossRef] [Green Version]
- Zheng, M.; Zhang, L.; Tang, M.; Liu, J.; Liu, H.; Yang, H.; Fan, S.; Terzaghi, W.; Wang, H.; Hua, W. Knockout of two BnaMAX1 homologs by CRISPR/Cas9-targeted mutagenesis improves plant architecture and increases yield in rapeseed (Brassica napus L.). Plant Biotechnol. J. 2020, 18, 644–654. [Google Scholar] [CrossRef] [Green Version]
- Botella, J.R. Now for the hard ones: Is there a limit on CRISPR genome editing in crops? J. Exp. Bot. 2019, 70, 785–793. [Google Scholar] [CrossRef]
- Braatz, J.; Harloff, H.J.; Mascher, M.; Stein, N.; Himmelbach, A.; Jung, C. CRISPR-Cas9 targeted mutagenesis leads to simultaneous modification of different homoeologous gene copies in polyploid oilseed rape (Brassica napus). Plant Physiol. 2017, 174, 935–942. [Google Scholar] [CrossRef] [Green Version]
- Wolter, F.; Schindele, P.; Puchta, H. Plant breeding at the speed of light: The power of CRISPR/Cas to generate directed genetic diversity at multiple sites. BMC Plant Biol. 2019, 19, 176. [Google Scholar] [CrossRef] [Green Version]
- Zaman, Q.U.; Li, C.; Cheng, H.; Hu, Q. Genome editing opens a new era of genetic improvement in polyploid crops. Crop J. 2019, 7, 141–150. [Google Scholar] [CrossRef]
- Zsögön, A.; Čermák, T.; Naves, E.R.; Notini, M.M.; Edel, K.H.; Weinl, S.; Freschi, L.; Voytas, D.F.; Kudla, J.; Peres, L.E.P. De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 2018, 36, 1211–1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-Leal, D.; Lemmon, Z.H.; Man, J.; Bartlett, M.E.; Lippman, Z.B. Engineering Quantitative Trait Variation for Crop Improvement by Genome Editing. Cell 2017, 171, 470–480.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Li, J.; Zhou, J.; Wang, L.; Yang, S.; Hurst, L.D.; Li, W.H.; Tian, D. Identifying a large number of high-yield genes in rice by pedigree analysis, whole-genome sequencing, and CRISPR-Cas9 gene knockout. Proc. Natl. Acad. Sci. USA 2018, 115, E7559–E7567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J.L. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 2014, 32, 947–951. [Google Scholar] [CrossRef] [PubMed]
- Nekrasov, V.; Wang, C.; Win, J.; Lanz, C.; Weigel, D.; Kamoun, S. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 2017, 7, 482. [Google Scholar] [CrossRef] [Green Version]
- Tsanova, T.; Stefanova, L.; Topalova, L.; Atanasov, A.; Pantchev, I. DNA-free gene editing in plants: A brief overview. Biotechnol. Biotechnol. Equip. 2021, 35, 131–138. [Google Scholar] [CrossRef]
- Metje-Sprink, J.; Menz, J.; Modrzejewski, D.; Sprink, T. DNA-Free genome editing: Past, present and future. Front. Plant Sci. 2019, 9, 1957. [Google Scholar] [CrossRef] [PubMed]
- Maher, M.F.; Nasti, R.A.; Vollbrecht, M.; Starker, C.G.; Clark, M.D.; Voytas, D.F. Plant gene editing through de novo induction of meristems. Nat. Biotechnol. 2020, 38, 84–89. [Google Scholar] [CrossRef]
- Rhee, S.Y.; Mutwil, M. Towards revealing the functions of all genes in plants. Trends Plant Sci. 2014, 19, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Borrelli, V.M.G.; Brambilla, V.; Rogowsky, P.; Marocco, A.; Lanubile, A. The enhancement of plant disease resistance using crispr/cas9 technology. Front. Plant Sci. 2018, 9, 1245. [Google Scholar] [CrossRef]
- European Commission. EU Biodiversity Strategy for 2030. Communication from the Commission to the European Parliament, the Council, the European Economic and Social Committee and the Committee of the Regions. 2021. Available online: https://www.europarl.europa.eu/committees/en/eu-biodiversity-strategy-for-2030-/product-details/20201026CDT04342 (accessed on 16 December 2021).
- European Commission. Farm to Fork Strategy. Communication from the Commission to the Europe—An Parliament, the Council, the European Economic and Social Committee and the Committee of the Regions. 2020. Available online: https://ec.europa.eu/food/horizontal-topics/farm-fork-strategy_en (accessed on 16 December 2021).
- United Nations. Transforming our world: The 2030 Agenda for Sustainable Development. Gen. Assem. 2015, 16301, 1–35. [Google Scholar] [CrossRef]
- Zhu, X.G.; Ort, D.R.; Parry, M.A.J.; Von Caemmerer, S. A wish list for synthetic biology in photosynthesis research. J. Exp. Bot. 2020, 71, 2219–2225. [Google Scholar] [CrossRef] [Green Version]
- European Commission Commission Staff Working Document: Study on the Status of New Genomic Techniques under Union Law and in Light of the Court of Justice Ruling in Case C-528/16 (Brussels, 29.4.2021 SWD(2021) 92 Final). 2021. Available online: https://ec.europa.eu/food/plants/genetically-modified-organisms/new-techniques-biotechnology/ec-study-new-genomic-techniques_en (accessed on 16 December 2021).
- Kuckartz, U. Computergestützte Analyse Qualitativer Daten: Eine Einführung in Methoden und Arbeitstechniken; Springer: Berlin/Heidelberg, Germany, 2013; Volume 178, ISBN 3322865924. [Google Scholar]
- Charmaz, K. Constructing Grounded Theory: A Practical Guide through Qualitative Analysis; SAGE Publications Ltd.: Los Angeles, CA, USA, 2006; Volume 10, ISBN 9780761973522. [Google Scholar]
- FAO. FAO Success Stories on Climate-Smart Agriculture; FAO: Rome, Italy, 2014; Volume 28. [Google Scholar]
- Spielman, D.; Mayes, S.; Cook, D.; Penman, D. Summaries of presentations: Opening plenary session and parallel sessions. In Proceedings of the FAO International Symposium on “The Role of Agricultural Biotechnologies in Sustainable Food Systems and Nutrition”, Rome, Italy, 15–17 February 2016; pp. 1–106. [Google Scholar]
- Bundesministerium für Ernährung und Landwirtschaft (BMEL). Ackerbaustrategie 2035—Perspektiven für einen Produktiven und Vielfältigen Pflanzenbau; Open Government Deutschland: Berlin, Germany, 2019.
- Reidsma, P.; Wolf, J.; Kanellopoulos, A.; Schaap, B.F.; Mandryk, M.; Verhagen, J.; van Ittersum, M.K. Climate Change Impact and Adaptation Research Requires Farming Systems Analysis and Integrated Assessment: A Case Study in the Netherlands. Procedia Environ. Sci. 2015, 29, 286–287. [Google Scholar] [CrossRef] [Green Version]
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Modrzejewski, D.; Hartung, F.; Sprink, T.; Krause, D.; Kohl, C.; Wilhelm, R. What is the available evidence for the range of applications of genome-editing as a new tool for plant trait modification and the potential occurrence of associated off-target effects: A systematic map. Environ. Evid. 2019, 8, 27. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Sapkota, M.; van der Knaap, E. Perspectives of CRISPR/Cas-mediated cis-engineering in horticulture: Unlocking the neglected potential for crop improvement. Hortic. Res. 2020, 7, 36. [Google Scholar] [CrossRef] [Green Version]
- Metje-Sprink, J.; Sprink, T.; Hartung, F. Genome-edited plants in the field. Curr. Opin. Biotechnol. 2020, 61, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Sharma, N.; Prasad, M. CRISPR/Cas9: A novel weapon in the arsenal to combat plant diseases. Front. Plant Sci. 2019, 9, 2008. [Google Scholar] [CrossRef] [PubMed]
- Menz, J.; Modrzejewski, D.; Hartung, F.; Wilhelm, R.; Sprink, T. Genome Edited Crops Touch the Market: A View on the Global Development and Regulatory Environment. Front. Plant Sci. 2020, 11, 586027. [Google Scholar] [CrossRef] [PubMed]
- Pendergrass, A.G.; Meehl, G.A.; Pulwarty, R.; Hobbins, M.; Hoell, A.; AghaKouchak, A.; Bonfils, C.J.W.; Gallant, A.J.E.; Hoerling, M.; Hoffmann, D. Flash droughts present a new challenge for subseasonal-to-seasonal prediction. Nat. Clim. Chang. 2020, 10, 191–199. [Google Scholar] [CrossRef]
- Kumar, M. Impact of climate change on crop yield and role of model for achieving food security. Environ. Monit. Assess. 2016, 188, 465. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Liu, B.; Piao, S.; Wang, X.; Lobell, D.B.; Huang, Y.; Huang, M.; Yao, Y.; Bassu, S.; Ciais, P.; et al. Temperature increase reduces global yields of major crops in four independent estimates. Proc. Natl. Acad. Sci. USA 2017, 114, 9326–9331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, G.C.; Valin, H.; Sands, R.D.; Havlík, P.; Ahammad, H.; Deryng, D.; Elliott, J.; Fujimori, S.; Hasegawa, T.; Heyhoe, E. Climate change effects on agriculture: Economic responses to biophysical shocks. Proc. Natl. Acad. Sci. USA 2014, 111, 3274–3279. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Wang, C.; Chen, H.Y.H.; Ruan, H. Response of plants to water stress: A meta-analysis. Front. Plant Sci. 2020, 11, 978. [Google Scholar] [CrossRef]
- Tripathi, A.K.; Pareek, A.; Singla-Pareek, S.L. A NAP-family histone chaperone functions in abiotic stress response and adaptation. Plant Physiol. 2016, 171, 2854–2868. [Google Scholar] [CrossRef] [Green Version]
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.D.; Schroeder, J.I. Genetic strategies for improving crop yields. Nature 2019, 575, 109–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira, T.H.S.; Tsunada, M.S.; Bassi, D.; Araújo, P.; Mattiello, L.; Guidelli, G.V.; Righetto, G.L.; Gonçalves, V.R.; Lakshmanan, P.; Menossi, M. Sugarcane water stress tolerance mechanisms and its implications on developing biotechnology solutions. Front. Plant Sci. 2017, 8, 1077. [Google Scholar] [CrossRef]
- Zafar, S.A.; Zaidi, S.S.E.A.; Gaba, Y.; Singla-Pareek, S.L.; Dhankher, O.P.; Li, X.; Mansoor, S.; Pareek, A.; Foyer, C. Engineering abiotic stress tolerance via CRISPR/Cas-mediated genome editing. J. Exp. Bot. 2020, 71, 470–479. [Google Scholar] [CrossRef]
- Kuromori, T.; Seo, M.; Shinozaki, K. ABA Transport and Plant Water Stress Responses. Trends Plant Sci. 2018, 23, 513–522. [Google Scholar] [CrossRef]
- Evans, J.R. Improving photosynthesis. Plant Physiol. 2013, 162, 1780–1793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blum, A. Drought resistance, water-use efficiency, and yield potential—Are they compatible, dissonant, or mutually exclusive ? Aust. J. Agric. Res. 2005, 56, 1159–1168. [Google Scholar] [CrossRef]
- Xu, F.; Wang, K.; Yuan, W.; Xu, W.; Liu, S.; Kronzucker, H.J.; Chen, G.; Miao, R.; Zhang, M.; Ding, M.; et al. Overexpression of rice aquaporin OsPIP1;2 improves yield by enhancing mesophyll CO2 conductance and phloem sucrose transport. J. Exp. Bot. 2019, 70, 671–681. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Yuan, J.; Ma, H.; Song, J.; Wang, L.; Weng, Q. Characterization and functional analysis of a B3 domain factor from zea mays. J. Appl. Genet. 2015, 56, 427–438. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Fukumoto, T.; Gena, P.; Feng, P.; Sun, Q.; Li, Q.; Matsumoto, T.; Kaneko, T.; Zhang, H.; Zhang, Y.; et al. Ectopic expression of a rice plasma membrane intrinsic protein (OsPIP1;3) promotes plant growth and water uptake. Plant J. 2020, 102, 779–796. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Guo, Y.; Liu, Y.; Zhang, F.; Wang, Z.; Wang, H.; Wang, F.; Li, D.; Mao, D.; Luan, S.; et al. 9-Cis-Epoxycarotenoid Dioxygenase 3 Regulates Plant Growth and Enhances Multi-Abiotic Stress Tolerance in Rice. Front. Plant Sci. 2018, 9, 162. [Google Scholar] [CrossRef]
- Yang, H.; Wu, J.J.; Tang, T.; De Liu, K.; Dai, C. CRISPR/Cas9-mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus. Sci. Rep. 2017, 7, 7489. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Wang, X.; Luo, Y.; Zhang, L.; Yao, Y.; Han, L.; Chen, Z.; Wang, L.; Li, Y. OsABA8ox2, an ABA catabolic gene, suppresses root elongation of rice seedlings and contributes to drought response. Crop J. 2020, 8, 480–491. [Google Scholar] [CrossRef]
- Latef, A.A.H.A.; Hashem, A.; Rasool, S.; Abd-Allah, E.F.; Alqarawi, A.A.; Egamberdieva, D.; Jan, S.; Anjum, N.A.; Ahmad, P. Arbuscular mycorrhizal symbiosis and abiotic stress in plants: A review. J. Plant Biol. 2016, 59, 407–426. [Google Scholar] [CrossRef]
- Quiroga, G.; Erice, G.; Ding, L.; Chaumont, F.; Aroca, R.; Ruiz-Lozano, J.M. The arbuscular mycorrhizal symbiosis regulates aquaporins activity and improves root cell water permeability in maize plants subjected to water stress. Plant. Cell Environ. 2019, 42, 2274–2290. [Google Scholar] [CrossRef]
- Bohnert, H.J.; Jensen, R.G. Strategies for engineering water-stress tolerance in plants. Trends Biotechnol. 1996, 14, 89–97. [Google Scholar] [CrossRef]
- Blum, A. Drought resistance—Is it really a complex trait? Funct. Plant Biol. 2011, 38, 753–757. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Xiong, L. Genetic engineering and breeding of drought-resistant crops. Annu. Rev. Plant Biol. 2014, 65, 715–741. [Google Scholar] [CrossRef]
- Nakashima, K.; Yamaguchi-Shinozaki, K. ABA signaling in stress-response and seed development. Plant Cell Rep. 2013, 32, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Planas-Riverola, A.; Gupta, A.; Betegoń-Putze, I.; Bosch, N.; Ibanḛs, M.; Cano-Delgado, A.I. Brassinosteroid signaling in plant development and adaptation to stress. Development 2019, 146, dev151894. [Google Scholar] [CrossRef] [Green Version]
- Vishwakarma, K.; Upadhyay, N.; Kumar, N.; Yadav, G.; Singh, J.; Mishra, R.K.; Kumar, V.; Verma, R.; Upadhyay, R.G.; Pandey, M.; et al. Abscisic Acid Signaling and Abiotic Stress Tolerance in Plants: A Review on Current Knowledge and Future Prospects. Front. Plant Sci. 2017, 8, 161. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Aguirre, L.; Rodríguez-Leal, D.; Hendelman, A.; Benoit, M.; Lippman, Z.B. Dissecting cis-regulatory control of quantitative trait variation in a plant stem cell circuit. Nat. Plants 2021, 7, 419–427. [Google Scholar] [CrossRef]
- Li, X.; Xie, Y.; Zhu, Q.; Liu, Y.G. Targeted Genome Editing in Genes and cis-Regulatory Regions Improves Qualitative and Quantitative Traits in Crops. Mol. Plant 2017, 10, 1368–1370. [Google Scholar] [CrossRef] [Green Version]
- Jiang, W.; Zhou, H.; Bi, H.; Fromm, M.; Yang, B.; Weeks, D.P. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013, 41, e188. [Google Scholar] [CrossRef]
- Reis, R.S.; Hart-Smith, G.; Eamens, A.L.; Wilkins, M.R.; Waterhouse, P.M. MicroRNA Regulatory Mechanisms Play Different Roles in Arabidopsis. J. Proteome Res. 2015, 14, 4743–4751. [Google Scholar] [CrossRef]
- Curtin, S.J.; Xiong, Y.; Michno, J.M.; Campbell, B.W.; Stec, A.O.; Čermák, T.; Starker, C.; Voytas, D.F.; Eamens, A.L.; Stupar, R.M. CRISPR/Cas9 and TALENs generate heritable mutations for genes involved in small RNA processing of Glycine max and Medicago truncatula. Plant Biotechnol. J. 2018, 16, 1125–1137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, J.; Habben, J.E.; Archibald, R.L.; Drummond, B.J.; Chamberlin, M.A.; Williams, R.W.; Renee Lafitte, H.; Weers, B.P. Overexpression of ARGOS genes modifies plant sensitivity to ethylene, leading to improved drought tolerance in both arabidopsis and maize. Plant Physiol. 2015, 169, 266–282. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.; Qin, X.; Luo, L.; Han, Y.; Wang, X.; Usman, B.; Nawaz, G.; Zhao, N.; Liu, Y.; Li, R. CRISPR/Cas9-Induced Mutagenesis of Semi-Rolled Leaf1,2 Confers Curled Leaf Phenotype and Drought Tolerance by Influencing Protein Expression Patterns and ROS Scavenging in Rice (Oryza sativa L.). Agronomy 2019, 9, 728. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Yan, G.; Duan, Z.; Wang, Z.; Kang, C.; Guo, L.; Liu, K.; Tu, J.; Shen, J.; Yi, B.; et al. Roles of the Brassica napus DELLA Protein BnaA6.RGA, in Modulating Drought Tolerance by Interacting With the ABA Signaling Component BnaA10.ABF2. Front. Plant Sci. 2020, 11, 577. [Google Scholar] [CrossRef] [PubMed]
- Wenjing, W.; Chen, Q.; Singh, P.K.; Huang, Y.; Pei, D. CRISPR/Cas9 edited HSFA6a and HSFA6b of Arabidopsis thaliana offers ABA and osmotic stress insensitivity by modulation of ROS homeostasis. Plant Signal. Behav. 2020, 15, 1816321. [Google Scholar] [CrossRef]
- Du, Y.T.; Zhao, M.J.; Wang, C.T.; Gao, Y.; Wang, Y.X.; Liu, Y.W.; Chen, M.; Chen, J.; Zhou, Y.B.; Xu, Z.S.; et al. Identification and characterization of GmMYB118 responses to drought and salt stress. BMC Plant Biol. 2018, 18, 320. [Google Scholar] [CrossRef]
- Santosh Kumar, V.V.; Verma, R.K.; Yadav, S.K.; Yadav, P.; Watts, A.; Rao, M.V.; Chinnusamy, V. CRISPR-Cas9 mediated genome editing of drought and salt tolerance (OsDST) gene in indica mega rice cultivar MTU1010. Physiol. Mol. Biol. Plants 2020, 26, 1099–1110. [Google Scholar] [CrossRef]
- Wang, L.; Chen, L.; Li, R.; Zhao, R.; Yang, M.; Sheng, J.; Shen, L. Reduced drought tolerance by CRISPR/Cas9-mediated SlMAPK3 mutagenesis in tomato plants. J. Agric. Food Chem. 2017, 65, 8674–8682. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Wang, L.; Li, Y.; Zhao, R.; Zhang, Y.; Sheng, J.; Ma, P.; Shen, L. Knockout of SlNPR1 enhances tomato plants resistance against Botrytis cinerea by modulating ROS homeostasis and JA/ET signaling pathways. Physiol. Plant. 2020, 170, 569–579. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, J.; Xu, J.; Li, Y.; Guo, L.; Wang, Z.; Zhang, X.; Zhao, B.; Guo, Y.D.; Zhang, N. CRISPR/Cas9 targeted mutagenesis of SlLBD40, a lateral organ boundaries domain transcription factor, enhances drought tolerance in tomato. Plant Sci. 2020, 301, 110683. [Google Scholar] [CrossRef]
- Badhan, S.; Ball, A.S.; Mantri, N. First report of CRISPR/Cas9 mediated DNA-free editing of 4CL and RVE7 genes in chickpea protoplasts. Int. J. Mol. Sci. 2021, 22, 396. [Google Scholar] [CrossRef]
- Morran, S.; Eini, O.; Pyvovarenko, T.; Parent, B.; Singh, R.; Ismagul, A.; Eliby, S.; Shirley, N.; Langridge, P.; Lopato, S. Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol. J. 2011, 9, 230–249. [Google Scholar] [CrossRef]
- Cheng, S.; Zhou, D.X.; Zhao, Y. WUSCHEL-related homeobox gene WOX11 increases rice drought resistance by controlling root hair formation and root system development. Plant Signal. Behav. 2016, 11, e1130198. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Alptekin, B.; Budak, H. CRISPR/Cas9 genome editing in wheat. Funct. Integr. Genom. 2018, 18, 31–41. [Google Scholar] [CrossRef] [Green Version]
- Chandler, P.M.; Harding, C.A. “Overgrowth” mutants in barley and wheat: New alleles and phenotypes of the “Green Revolution” della gene. J. Exp. Bot. 2013, 64, 1603–1613. [Google Scholar] [CrossRef] [Green Version]
- Nie, J.; Wen, C.; Xi, L.; Lv, S.; Zhao, Q.; Kou, Y.; Ma, N.; Zhao, L.; Zhou, X. The AP2/ERF transcription factor CmERF053 of chrysanthemum positively regulates shoot branching, lateral root, and drought tolerance. Plant Cell Rep. 2018, 37, 1049–1060. [Google Scholar] [CrossRef]
- Nuccio, M.L.; Paul, M.; Bate, N.J.; Cohn, J.; Cutler, S.R. Where are the drought tolerant crops? An assessment of more than two decades of plant biotechnology effort in crop improvement. Plant Sci. 2018, 273, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Mickelbart, M.V.; Hasegawa, P.M.; Bailey-Serres, J. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat. Rev. Genet. 2015, 16, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Semagn, K.; Beyene, Y.; Warburton, M.L.; Tarekegne, A.; Mugo, S.; Meisel, B.; Sehabiague, P.; Prasanna, B.M. Meta-analyses of QTL for grain yield and anthesis silking interval in 18 maize populations evaluated under water-stressed and well-watered environments. BMC Genom. 2013, 14, 313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harfouche, A.L.; Jacobson, D.A.; Kainer, D.; Romero, J.C.; Harfouche, A.H.; Scarascia Mugnozza, G.; Moshelion, M.; Tuskan, G.A.; Keurentjes, J.J.B.; Altman, A. Accelerating Climate Resilient Plant Breeding by Applying Next-Generation Artificial Intelligence. Trends Biotechnol. 2019, 37, 1217–1235. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Li, C.; Zhang, X.; Li, Y.; Zhang, D.; Shi, Y.; Song, Y.; Li, Y.; Yang, D.; Wang, T. Transcriptome and GWAS analyses reveal candidate gene for seminal root length of maize seedlings under drought stress. Plant Sci. 2020, 292, 110380. [Google Scholar] [CrossRef]
- Wang, X.; Wang, H.; Liu, S.; Ferjani, A.; Li, J.; Yan, J.; Yang, X.; Qin, F. Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat. Genet. 2016, 48, 1233–1241. [Google Scholar] [CrossRef]
- Cowger, C.; Brown, J.K.M. Durability of Quantitative Resistance in Crops: Greater Than We Know? Annu. Rev. Phytopathol. 2019, 57, 253–277. [Google Scholar] [CrossRef] [PubMed]
- Fawke, S.; Doumane, M.; Schornack, S. Oomycete Interactions with Plants: Infection Strategies and Resistance Principles. Microbiol. Mol. Biol. Rev. 2015, 79, 263–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saur, I.M.L.; Hückelhoven, R. Recognition and defence of plant-infecting fungal pathogens. J. Plant Physiol. 2021, 256, 153324. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.K.M. Durable Resistance of Crops to Disease: A Darwinian Perspective. Annu. Rev. Phytopathol. 2015, 53, 513–539. [Google Scholar] [CrossRef] [PubMed]
- Eckardt, N.A. Plant disease susceptibility genes? Plant Cell 2002, 14, 1983–1986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pavan, S.; Jacobsen, E.; Visser, R.G.F.; Bai, Y. Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol. Breed. 2010, 25, 1. [Google Scholar] [CrossRef] [Green Version]
- Corredor-Moreno, P.; Minter, F.; Davey, P.E.; Wegel, E.; Kular, B.; Brett, P.; Lewis, C.M.; Morgan, Y.M.L.; Macías Pérez, L.A.; Korolev, A.V.; et al. The branched-chain amino acid aminotransferase TaBCAT1 modulates amino acid metabolism and positively regulates wheat rust susceptibility. Plant Cell 2021, 33, 1728–1747. [Google Scholar] [CrossRef]
- Pandey, P.; Irulappan, V.; Bagavathiannan, M.V.; Senthil-Kumar, M. Impact of combined abiotic and biotic stresses on plant growth and avenues for crop improvement by exploiting physio-morphological traits. Front. Plant Sci. 2017, 8, 537. [Google Scholar] [CrossRef] [Green Version]
- Ellis, J.G.; Lagudah, E.S.; Spielmeyer, W.; Dodds, P.N. The past, Present and future of breeding rust resistant wheat. Front. Plant Sci. 2014, 5, 641. [Google Scholar] [CrossRef] [Green Version]
- McIntosh, R.A.; Wellings, C.R.; Park, R.F. Wheat Rusts: An Atlas of Resistance Genes; CSIRO Publishing: Clayton, Australia, 1995. [Google Scholar]
- Wang, F.; Wang, C.; Liu, P.; Lei, C.; Hao, W.; Gao, Y.; Liu, Y.G.; Zhao, K. Enhanced rice blast resistance by CRISPR/ Cas9-Targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS ONE 2016, 11, e0154027. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Chen, X.; Liu, J.; Ye, J.; Guo, Z. The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J. Exp. Bot. 2012, 63, 3899–3912. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Guo, R.; Tu, M.; Wang, D.; Guo, C.; Wan, R.; Li, Z.; Wang, X. Ectopic expression of the wild grape WRKY transcription factor vqWRKY52 in arabidopsis thaliana enhances resistance to the biotrophic pathogen powdery mildew but not to the necrotrophic pathogen botrytis cinerea. Front. Plant Sci. 2017, 8, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Liu, Q.; Yang, X.; Xu, J.; Liu, G.; Yao, X.; Ren, R.; Xu, J.; Lou, L. CRISPR/Cas9-mediated mutagenesis of Clpsk1 in watermelon to confer resistance to Fusarium oxysporum f.sp. niveum. Plant Cell Rep. 2020, 39, 589–595. [Google Scholar] [CrossRef]
- Paula de Toledo Thomazella, D.; Brail, Q.; Dahlbeck, D.; Staskawicz, B. CRISPR-Cas9 mediated mutagenesis of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. bioRxiv 2016, 064824. [Google Scholar] [CrossRef] [Green Version]
- Santillán Martínez, M.I.; Bracuto, V.; Koseoglou, E.; Appiano, M.; Jacobsen, E.; Visser, R.G.F.; Wolters, A.M.A.; Bai, Y. CRISPR/Cas9-targeted mutagenesis of the tomato susceptibility gene PMR4 for resistance against powdery mildew. BMC Plant Biol. 2020, 20, 284. [Google Scholar] [CrossRef] [PubMed]
- Koseoglou, E. The study of SlPMR4 CRISPR/Cas9- Mediated Tomato Allelic Series for Resistance against Powdery Mildew Eleni Koseoglou. Master’s Thesis, Wageningen University and Research, Wageningen, The Netherlands, 2017. [Google Scholar]
- Zhang, Y.; Bai, Y.; Wu, G.; Zou, S.; Chen, Y.; Gao, C.; Tang, D. Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 2017, 91, 714–724. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Tu, M.; Wang, D.; Liu, J.; Li, Y.; Li, Z.; Wang, Y.; Wang, X. CRISPR/Cas9-mediated efficient targeted mutagenesis in grape in the first generation. Plant Biotechnol. J. 2018, 16, 844–855. [Google Scholar] [CrossRef] [Green Version]
- Büschges, R.; Hollricher, K.; Panstruga, R.; Simons, G.; Wolter, M.; Frijters, A.; Van Daelen, R.; Van der Lee, T.; Diergaarde, P.; Groenendijk, J.; et al. The barley Mlo gene: A novel control element of plant pathogen resistance. Cell 1997, 88, 695–705. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Li, D.; Wang, G.; Wang, F.; Kunjal, M.; Joldersma, D.; Liu, Z. Application and future perspective of CRISPR/Cas9 genome editing in fruit crops. J. Integr. Plant Biol. 2020, 62, 269–286. [Google Scholar] [CrossRef] [Green Version]
- Nekrasov, V. Sequence-specific nucleases as tools for enhancing disease resistance in crops. Transgenic Res. 2019, 28, 75–80. [Google Scholar] [CrossRef]
- Blanvillain-Baufumé, S.; Reschke, M.; Solé, M.; Auguy, F.; Doucoure, H.; Szurek, B.; Meynard, D.; Portefaix, M.; Cunnac, S.; Guiderdoni, E.; et al. Targeted promoter editing for rice resistance to Xanthomonas oryzae pv. oryzae reveals differential activities for SWEET14-inducing TAL effectors. Plant Biotechnol. J. 2017, 15, 306–317. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Liu, B.; Spalding, M.H.; Weeks, D.P.; Yang, B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 2012, 30, 390–392. [Google Scholar] [CrossRef]
- von Schell, T.; Mohr, H. Biotechnologie—Gentechnik Eine Chance für Neue Industrien; von Schell, T., Ed.; Auflage; Springer: Berlin/Heidelberg, Germany, 1995; ISBN 9783642793882. [Google Scholar]
- Brandt, P.; Gassen, H.G.; Bangsow, T.; Hektor, T.; König, B.; Meyer, G.; Mohr, H.; Müllner, H.; Donn, G.; Rasche, E.; et al. Zukunft der Gentechnik; Springer Basel AG: Basel, Switzerland, 1997; ISBN 978-3-7643-5662-0. [Google Scholar]
- Parisi, C.; Rodríguez-Cerezo, E. Current and Future Market Applications of New Genomic Techniques; Publications Office of the European Union: Luxembourg, 2021. [Google Scholar]
- Ricroch, A.E.; Martin-Laffon, J.; Rault, B.; Pallares, V.C.; Kuntz, M. Next biotechnological plants for addressing global challenges: The contribution of transgenesis and new breeding techniques. N Biotechnol. 2021, 66, 25–35. [Google Scholar] [CrossRef]
- Ishii, T.; Araki, M. Consumer acceptance of food crops developed by genome editing. Plant Cell Rep. 2016, 35, 1507–1518. [Google Scholar] [CrossRef] [PubMed]
- Miladinovic, D.; Antunes, D.; Yildirim, K.; Bakhsh, A.; Cvejić, S.; Kondić-Špika, A.; Marjanovic Jeromela, A.; Opsahl-Sorteberg, H.-G.; Zambounis, A.; Hilioti, Z. Targeted plant improvement through genome editing: From laboratory to field. Plant Cell Rep. 2021, 40, 935–951. [Google Scholar] [CrossRef] [PubMed]
- Jorasch, P. Potential, Challenges, and Threats for the Application of New Breeding Techniques by the Private Plant Breeding Sector in the EU. Front. Plant Sci. 2020, 11, 582011. [Google Scholar] [CrossRef] [PubMed]
- Langner, T.; Kamoun, S.; Belhaj, K. CRISPR Crops: Plant genome editing toward disease resistance. Annu. Rev. Phytopathol. 2018, 56, 479–512. [Google Scholar] [CrossRef] [Green Version]
- Cheng, C.; Gao, X.; Feng, B.; Sheen, J.; Shan, L.; He, P. Plant immune response to pathogens differs with changing temperatures. Nat. Commun. 2013, 4, 2530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velásquez, A.C.; Castroverde, C.D.M.; He, S.Y. Plant-Pathogen Warfare under Changing Climate Conditions. Curr. Biol. 2018, 28, R619–R634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- EFSA Panel on Genetically Modified Organisms (GMO). Guidance on the environmental risk assessment of genetically modified plants. EFSA J. 2010, 8, 1879. [Google Scholar] [CrossRef]
- Nemali, K.S.; Bonin, C.; Dohleman, F.G.; Stephens, M.; Reeves, W.R.; Nelson, D.E.; Castiglioni, P.; Whitsel, J.E.; Sammons, B.; Silady, R.A.; et al. Physiological responses related to increased grain yield under drought in the first biotechnology-derived drought-tolerant maize. Plant. Cell Environ. 2015, 38, 1866–1880. [Google Scholar] [CrossRef] [Green Version]
- Castiglioni, P.; Warner, D.; Bensen, R.J.; Anstrom, D.C.; Harrison, J.; Stoecker, M.; Abad, M.; Kumar, G.; Salvador, S.; D’Ordine, R.; et al. Bacterial RNA chaperones confer abiotic stress tolerance in plants and improved grain yield in maize under water-limited conditions. Plant Physiol. 2008, 147, 446–455. [Google Scholar] [CrossRef] [Green Version]
- Tardieu, F.; Simonneau, T.; Muller, B. The Physiological Basis of Drought Tolerance in Crop Plants: A Scenario-Dependent Probabilistic Approach. Annu. Rev. Plant Biol. 2018, 69, 733–759. [Google Scholar] [CrossRef] [Green Version]
- Kuckartz, U.; Rädiker, S. Analyzing Qualitative Data with MAXQDA; Springer International Publishing: Basel, Switzerland, 2019; pp. 1–290. [Google Scholar] [CrossRef]
- Creswell, J.W.; Creswell, J.D. Research Design: Qualitative, Quantitative, and Mixed Methods Approaches; Sage Publications: Southend Oaks, CA, USA, 2017; ISBN 1506386717. [Google Scholar]
- Luborsky, M.R.; Rubinstein, R.L. Sampling in Qualitative Research: Rationale, Issues, and Methods. Res. Aging 1995, 17, 89–113. [Google Scholar] [CrossRef]
- Yin, R.K. Case Study Reserach—Design and Methods. Clin. Res. 2006, 2, 8–13. [Google Scholar] [CrossRef]
- Zeug, W.; Bezama, A.; Moesenfechtel, U.; Jähkel, A.; Thrän, D. Stakeholders’ interests and perceptions of bioeconomy monitoring using a sustainable development goal framework. Sustainability 2019, 11, 1511. [Google Scholar] [CrossRef] [Green Version]
- Biber-Freudenberger, L.; Ergeneman, C.; Förster, J.J.; Dietz, T.; Börner, J. Bioeconomy futures: Expectation patterns of scientists and practitioners on the sustainability of bio-based transformation. Sustain. Dev. 2020, 28, 1220–1235. [Google Scholar] [CrossRef]
- Allen, C.; Metternicht, G.; Wiedmann, T. Prioritising SDG targets: Assessing baselines, gaps and interlinkages. Sustain. Sci. 2019, 14, 421–438. [Google Scholar] [CrossRef]
- Sedeek, K.E.M.; Mahas, A.; Mahfouz, M. Plant genome engineering for targeted improvement of crop traits. Front. Plant Sci. 2019, 10, 114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eckerstorfer, M.F.; Grabowski, M.; Lener, M.; Engelhard, M.; Simon, S.; Dolezel, M.; Heissenberger, A.; Lüthi, C. Biosafety of Genome Editing Applications in Plant Breeding: Considerations for a Focused Case-Specific Risk Assessment in the EU. BioTech 2021, 10, 10. [Google Scholar] [CrossRef]
- Challinor, A.J.; Koehler, A.-K.; Ramirez-Villegas, J.; Whitfield, S.; Das, B. Current warming will reduce yields unless maize breeding and seed systems adapt immediately. Nat. Clim. Chang. 2016, 6, 954–958. [Google Scholar] [CrossRef]

| Resilience | Salt Tolerance | Drought Tolerance | Extreme Temperatures | Pathogens | Plant Nutrition | Weed Resistance | Yield | Nutritional Capacity | |
|---|---|---|---|---|---|---|---|---|---|
| Total occurrence | 632 | 22 | 106 | 51 | 227 | 186 | 134 | 331 | 245 |
| Political documents Germany (12) | 10 | 2 | 17 | 5 | 2 | 69 | 3 | 13 | 14 |
| Political documents EU (6) | 29 | 0 | 1 | 0 | 7 | 9 | 7 | 9 | 22 |
| Political documents international organisations (10) | 190 | 5 | 52 | 19 | 46 | 70 | 31 | 124 | 134 |
| Scientist organisations and associations (8) | 33 | 2 | 14 | 9 | 28 | 7 | 12 | 25 | 22 |
| Peer-reviewed, scientific reviews (27) | 370 | 13 | 22 | 18 | 144 | 31 | 81 | 160 | 53 |
| SDG 1 no poverty | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| 1.4 equal access to resources | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| SDG 2 zero hunger | 14 | 1 | 2 | 4 | 5 | 10 | 1 | 19 | 9 |
| 2.1 nutrition quantity and food security | 9 | 1 | 2 | 2 | 3 | 4 | 1 | 17 | 5 |
| 2.3 improve smallholder situation | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 3 |
| 2.5 ensure agricultural genetic diversity | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2.4 sustainable and resilient agriculture | 8 | 0 | 0 | 2 | 4 | 5 | 1 | 7 | 3 |
| 2.2 nutrition improvement | 7 | 1 | 2 | 1 | 3 | 7 | 1 | 10 | 5 |
| SDG 3 good health and well-being | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | 2 |
| 3.9 reduce illness from contamination/allergy | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | 2 |
| SDG 7 affordable and clean energy | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7.2 renewable energy | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| SDG 8 decent work and economic growth | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 2 |
| 8.1 inclusive economic growth | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 2 |
| 8.5 employment | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8.2 innovation | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| SDG 12 responsible consumption and production | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12.5 reduce waste, recycle | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| SDG 13 climate action | 14 | 3 | 8 | 8 | 4 | 9 | 1 | 12 | 1 |
| 13.2 mitigation actions | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 13.1 adaptation actions | 14 | 3 | 8 | 8 | 4 | 9 | 1 | 12 | 1 |
| SDG 14 life below water | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14.1 avoid pollution and overnutrition | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| SDG 15 life on land | 3 | 0 | 0 | 0 | 1 | 1 | 0 | 2 | 0 |
| 15.1 ecosystem conservation | 3 | 0 | 0 | 0 | 1 | 1 | 0 | 2 | 0 |
| SUM | 34 | 6 | 16 | 16 | 10 | 22 | 2 | 30 | 6 |
| Plant | Intended Trait | Loci | Genetic Changes | Method | Development Stage | Reported Effect | References |
|---|---|---|---|---|---|---|---|
| Glycine max, soybean | Drought and salt tolerance | GmDrb2a and GmDrb2b | Knock-out mutations in GmDrb2a and GmDrb2b (SDN1) | CRISPR/Cas | Field trial registered (USDA) | Not described—in Arabidopsis AtDRB2 dependent micro-RNAs are involved in the abiotic stress response | [100,101]; USDA 17-219-01 |
| Zea mays, maize | Improved drought tolerance and yield stability | Confidential information deleted | Base editing in not-specified genes (SDN2) | CRISPR/Cas | Field trial registered (USDA) | Not described in detail: plants with improved drought tolerance and yield stability | USDA 20-168-23 |
| Zea mays, maize | Improved drought tolerance and corn yield | Cis-regulatory region of ARGOS8 | Exchange of the promoter (SDN3) -> change in the expression of the transcription factor ARGOS8 | CRISPR/Cas | Field trials 2015; 8 locations in the US in total, each with random block design | Increase in grain yield by 2–3% under drought stress at flowering time. No increase (slight decrease 2–3%) under drought stress during grain ripening | [26,102] |
| Oryza sativa, rice | Drought tolerance | OsABA8ox2 | Knock-out mutation in OsABA8ox2 (SDN1) | CRISPR/Cas | Crop—greenhouse/lab trial | Improved drought tolerance through increased ABA sensitivity, reduced ABA degradation and vertical root growth | [88] |
| Oryza sativa, rice | Drought tolerance | OsSRL1 and OsSRL2 | Knock-out mutation in OsSRL1 and OsSRL2 (SDN1); subsequent hybridisation with wild type | CRISPR/Cas | Crop—greenhouse/lab trial | Increased survival rate under drought stress, but slightly lower yield under unstressed conditions; in hybrid plants with half-rolled leaves the yield was slightly higher than that of wild-type lines | [103] |
| Brassica napus, canola | Drought tolerance | BnaRGA, BnaA6.RGA | Quadruple knock-out mutant of the BnaRGA gene and simple gain-of-function mutant in the BnaA6.RGA gene (SDN1) | CRISPR/Cas | Crop—greenhouse/lab trial | Gain-of-function mutant with increased drought tolerance and higher ABA sensitivity than wild type, quadruple mutant with low drought tolerance | [38,104] |
| Plant | Intended Trait | Loci | Genetic Changes | Method | Development Stage | Reported Effect | References |
|---|---|---|---|---|---|---|---|
| Arabidopsis | Drought tolerance | cis-regulatory region of AtAREB1 | Activation of gene expression through modification of the chromatin status by AtHAT1 (SDN2) in the cis-regulatory region of AtAREB1 | CRISPR- dCas9HAT | In model organism | Higher gene expression of AtAREB1; dwarf phenotype; faster stomatal closure and better survival rate under drought stress | [22] |
| Arabidopsis | Functional analysis under abiotic stress | HSFA6a und HSFA6b | Knock-out mutations in HSFA6a und HSFA6b (SDN1) | CRISPR/Cas | In model organism | Double mutant with abiotic and osmotic stress tolerance | [105] |
| Glycine max, soybean | Functional analysis under abiotic stress | GmMYB118 | Knock-out mutation in GmMYB118 (SDN1) and overexpression | CRISPR/Cas and genetic engineering | Crop—greenhouse/lab trial | Reduced tolerance and lower proline and chlorophyll content in the knock-out—improved properties in the overexpression | [106] |
| Oryza sativa, rice | Functional analysis under abiotic stress | OsNCED3 | Knock-out mutation in OsNCED3 (SDN1) and overexpression | CRISPR/Cas and genetic engineering | Crop—greenhouse/lab trial | Reduced tolerance to drought, longer growth, more open stomata due to lower ABA levels in the knock-out—improvement compared to wild type in the overexpression | [86] |
| Oryza sativa rice | Functional analysis under abiotic stress | OsDST | Knock-out mutation in OsDST (SDN1) | CRISPR/Cas | Crop—greenhouse/lab trial | Lower stomatal density and improved water balance under drought stress; high salt stress tolerance; no noticeable phenotype under normal conditions | [107] |
| Solanum lycopersicum, tomato | Functional analysis under abiotic stress | SlMAPK3 | Knock-out mutation in SlMPAK3 (SDN1) | CRISPR/Cas | Crop—greenhouse/lab trial | Lower drought tolerance and stronger wilt syndrome in knock-out plants | [108] |
| Solanum lycopersicum, tomato | Functional analysis under abiotic stress | SlNPR1 | Knock-out mutation in SlNPR1 (SDN1) | CRISPR/Cas | Crop—greenhouse/lab trial | Lower drought tolerance and more open stomata in knock-out plants | [109] |
| Solanum lycopersicum, tomato | Functional analysis under abiotic stress | SlLBD40 | Knock-out mutation in SlBD40 (SDN1) & overexpression | CRISPR/Cas and genetic engineering | Crop—greenhouse/lab trial | Improved drought tolerance in knock-out lines due to increased water retention capacity—overexpression led to a lower drought tolerance | [110] |
| Cicer arietinum, chickpea | Functional analysis under drought stress and method | Ca4CL, CaRVE1 | Knock-out mutations in Ca4CL, CaRVE1 | CRISPR/Cas | Crop—greenhouse/lab trial | Validation of genome-editing method in chickpea using protoplast transfection | [111] |
| Triticum aestivum, wheat | Drought tolerance | TaERF3 and TaDREB2 | Knock-out mutations in TaERF3 and TaDREB2 (SDN1) | CRISPR/Cas | Crop—greenhouse/lab trial | DREB2 and ERF3 were identified in wheat and rice as important genes in the drought stress response; in wheat, the expression of TaERF3 and TaDREB2 reacts to drought stress | [112,113,114] |
| Plant | Intended Trait | Loci | Genetic Changes | Method | Development Stage | Reported Effect | References |
|---|---|---|---|---|---|---|---|
| Brassica napus, canola | Fungi pathogen resistance | Confidential information deleted | Confidential information deleted | Gene editing, not specified | Registered for commercialisation | Resistance to fungal pathogens | USDA 20-168-24 |
| Oryza sativa japonica, rice | Resistance to rice blast | OsERF922 | Knock-out mutation (SDN1) | CRISPR/Cas | Crop—field trial/greenhouse | ~50–70% higher resistance | [133] |
| Citrullus lanatus, water melon | Resistance to Fusarium oxysporum | ClPSK1 | Knock-out mutation (SDN1) | CRISPR/Cas | Crop—greenhouse/lab trial | 19–60% higher resistance | [136] |
| Solanum lycopersicum, tomato | Multi-resistance | SlDMR6 | Knock-out mutation (SDN1) | CRISPR/Cas | Crop—greenhouse/lab trial | ~20% higher resistance to 3 pathogens | [137,138] |
| Solanum lycopersicum, tomato | Resistance to powdery mildew | SlMLO1 | Knock-out mutation (SDN1) | CRISPR/Cas | Crop—greenhouse/lab trial | Complete resistance | [48] |
| Solanum lycopersicum, tomato | Resistance to powdery mildew | PMR4 | Knock-out mutation (SDN1) | CRISPR/Cas | Crop—greenhouse/lab trial | Higher resistance to powdery mildew | [139] |
| Solanum lycopersicum, tomato | Resistance to Botrytis cinerea | SlNPR1 | Knock-out mutation (SDN1) | CRISPR/Cas | Crop—greenhouse/lab trial | 33–40% higher resistance | [109] |
| Triticum aestivum, wheat | Resistance to powdery mildew | TaEDR1-A, -B und -D | simultaneous Knock-out in 3 loci (SDN1) | CRISPR/Cas | Crop—greenhouse/lab trial | Reduction of infection by ~50% | [140] |
| Triticum aestivum, wheat | Resistance to powdery mildew | TaMLO-A, -B, D | Knock-out mutation (SDN1) | TALEN | Crop—greenhouse/lab trial, 2 varieties | Complete resistance | [47], USDA 15-238-01 |
| Vitis vinifera, grape | Resistance to Botrytis cinerea | VvWRKY52 | Knock-out mutation (SDN1) | CRISPR/Cas | Crop—greenhouse/lab trial | 50% higher resistance | [141] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hüdig, M.; Laibach, N.; Hein, A.-C. Genome Editing in Crop Plant Research—Alignment of Expectations and Current Developments. Plants 2022, 11, 212. https://doi.org/10.3390/plants11020212
Hüdig M, Laibach N, Hein A-C. Genome Editing in Crop Plant Research—Alignment of Expectations and Current Developments. Plants. 2022; 11(2):212. https://doi.org/10.3390/plants11020212
Chicago/Turabian StyleHüdig, Meike, Natalie Laibach, and Anke-Christiane Hein. 2022. "Genome Editing in Crop Plant Research—Alignment of Expectations and Current Developments" Plants 11, no. 2: 212. https://doi.org/10.3390/plants11020212
APA StyleHüdig, M., Laibach, N., & Hein, A.-C. (2022). Genome Editing in Crop Plant Research—Alignment of Expectations and Current Developments. Plants, 11(2), 212. https://doi.org/10.3390/plants11020212






