The Potential of Molecular Indicators of Plant Virus Infection: Are Plants Able to Tell Us They Are Infected?
Abstract
:1. Introduction
2. Results
2.1. The First Tool: Elevated sRNA Levels Correlate with Virus Infection
2.1.1. Assessment of the Bioanalyzer Method
2.1.2. Analysis of the Proportion of LMW-RNA Components from Total LMW-RNA
2.1.3. Ratio of sRNA/rRNA Accumulation as a Tool Predictive of Virus Infection
2.2. The Second Tool: Transcript Accumulation Changes of Virus-Responsive Genes
2.2.1. AtSGS3 Transcript Accumulation Decreases in Response to Virus Infection
2.2.2. AtCPK3 Transcript Accumulation Increases in Response to Virus Infection
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. sRNA Isolation
4.3. Total RNA Extraction, Quality Analysis and cDNA Synthesis
4.4. Small RNA Lab on Chip Protocol
4.5. RT-qPCR
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gimenez, E.; Salinas, M.; Manzano-Agugliaro, F. Worldwide research on plant defense against biotic stresses as improvement for sustainable agriculture. Sustainability 2018, 10, 391. [Google Scholar] [CrossRef] [Green Version]
- Rubio, L.; Galipienso, L.; Ferriol, I. Detection of plant viruses and disease management: Relevance of genetic diversity and evolution. Front. Plant Sci. 2020, 11, 1092. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; McKirdy, S.; Macbeth, F. The biosecurity ontinuum and trade: Tools for post-border biosecurity. In The Handbook of Plant Biosecurity: Principles and Practices for The Identification, Containment and Control of Organisms That Threaten Agriculture and The Environment Globally; Gordh, G., McKirdy, S., Eds.; Springer: Dordrecht, The Netherlands, 2014; pp. 189–206. [Google Scholar]
- Barrero, R.A.; Napier, K.R.; Cunnington, J.; Liefting, L.; Keenan, S.; Frampton, R.A.; Szabo, T.; Bulman, S.; Hunter, A.; Ward, L.; et al. An internet-based bioinformatics toolkit for plant biosecurity diagnosis and surveillance of viruses and viroids. BMC Bioinform. 2017, 18, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehetre, G.T.; Leo, V.V.; Singh, G.; Sorokan, A.; Maksimov, I.; Yadav, M.K.; Upadhyaya, K.; Hashem, A.; Alsaleh, A.N.; Dawoud, T.M.; et al. Current developments and challenges in plant viral diagnostics: A systematic review. Viruses 2021, 13, 412. [Google Scholar] [CrossRef] [PubMed]
- Neethirajan, S.; Ragavan, K.V.; Weng, X. Agro-defense: Biosensors for food from healthy crops and animals. Trends Food Sci. Technol. 2018, 73, 25–44. [Google Scholar] [CrossRef]
- Katsarou, K.; Bardani, E.; Kallemi, P.; Kalantidis, K. Viral detection: Past, present, and future. BioEssays 2019, 41, 1900049. [Google Scholar] [CrossRef]
- Jones, S.; Baizan-Edge, A.; MacFarlane, S.; Torrance, L. Viral diagnostics in plants using next generation sequencing: Computational analysis in practice. Front. Plant Sci. 2017, 8, 1770. [Google Scholar] [CrossRef] [PubMed]
- Boonham, N.; Kreuze, J.; Winter, S.; van der Vlugt, R.; Bergervoet, J.; Tomlinson, J.; Mumford, R. Methods in virus diagnostics: From ELISA to next generation sequencing. Virus Res. 2014, 186, 20–31. [Google Scholar] [CrossRef]
- Villamor, D.E.V.; Ho, T.; Al Rwahnih, M.; Martin, R.R.; Tzanetakis, I.E. High throughput sequencing for plant virus detection and discovery. Phytopathology 2019, 109, 716–725. [Google Scholar] [CrossRef]
- McClain, M.T.; Constantine, F.J.; Nicholson, B.P.; Nichols, M.; Burke, T.W.; Henao, R.; Jones, D.C.; Hudson, L.L.; Jaggers, L.B.; Veldman, T.; et al. A blood-based host gene expression assay for early detection of respiratory viral infection: An index-cluster prospective cohort study. Lancet Infect. Dis. 2021, 21, 396–404. [Google Scholar] [CrossRef]
- Hjertner, B.; Lützelschwab, C.; Schieck, E.; Nzau, B.; Henson, S.; Sjölund, M.; Fossum, C.; Magnusson, U. Development of a 3-transcript host expression assay to differentiate between viral and bacterial infections in pigs. PLoS ONE 2021, 16, e0256106. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Peterson, D.R.; Baran, A.M.; Bhattacharya, S.; Wylie, T.N.; Falsey, A.R.; Mariani, T.J.; Storch, G.A. Host gene expression in nose and blood for the diagnosis of viral respiratory infection. J. Infect. Dis. 2019, 219, 1151–1161. [Google Scholar] [CrossRef]
- Holcomb, Z.E.; Tsalik, E.L.; Woods, C.W.; McClain, M.T. Host-based peripheral blood gene expression analysis for diagnosis of infectious diseases. J. Clin. Microbiol. 2017, 55, 360–368. [Google Scholar] [CrossRef] [Green Version]
- van Engelen, T.S.R.; Wiersinga, W.J.; Scicluna, B.P.; van der Poll, T. Biomarkers in Sepsis. Crit. Care Clin. 2018, 34, 139–152. [Google Scholar] [CrossRef]
- Rosas-Diaz, T.; Zhang, D.; Fan, P.; Wang, L.; Ding, X.; Jiang, Y.; Jimenez-Gongora, T.; Medina-Puche, L.; Zhao, X.; Feng, Z.; et al. A virus-targeted plant receptor-like kinase promotes cell-to-cell spread of RNAi. Proc. Natl. Acad. Sci. USA 2018, 115, 1388–1393. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, A.B.; Aragão, F.J.L. RNAi-mediated resistance to viruses in genetically engineered plants. Methods Mol. Biol. 2015, 1287, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.-W.; Voinnet, O. Antiviral immunity directed by small RNAs. Cell 2007, 130, 413–426. [Google Scholar] [CrossRef] [Green Version]
- Vivek, A.T.; Zahra, S.; Kumar, S. From current knowledge to best practice: A primer on viral diagnostics using deep sequencing of virus-derived small interfering RNAs (vsiRNAs) in infected plants. Methods 2020, 183, 30–37. [Google Scholar] [CrossRef]
- Zhang, B.; Li, W.; Zhang, J.; Wang, L.; Wu, J. Roles of small RNAs in virus-plant interactions. Viruses 2019, 11, 827. [Google Scholar] [CrossRef] [Green Version]
- Csorba, T.; Kontra, L.; Burgyán, J. Viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology 2015, 479–480, 85–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, S.-W. RNA-based antiviral immunity. Nat. Rev. Immunol. 2010, 10, 632–644. [Google Scholar] [CrossRef] [PubMed]
- Bazzini, A.A.; Almasia, N.I.; Manacorda, C.A.; Mongelli, V.C.; Conti, G.; Maroniche, G.A.; Rodriguez, M.C.; Distéfano, A.J.; Hopp, H.E.; del Vas, M.; et al. Virus infection elevates transcriptional activity of miR164a promoter in plants. BMC Plant Biol. 2009, 9, 152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blevins, T.; Rajeswaran, R.; Aregger, M.; Borah, B.K.; Schepetilnikov, M.; Baerlocher, L.; Farinelli, L.; Meins, F.; Hohn, T.; Pooggin, M.M. Massive production of small RNAs from a non-coding region of Cauliflower mosaic virus in plant defense and viral counter-defense. Nucleic Acids Res. 2011, 39, 5003–5014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González, L.E.; Peiró, R.; Rubio, L.; Galipienso, L. Persistent southern tomato virus (Stv) interacts with cucumber mosaic and/or pepino mosaic virus in mixed-infections modifying plant symptoms, viral titer and small RNA accumulation. Microorganisms 2021, 9, 689. [Google Scholar] [CrossRef] [PubMed]
- Shidore, T.; Zuverza-Mena, N.; da Silva, W. Small RNA profiling analysis of two recombinant strains of potato virus Y in infected tobacco plants. Virus Res. 2020, 288, 198125. [Google Scholar] [CrossRef]
- Wu, G.; Hu, Q.; Du, J.; Li, K.; Sun, M.; Jing, C.; Li, M.; Li, J.; Qing, L. Molecular characterization of virus-derived small RNAs in Nicotiana benthamiana plants infected with tobacco curly shoot virus and its β satellite. Virus Res. 2019, 265, 10–19. [Google Scholar] [CrossRef]
- Donaire, L.; Llave, C. Computational workflow for small RNA profiling in virus-infected plants. Methods Mol. Biol. 2019, 2028, 185–214. [Google Scholar] [CrossRef]
- Santala, J.; Valkonen, J.P.T. Sensitivity of small RNA-based detection of plant viruses. Front. Microbiol. 2018, 9, 939. [Google Scholar] [CrossRef] [Green Version]
- Pooggin, M.M. Small RNA-omics for plant virus identification, virome reconstruction, and antiviral defense characterization. Front. Microbiol. 2018, 9, 2779. [Google Scholar] [CrossRef]
- Niu, X.; Sun, Y.; Chen, Z.; Li, R.; Padmanabhan, C.; Ruan, J.; Kreuze, J.F.; Ling, K.; Fei, Z.; Gao, S. Using small RNA-seq data to detect siRNA duplexes induced by plant viruses. Genes 2017, 8, 163. [Google Scholar] [CrossRef] [Green Version]
- Pecman, A.; Kutnjak, D.; Gutiérrez-Aguirre, I.; Adams, I.; Fox, A.; Boonham, N.; Ravnikar, M. Next generation sequencing for detection and discovery of plant viruses and viroids: Comparison of two approaches. Front. Microbiol. 2017, 8, 1998. [Google Scholar] [CrossRef] [Green Version]
- Gaafar, Y.Z.A.; Ziebell, H. Comparative study on three viral enrichment approaches based on RNA extraction for plant virus/viroid detection using high-throughput sequencing. PLoS ONE 2020, 15, e0237951. [Google Scholar] [CrossRef]
- Die, J.V.; Román, B. RNA quality assessment: A view from plant qPCR studies. J. Exp. Bot. 2012, 63, 6069–6077. [Google Scholar] [CrossRef] [Green Version]
- Postnikova, O.A.; Nemchinov, L.G. Comparative analysis of microarray data in Arabidopsis transcriptome during compatible interactions with plant viruses. Virol. J. 2012, 9, 101. [Google Scholar] [CrossRef] [Green Version]
- Slavokhotova, A.; Korostyleva, T.; Shelenkov, A.; Pukhalskiy, V.; Korottseva, I.; Slezina, M.; Istomina, E.; Odintsova, T. Transcriptomic analysis of genes involved in plant defense response to the cucumber green mottle mosaic virus infection. Life 2021, 11, 1064. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.; Yu, J.; Kim, K.H. Pepper mottle virus and its host interactions: Current state of knowledge. Viruses 2021, 13, 1930. [Google Scholar] [CrossRef] [PubMed]
- Akbar, S.; Wei, Y.; Yuan, Y.; Khan, M.T.; Qin, L.; Powell, C.A.; Chen, B.; Zhang, M. Gene expression profiling of reactive oxygen species (ROS) and antioxidant defense system following Sugarcane mosaic virus (SCMV) infection. BMC Plant Biol. 2020, 20, 532. [Google Scholar] [CrossRef] [PubMed]
- Zanardo, L.G.; de Souza, G.B.; Alves, M.S. Transcriptomics of plant–virus interactions: A review. Theor. Exp. Plant Physiol. 2019, 31, 103–125. [Google Scholar] [CrossRef]
- Sun, Y.; Fan, M.; He, Y. Transcriptome analysis of watermelon leaves reveals candidate genes responsive to cucumber green mottle mosaic virus infection. Int. J. Mol. Sci. 2019, 20, 610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheng, Y.; Yang, L.; Li, C.; Wang, Y.; Guo, H. Transcriptomic changes in Nicotiana tabacum leaves during mosaic virus infection. 3 Biotech. 2019, 9, 220. [Google Scholar] [CrossRef]
- Osmani, Z.; Sabet, M.S.; Shams-Bakhsh, M.; Moieni, A.; Vahabi, K. Virus-specific and common transcriptomic responses of potato (Solanum tuberosum) against PVY, PVA and PLRV using microarray meta-analysis. Plant Breed. 2019, 138, 216–228. [Google Scholar] [CrossRef]
- Mourrain, P.; Béclin, C.; Elmayan, T.; Feuerbach, F.; Godon, C.; Morel, J.-B.; Jouette, D.; Lacombe, A.-M.; Nikic, S.; Picault, N.; et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 2000, 101, 533–542. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Wang, Y.; Zhou, X. SGS3 cooperates with RDR6 in triggering Geminivirus-induced gene silencing and in suppressing Geminivirus infection in Nicotiana benthamiana. Viruses 2017, 9, 247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, X.; Liu, S.-Y.; Zou, J.-Z.; Zhao, J.-J.; Zhu, F.-F.; Chai, L.-X.; Wang, Y.; Han, C.; Wang, X.-B. A small peptide inhibits siRNA amplification in plants by mediating autophagic degradation of SGS3/RDR6 bodies. EMBO J. 2021, 40, e108050. [Google Scholar] [CrossRef] [PubMed]
- Rajamäki, M.-L.; Streng, J.; Valkonen, J.P.T. Silencing suppressor protein VPg of a potyvirus interacts with the plant silencing-related protein SGS3. Mol. Plant-Microbe Interact. 2014, 27, 1199–1210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valmonte, G.R.; Arthur, K.; Higgins, C.M.; MacDiarmid, R.M. Calcium-dependent protein kinases in plants: Evolution, expression and function. Plant Cell Physiol. 2014, 55, 551–569. [Google Scholar] [CrossRef] [Green Version]
- Zhao, P.; Liu, Y.; Kong, W.; Ji, J.; Cai, T.; Guo, Z. Genome-wide identification and characterization of calcium-dependent protein kinase (CDPK) and CDPK-related kinase (CRK) gene families in Medicago truncatula. Int. J. Mol. Sci. 2021, 22, 1044. [Google Scholar] [CrossRef]
- Zhu, L.; Zheng, B.; Song, W.; Li, H.; Jin, X. Evolutionary analysis of calcium-dependent protein kinase in five asteraceae species. Plants 2020, 9, 32. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Liu, Y.; He, Q.; Chai, M.; Huang, Y.; Chen, F.; Wang, X.; Liu, Y.; Cai, H.; Qin, Y. Genome-wide investigation of calcium-dependent protein kinase gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genom. 2020, 21, 72. [Google Scholar] [CrossRef]
- Yadav, A.; Garg, T.; Singh, H.; Yadav, S.R. Tissue-specific expression pattern of calcium-dependent protein kinases-related kinases (CRKs) in rice. Plant Signal. Behav. 2020, 15, 1809846. [Google Scholar] [CrossRef]
- Li, M.; Hu, W.; Ren, L.; Jia, C.; Liu, J.; Miao, H.; Guo, A.; Xu, B.; Jin, Z. Identification, expression, and interaction network analyses of the CDPK gene family reveal their involvement in the development, ripening, and abiotic stress response in banana. Biochem. Genet. 2020, 58, 40–62. [Google Scholar] [CrossRef] [PubMed]
- Atif, R.M.; Shahid, L.; Waqas, M.; Ali, B.; Rashid, M.A.R.; Azeem, F.; Nawaz, M.A.; Wani, S.H.; Chung, G. Insights on calcium-dependent protein kinases (CPKs) signaling for abiotic stress tolerance in plants. Int. J. Mol. Sci. 2019, 20, 5298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, S.; Li, S.; Asim, M.; Mao, J.; Xu, D.; Ullah, Z.; Liu, G.; Wang, Q.; Liu, H. The arabidopsis calcium-dependent protein kinases (CDPKs) and their roles in plant growth regulation and abiotic stress responses. Int. J. Mol. Sci. 2018, 19, 1900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyodo, K.; Suzuki, N.; Okuno, T. Hijacking a host scaffold protein, RACK1, for replication of a plant RNA virus. New Phytol. 2019, 221, 935–945. [Google Scholar] [CrossRef]
- Romeis, T.; Ludwig, A.A.; Martin, R.; Jones, J.D.G. Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J. 2001, 20, 5556–5567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Li, Y. Dissection of RNAi-based antiviral immunity in plants. Curr. Opin. Virol. 2018, 32, 88–99. [Google Scholar] [CrossRef]
- Leonetti, P.; Stuttmann, J.; Pantaleo, V. Regulation of plant antiviral defense genes via host RNA-silencing mechanisms. Virol. J. 2021, 18, 194. [Google Scholar] [CrossRef]
- Hamilton, A.; Voinnet, O.; Chappell, L.; Baulcombe, D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002, 21, 4671–4679. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Zhao, N.; Li, Z.; Xu, X.; Wang, Y.; Yang, X.; Liu, S.S.; Wang, A.; Zhou, X. A calmodulin-like protein suppresses RNA silencing and promotes geminivirus infection by degrading SGS3 via the autophagy pathway in Nicotiana benthamiana. PLoS Pathog. 2017, 13, e1006213. [Google Scholar] [CrossRef]
- Qi, J.; Song, C.P.; Wang, B.; Zhou, J.; Kangasjärvi, J.; Zhu, J.K.; Gong, Z. Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J. Integr. Plant Biol. 2018, 60, 805–826. [Google Scholar] [CrossRef] [Green Version]
- Mott, G.A.; Desveaux, D.; Guttman, D.S. A high-sensitivity, microtiter-based plate assay for plant pattern-triggered immunity. Mol. Plant-Microbe Interact. 2018, 31, 499–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Gresa, M.P.; Lisón, P.; Kim, H.K.; Choi, Y.H.; Verpoorte, R.; Rodrigo, I.; Conejero, V.; Bellés, J.M. Metabolic fingerprinting of tomato mosaic virus infected Solanum lycopersicum. J. Plant Physiol. 2012, 169, 1586–1596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prasch, C.M.; Sonnewald, U. Simultaneous application of heat, drought, and virus to Arabidopsis plants reveals significant shifts in signaling networks. Plant Physiol. 2013, 162, 1849–1866. [Google Scholar] [CrossRef] [PubMed]
- Moore, C.J.; Sutherland, P.W.; Forster, R.L.; Gardner, R.C.; MacDiarmid, R.M. Dark green islands in plant virus infection are the result of posttranscriptional gene silencing. Mol. Plant Microbe Interact. 2001, 14, 939–946. [Google Scholar] [CrossRef] [Green Version]
- Pesti, R.; Kontra, L.; Paul, K.; Vass, I.; Csorba, T.; Havelda, Z.; Várallyay, É. Differential gene expression and physiological changes during acute or persistent plant virus interactions may contribute to viral symptom differences. PLoS ONE 2019, 14, e0216618. [Google Scholar] [CrossRef]
- Alexander, M.M.; Cilia, M. A molecular tug-of-war: Global plant proteome changes during viral infection. Curr. Plant Biol. 2016, 5, 13–24. [Google Scholar] [CrossRef] [Green Version]
- Jang, H.; Hwang, E.Y.; Kim, Y.; Choo, J.; Jeong, J.; Lim, D.W. Surface-enhanced raman scattering and fluorescence-based dual nanoprobes for multiplexed detection of bacterial pathogens. J. Biomed. Nanotechnol. 2016, 12, 1938–1951. [Google Scholar] [CrossRef]
- Park, K.S. Nucleic acid aptamer-based methods for diagnosis of infections. Biosens. Bioelectron. 2018, 102, 179–188. [Google Scholar] [CrossRef]
- Leader, D.J.; Sanders, J.F.; Waugh, R.; Shaw, P.; Brown, J.W.S. Molecular characterisation of plant U14 small nucleolar RNA genes: Closely linked genes are transcribed as polycistronic U14 transcripts. Nucleic Acids Res. 1994, 22, 5196–5203. [Google Scholar] [CrossRef] [Green Version]
- Wei, L.; An, Z.; Mason, A.S.; Xiao, M.; Guo, Y.; Yin, J.; Li, J.; Fu, D. Extensive tRNA gene changes in synthetic Brassica napus. J. Mol. Evol. 2014, 78, 38–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lilly, S.T.; Drummond, R.S.M.; Pearson, M.N.; MacDiarmid, R.M. Identification and validation of reference genes for normalization of transcripts from virus-infected Arabidopsis thaliana. Mol. Plant-Microbe Interact. 2011, 24, 294–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willems, E.; Leyns, L.; Vandesompele, J. Standardization of real-time PCR gene expression data from independent biological replicates. Anal. Biochem. 2008, 379, 127–129. [Google Scholar] [CrossRef] [PubMed]



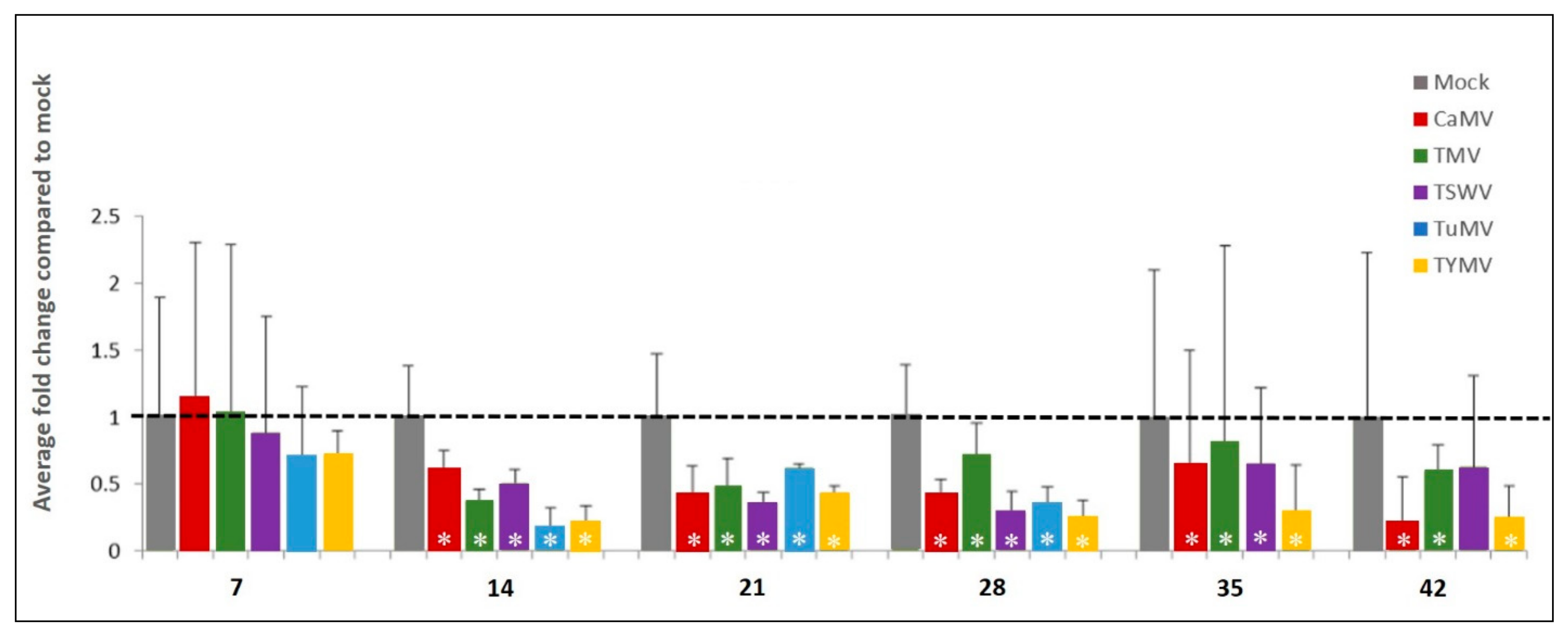
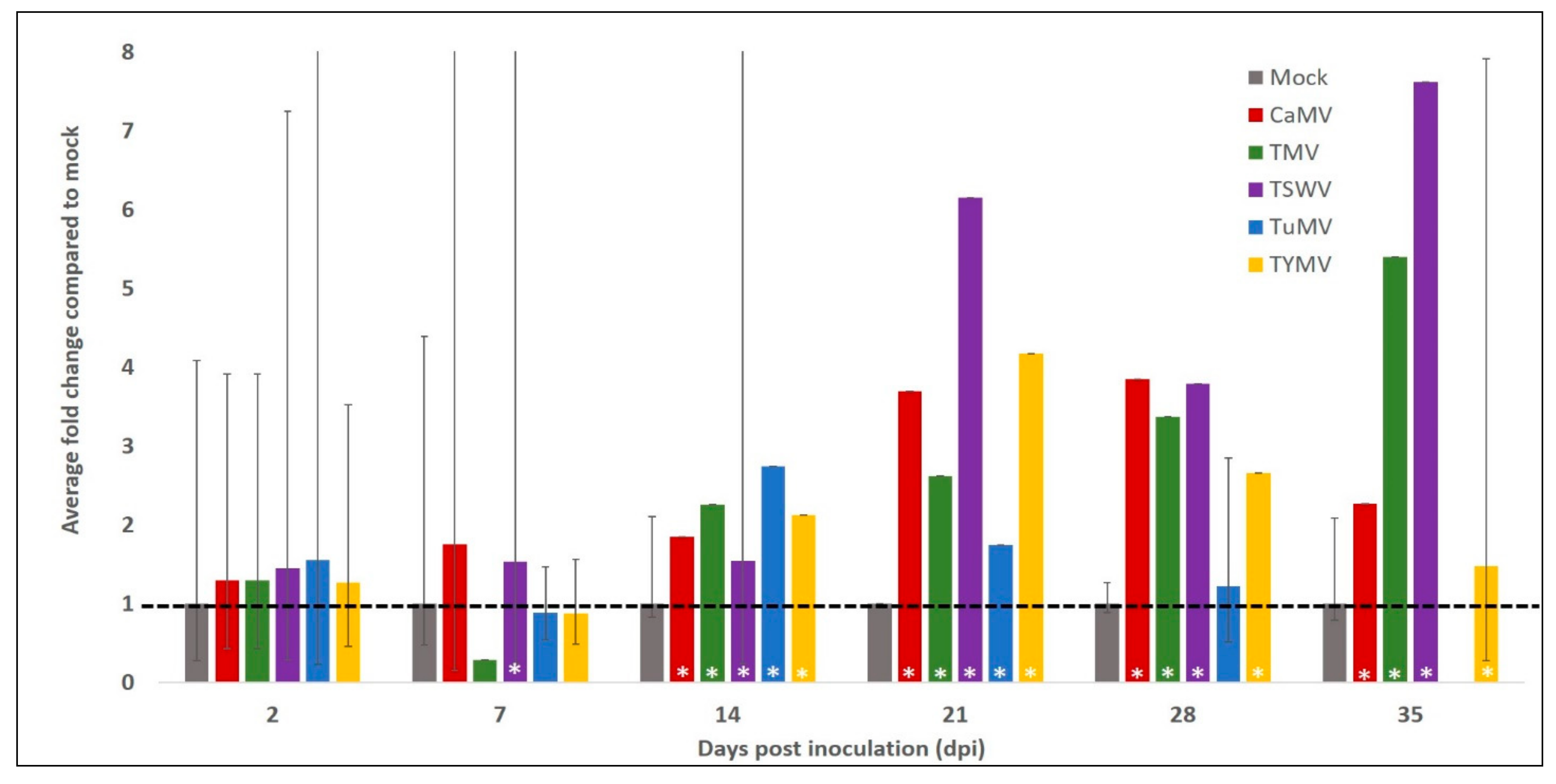

| Treatments | AtSGS3 Transcript Accumulation | AtCPK3 Transcript Accumulation |
|---|---|---|
| Viruses | Decrease by less than 2.0-fold | Increase by about 1.6 to 7-fold |
| Pseudomonas syringae DC3000 | Increase by less than 2-fold, or no change | Decrease by about 1.9-fold |
| Botrytis cinerea | Not tested | Decrease by about 2.9-fold |
| Drought | Decrease by greater than 2-fold | Decrease by about 1.5-1.7-fold |
| Salt | Decrease by greater than 2-fold | Decrease by less than 2-fold |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valmonte-Cortes, G.R.; Lilly, S.T.; Pearson, M.N.; Higgins, C.M.; MacDiarmid, R.M. The Potential of Molecular Indicators of Plant Virus Infection: Are Plants Able to Tell Us They Are Infected? Plants 2022, 11, 188. https://doi.org/10.3390/plants11020188
Valmonte-Cortes GR, Lilly ST, Pearson MN, Higgins CM, MacDiarmid RM. The Potential of Molecular Indicators of Plant Virus Infection: Are Plants Able to Tell Us They Are Infected? Plants. 2022; 11(2):188. https://doi.org/10.3390/plants11020188
Chicago/Turabian StyleValmonte-Cortes, Gardette R., Sonia T. Lilly, Michael N. Pearson, Colleen M. Higgins, and Robin M. MacDiarmid. 2022. "The Potential of Molecular Indicators of Plant Virus Infection: Are Plants Able to Tell Us They Are Infected?" Plants 11, no. 2: 188. https://doi.org/10.3390/plants11020188
APA StyleValmonte-Cortes, G. R., Lilly, S. T., Pearson, M. N., Higgins, C. M., & MacDiarmid, R. M. (2022). The Potential of Molecular Indicators of Plant Virus Infection: Are Plants Able to Tell Us They Are Infected? Plants, 11(2), 188. https://doi.org/10.3390/plants11020188







