Binding Properties of Odorant-Binding Protein 4 of Tirathaba rufivena to Areca catechu Volatiles
Abstract
1. Introduction
2. Results
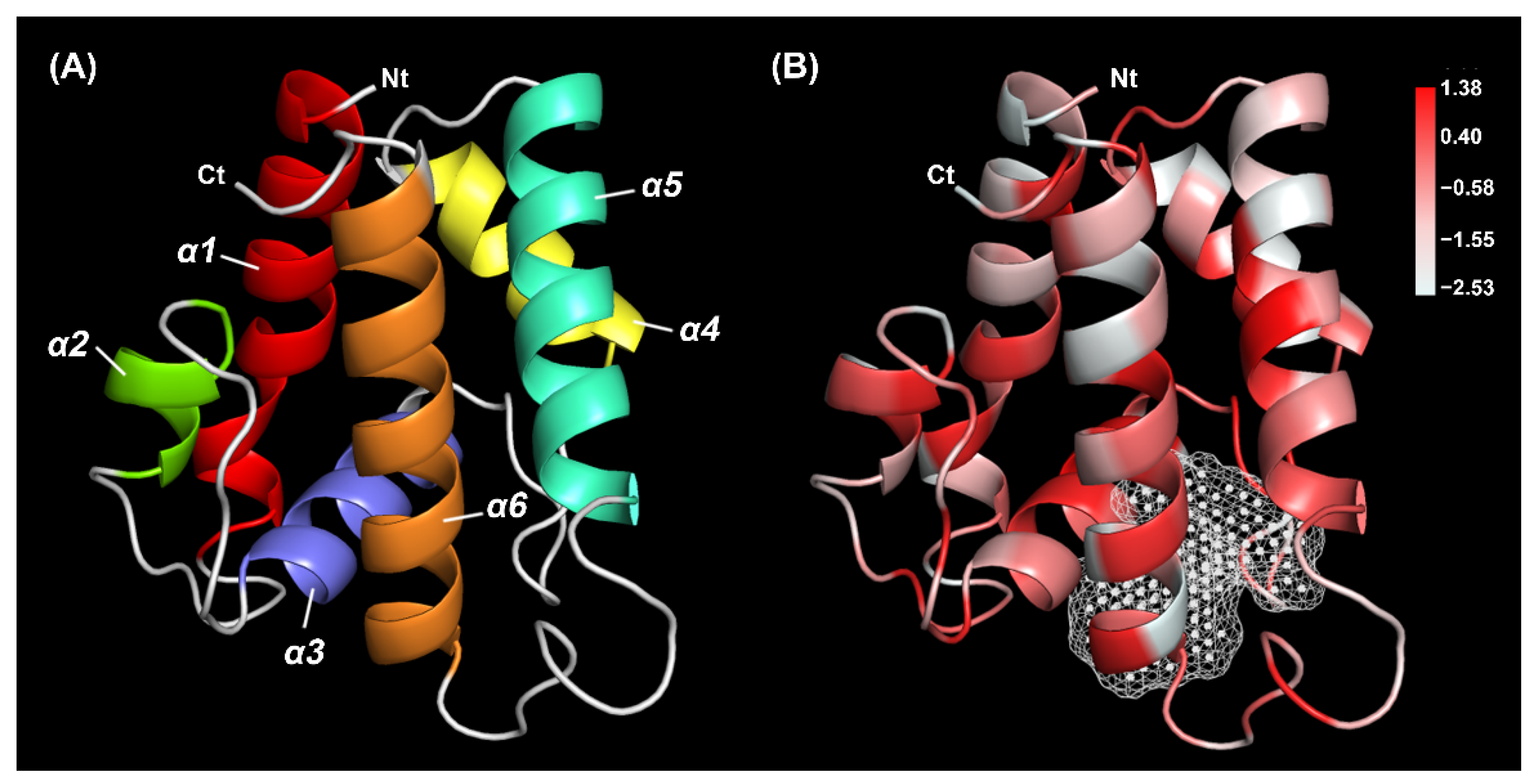
2.1. Sequence Analysis of TrufOBP4
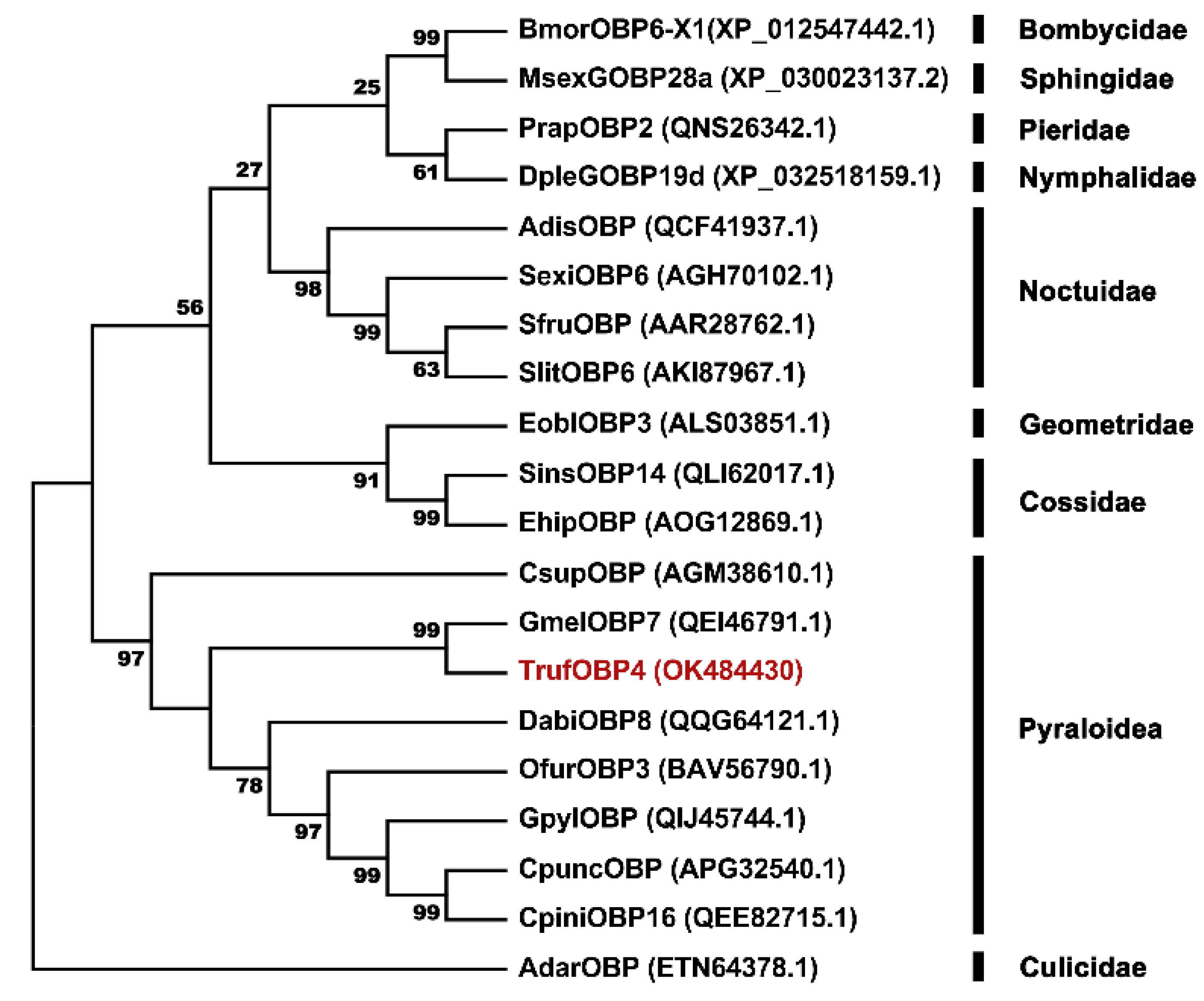
2.2. Phylogenetic Tree Construction of TrufOBP4
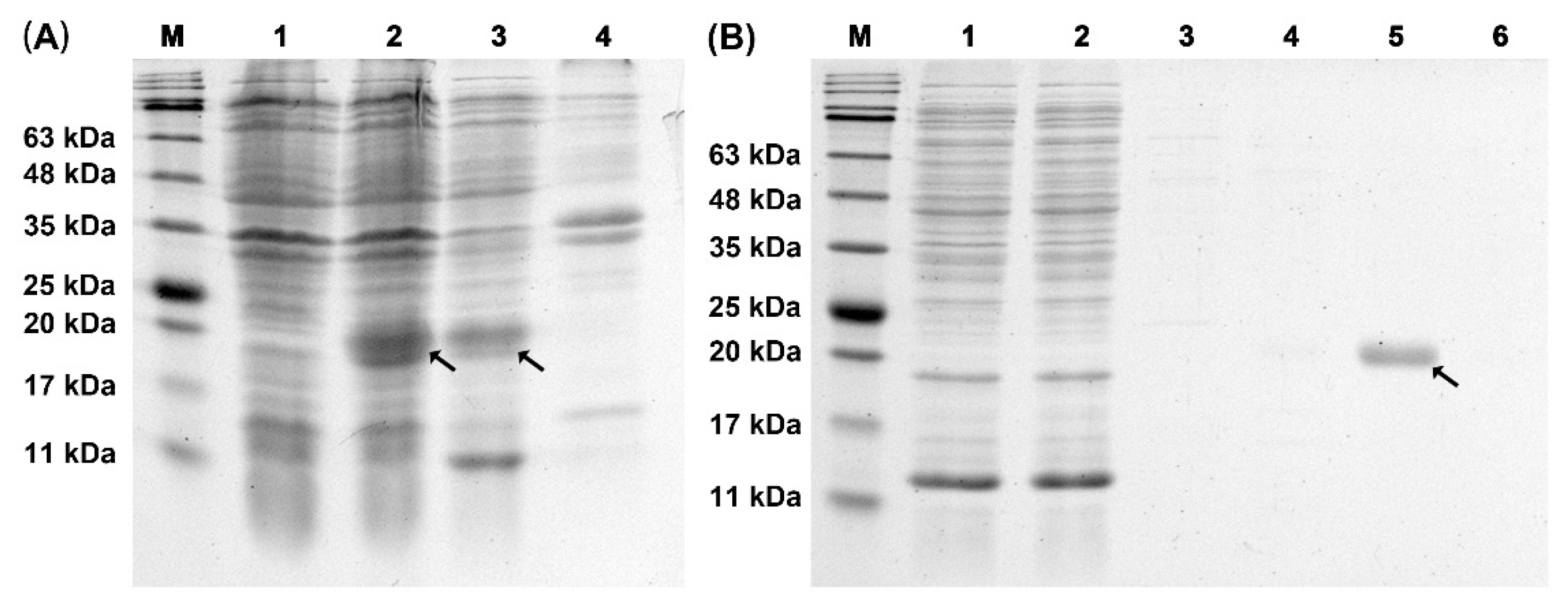
2.3. Expression and Purification of Recombinant TrufOBP4
2.4. Fluorescence Competitive Binding Assays
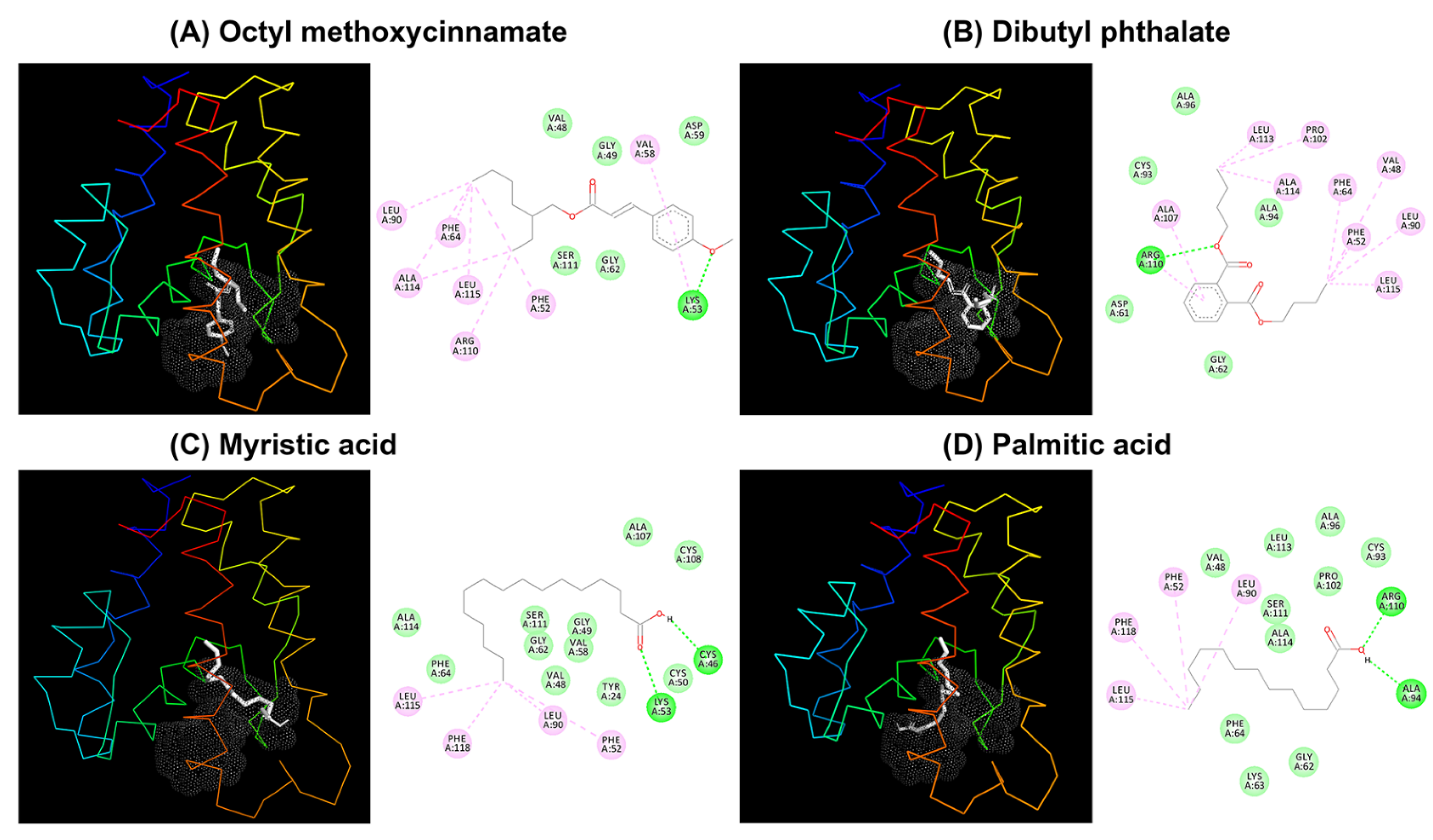
2.5. Molecular Docking
3. Discussion
4. Materials and Methods
4.1. Insect Rearing
4.2. RNA Extraction and cDNA Synthesis
4.3. Cloning and Sequencing Analysis
4.4. Expression and Purification of Recombinant TrufOBP4
4.5. Fluorescence Competitive Binding Assays
4.6. Structure Modelling and Molecular Docking
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhou, J.J. Odorant-binding proteins in insects. Vitam. Horm. 2010, 83, 241–272. [Google Scholar] [CrossRef] [PubMed]
- Hansson, B.S.; Stensmyr, M.C. Evolution of insect olfaction. Neuron 2011, 72, 698–711. [Google Scholar] [CrossRef] [PubMed]
- Mohapatra, P.; Menuz, K. Molecular profiling of the Drosophila antenna reveals conserved genes underlying olfaction in Insects. G3-Genes Genomes Genet. 2019, 9, 3753–3771. [Google Scholar] [CrossRef] [PubMed]
- Sachse, S.; Krieger, J. Olfaction in insects. e-Neuroforum 2011, 2, 49. [Google Scholar] [CrossRef]
- Wei, H.; Tan, S.; Li, Z.; Li, J.; Moural, T.W.; Zhu, F.; Liu, X. Odorant degrading carboxylesterases modulate foraging and mating behaviors of Grapholita molesta. Chemosphere 2021, 270, 128647. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, P.; Zhou, J.J.; Ban, L.P.; Calvello, M. Soluble proteins in insect chemical communication. Cell. Mol. Life Sci. 2006, 63, 1658–1676. [Google Scholar] [CrossRef]
- Yang, K.; Liu, Y.; Niu, D.J.; Wei, D.; Li, F.; Wang, G.R.; Dong, S.L. Identification of novel odorant binding protein genes and functional characterization of OBP8 in Chilo suppressalis (Walker). Gene 2016, 591, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, P.; Iovinella, I.; Felicioli, A.; Dani, F.R. Soluble proteins of chemical communication: An overview across arthropods. Front. Physiol. 2014, 5, 320. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Guo, J.; Zhang, M.; Bai, C.; Wang, Z.; Li, Z. Antennal transcriptome analysis and candidate olfactory genes in Crematogaster rogenhoferi. Bull. Entomol. Res. 2021, 111, 464–475. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhang, N.; Wang, P.; Zhang, S.; Li, D.; Liu, K.; Wang, G.; Wang, X.; Ai, H. Identification of host-plant volatiles and characterization of two novel general odorant-binding proteins from the legume pod borer, Maruca vitrata Fabricius (Lepidoptera: Crambidae). PLoS ONE 2015, 10, e0141208. [Google Scholar]
- Khuhro, S.A.; Liao, H.; Dong, X.-T.; Yu, Q.; Yan, Q.; Dong, S.-L. Two general odorant binding proteins display high bindings to both host plant volatiles and sex pheromones in a pyralid moth Chilo suppressalis (Lepidoptera: Pyralidae). J. Asia-Pac. Entomol. 2017, 20, 521–528. [Google Scholar] [CrossRef]
- Campanacci, V.; Krieger, J.; Bette, S.; Sturgis, J.N.; Lartigue, A.; Cambillau, C.; Breer, H.; Tegoni, M. Revisiting the specificity of Mamestra brassicae and Antheraea polyphemus pheromone-binding proteins with a fluorescence binding assay. J. Biol. Chem. 2001, 276, 20078–20084. [Google Scholar] [CrossRef]
- Gong, D.P.; Zhang, H.J.; Zhao, P.; Xia, Q.Y.; Xiang, Z.H. The odorant binding protein gene family from the genome of silkworm, Bombyx mori. BMC Genom. 2009, 10, 332. [Google Scholar] [CrossRef]
- Gu, S.H.; Sun, L.; Yang, R.N.; Wu, K.M.; Guo, Y.Y.; Li, X.C.; Zhou, J.J.; Zhang, Y.J. Molecular characterization and differential expression of olfactory genes in the antennae of the black cutworm moth Agrotis ipsilon. PLoS ONE 2014, 9, e103420. [Google Scholar] [CrossRef]
- Zhang, T.T.; Wang, W.X.; Gu, S.H.; Zhang, Z.D.; Wu, K.M.; Zhang, Y.J.; Guo, Y.Y. Structure, binding characteristics, and 3D model prediction of a newly identified odorant-binding protein from the cotton bollworm, Helicoverpa armigera (Hübner). J. Integr. Agric. 2012, 11, 430–438. [Google Scholar] [CrossRef]
- Vogt, R.G.; Grosse-Wilde, E.; Zhou, J.J. The Lepidoptera odorant binding protein gene family: Gene gain and loss within the GOBP/PBP complex of moths and butterflies. Insect Biochem. Mol. Biol. 2015, 62, 142–153. [Google Scholar] [CrossRef]
- Tang, B.; Tai, S.; Dai, W.; Zhang, C. Expression and functional analysis of two odorant-binding proteins from Bradysia odoriphaga (Diptera: Sciaridae). J. Agric. Food Chem. 2019, 67, 3565–3574. [Google Scholar] [CrossRef]
- Gong, Z.J.; Zhou, W.W.; Yu, H.Z.; Mao, C.G.; Zhang, C.X.; Cheng, J.A.; Zhu, Z.R. Cloning, expression and functional analysis of a general odorant-binding protein 2 gene of the rice striped stem borer, Chilo suppressalis (Walker) (Lepidoptera: Pyralidae). Insect. Mol. Biol. 2009, 18, 405–417. [Google Scholar] [CrossRef]
- Jing, D.; Zhang, T.; Prabu, S.; Bai, S.; He, K.; Wang, Z. Molecular characterization and volatile binding properties of pheromone binding proteins and general odorant binding proteins in Conogethes pinicolalis (Lepidoptera: Crambidae). Int. J. Biol. Macromol. 2020, 146, 263–272. [Google Scholar] [CrossRef]
- Zhong, B.; Lv, C.; Qin, W. Preliminary study on biology and feeding capacity of Chelisoches morio (Fabricius) (Dermaptera:Chelisochidae) on Tirathaba rufivena (Walker). SpringerPlus 2016, 5, 1944. [Google Scholar] [CrossRef]
- Zhong, B.; Lv, C.; Qin, W. Effectiveness of the botanical insecticide azadirachtin against Tirathaba rufivena (Lepidoptera: Pyralidae). Fla Entomol. 2017, 100, 215–218. [Google Scholar] [CrossRef][Green Version]
- Venthur, H.; Zhou, J.-J. Odorant receptors and odorant-binding proteins as insect pest control targets: A comparative analysis. Front. Physiol. 2018, 9, 1163. [Google Scholar] [CrossRef]
- Karlsson, M.F.; Proffit, M.; Birgersson, G. Host-plant location by the Guatemalan potato moth Tecia solanivora is assisted by floral volatiles. Chemoecology 2017, 27, 187–198. [Google Scholar] [CrossRef]
- Bai, P.H.; Wang, H.M.; Liu, B.S.; Li, M.; Liu, B.M.; Gu, X.S.; Tang, R. Botanical volatiles selection in mediating electrophysiological responses and reproductive behaviors for the fall webworm Moth Hyphantria cunea. Front. Physiol. 2020, 11, 486. [Google Scholar] [CrossRef]
- Wei, H.S.; Qin, J.H.; Cao, Y.Z.; Li, K.B.; Yin, J. Two classic OBPs modulate the responses of female Holotrichia oblita to three major ester host plant volatiles. Insect Mol. Biol. 2021, 30, 390–399. [Google Scholar] [CrossRef]
- Zhang, M.; Huang, Y.L.; Song, F.; Chen, W.J.; Zhao, S.L.; Deng, F.M. Analysis of aromatic components in areca inflorescence by SPME coupled with GC-MS. Chin. J. Trop. Crops 2014, 35, 1244–1249. [Google Scholar]
- Li, Z.D. Preliminary study on the grading of ovarian development characteristics and volatile activity of betel flower to the Tirathaba rufivena Walker. Master’s Thesis, Hainan University, Haikou, China, 2021; pp. 20–23. [Google Scholar]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Liu, H.; Duan, H.; Wang, Q.; Xiao, Y.; Wang, Q.; Xiao, Q.; Sun, L.; Zhang, Y. Key Amino Residues Determining Binding Activities of the Odorant Binding Protein AlucOBP22 to Two Host Plant Terpenoids of Apolygus lucorum. J. Agric. Food Chem. 2019, 67, 5949–5956. [Google Scholar] [CrossRef]
- Wang, C.Q.; Li, J.Q.; Li, E.T.; Nyamwasa, I.; Li, K.B.; Zhang, S.; Peng, Y.; Wei, Z.J.; Yin, J. Molecular and functional characterization of odorant-binding protein genes in Holotrichia oblita Faldermann. Int. J. Biol. Macromol. 2019, 136, 359–367. [Google Scholar] [CrossRef]
- Xu, Y.L.; He, P.; Zhang, L.; Fang, S.Q.; Dong, S.L.; Zhang, Y.J.; Li, F. Large-scale identification of odorant-binding proteins and chemosensory proteins from expressed sequence tags in insects. BMC Genom. 2009, 10, 632. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.-J.; Zhang, G.-A.; Huang, W.; Birkett, M.A.; Field, L.M.; Pickett, J.A.; Pelosi, P. Revisiting the odorant-binding protein LUSH of Drosophila melanogaster: Evidence for odour recognition and discrimination. FEBS Lett. 2004, 558, 23–26. [Google Scholar] [CrossRef]
- Ullah, R.M.K.; Quershi, S.R.; Adeel, M.M.; Abdelnabby, H.; Waris, M.I.; Duan, S.G.; Wang, M.Q. An odorant binding protein (SaveOBP9) involved in chemoreception of the wheat aphid Sitobion avenae. Int. J. Mol. Sci. 2020, 21, 8331. [Google Scholar] [CrossRef] [PubMed]
- Yao, R.; Zhao, M.; Zhong, L.; Li, Y.; Li, D.; Deng, Z.; Ma, X. Characterization of the binding ability of the odorant binding protein BminOBP9 of Bactrocera minax to citrus volatiles. Pest. Manag. Sci. 2021, 77, 1214–1225. [Google Scholar] [CrossRef] [PubMed]
- Rizwangul, A.; Cao, Y.; Zhang, S.; Yin, J.; Li, X.; Li, K. Feeding preference and taxis behavior of adult Holotrichia oblita (Coleoptera: Scarabaeidae) on three plants. Acta Entomol. Sin. 2018, 61, 585–595. [Google Scholar]
- Huang, G.Z.; Liu, J.T.; Zhou, J.J.; Wang, Q.; Dong, J.Z.; Zhang, Y.J.; Li, X.C.; Li, J.; Gu, S.H. Expressional and functional comparisons of two general odorant binding proteins in Agrotis ipsilon. Insect Biochem. Mol. Biol. 2018, 98, 34–47. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Fu, X.B.; Cui, H.C.; Zhao, L.; Yu, J.Z.; Li, H.L. Functional Characteristics, Electrophysiological and antennal immunolocalization of general odorant-binding protein 2 in tea geometrid, Ectropis obliqua. Int. J. Mol. Sci. 2018, 19, 875. [Google Scholar] [CrossRef] [PubMed]
- Ishida, Y.; Ishibashi, J.; Leal, W.S. Fatty acid solubilizer from the oral disk of the blowfly. PLoS ONE 2013, 8, e51779. [Google Scholar] [CrossRef]
- Chen, D.; Zhang, L.; Xing, Z.; Lei, Z. Identification and function of the OBP13 protein from the leafminer (Liriomyza sativae). Sci. Agric. Sin. 2018, 51, 893–904. [Google Scholar]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Ahmed, T.; Zhang, T.; Wang, Z.; He, K.; Bai, S. Molecular cloning, expression profile, odorant affinity, and stability of two odorant-binding proteins in Macrocentrus cingulum Brischke (Hymenoptera: Braconidae). Arch. Insect Biochem. Physiol. 2017, 94, e21374. [Google Scholar] [CrossRef]
- Ma, L.; Li, Z.; Zhang, W.; Cai, X.; Luo, Z.; Zhang, Y.; Chen, Z. The Odorant Binding Protein 6 Expressed in Sensilla Chaetica Displays Preferential Binding Affinity to Host Plants Volatiles in Ectropis obliqua. Front. Physiol. 2018, 9, 534. [Google Scholar] [CrossRef]
- Ju, Q.; Qu, M.J.; Wang, Y.; Jiang, X.J.; Li, X.; Dong, S.L.; Han, Z.J. Molecular and biochemical characterization of two odorant-binding proteins from dark black chafer, Holotrichia parallela. Genome 2012, 55, 537–546. [Google Scholar] [CrossRef]
- Lautenschlager, C.; Leal, W.S.; Clardy, J. Bombyx mori pheromone-binding protein binding nonpheromone ligands: Implications for pheromone recognition. Structure 2007, 15, 1148–1154. [Google Scholar] [CrossRef][Green Version]



| No. | Ligands | Formula | CAS No# | IC50 (μM) | Ki (μM) |
|---|---|---|---|---|---|
| 1 | (R)-(+)-limonene | C10H16 | 5989-27-5 | 25.32 | 22.07 |
| 2 | (S)-(−)-Limonene | C10H16 | 5989-54-8 | 33.36 | 29.08 |
| 3 | 2-Ethylhexanol | C8H18O | 104-76-7 | 24.08 | 20.99 |
| 4 | Octyl methoxycinnamate | C18H26O3 | 83834-59-7 | 1.33 | 1.16 |
| 5 | 3-Carene | C10H16 | 13466-78-9 | 22.41 | 19.53 |
| 6 | Methyl salicylate | C8H8O3 | 119-36-8 | >50 | >50 |
| 7 | Cinene | C10H16 | 138-86-3 | >50 | >50 |
| 8 | Cineole | C10H18O | 470-82-6 | >50 | >50 |
| 9 | 3-Hexenyl acetate | C8H14O2 | 3681-71-8 | 46.59 | 40.61 |
| 10 | Dibutyl phthalate | C16H22O4 | 84-74-2 | 5.29 | 4.61 |
| 11 | Hexyl acetate | C8H16O2 | 142-92-7 | >50 | >50 |
| 12 | Isoanyl acetate | C7H14O2 | 123-92-2 | 24.11 | 21.02 |
| 13 | 3-Methylbutyl 2-methylbutanoate | C10H20O2 | 27625-35-0 | >50 | 45.83 |
| 14 | Linalol | C10H18O | 78-70-6 | >50 | >50 |
| 15 | Myrcene | C10H16 | 123-35-3 | 19.80 | 17.26 |
| 16 | Myristic acid | C14H28O2 | 544-63-8 | 4.04 | 3.52 |
| 17 | Palmitic acid | C16H32O2 | 57-10-3 | 6.06 | 5.28 |
| 18 | Styrene | C8H8 | 100-42-5 | 41.50 | 36.17 |
| 19 | β-Pinene | C10H16 | 18172-67-3 | 33.29 | 29.02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, X.; Wang, Z.; Cui, G.; Du, Z.; Qian, Y.; Yang, S.; Liu, M.; Guo, J. Binding Properties of Odorant-Binding Protein 4 of Tirathaba rufivena to Areca catechu Volatiles. Plants 2022, 11, 167. https://doi.org/10.3390/plants11020167
Zhou X, Wang Z, Cui G, Du Z, Qian Y, Yang S, Liu M, Guo J. Binding Properties of Odorant-Binding Protein 4 of Tirathaba rufivena to Areca catechu Volatiles. Plants. 2022; 11(2):167. https://doi.org/10.3390/plants11020167
Chicago/Turabian StyleZhou, Xiang, Zheng Wang, Guangchao Cui, Zimeng Du, Yunlong Qian, Shumei Yang, Minghui Liu, and Jixing Guo. 2022. "Binding Properties of Odorant-Binding Protein 4 of Tirathaba rufivena to Areca catechu Volatiles" Plants 11, no. 2: 167. https://doi.org/10.3390/plants11020167
APA StyleZhou, X., Wang, Z., Cui, G., Du, Z., Qian, Y., Yang, S., Liu, M., & Guo, J. (2022). Binding Properties of Odorant-Binding Protein 4 of Tirathaba rufivena to Areca catechu Volatiles. Plants, 11(2), 167. https://doi.org/10.3390/plants11020167






