The S-Gene YUC6 Pleiotropically Determines Male Mating Type and Pollen Size in Heterostylous Turnera (Passifloraceae): A Novel Neofunctionalization of the YUCCA Gene Family
Abstract
1. Introduction
2. Results
2.1. Knockdown of TjYUC6 in T. joelii
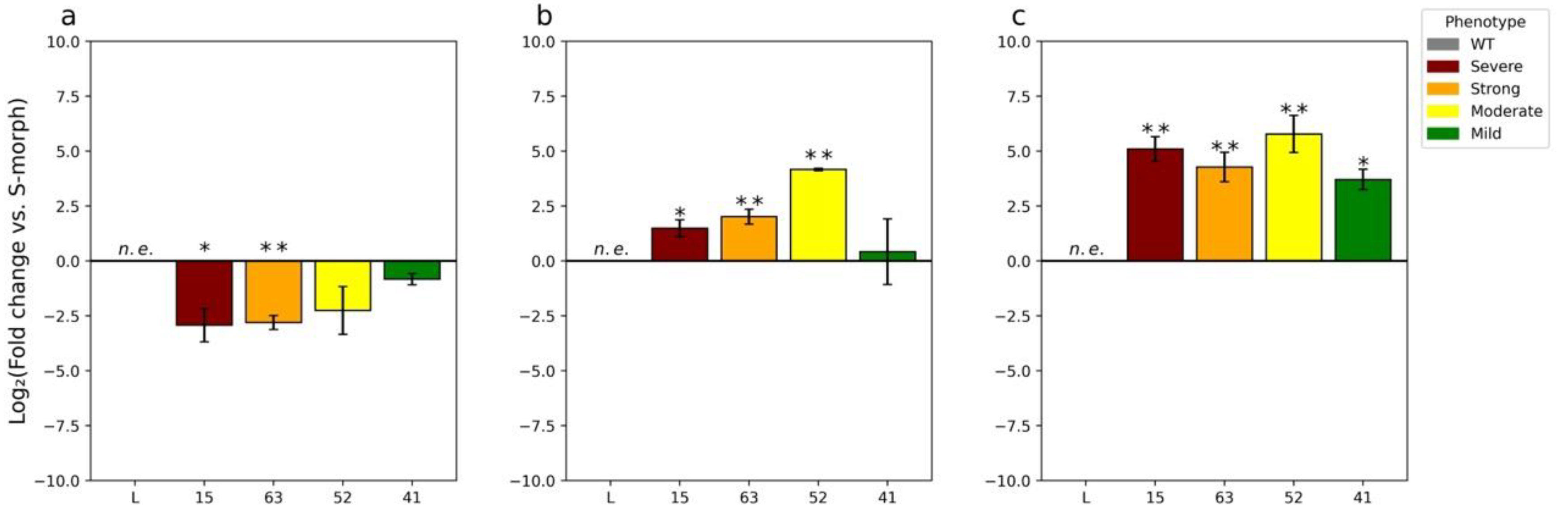
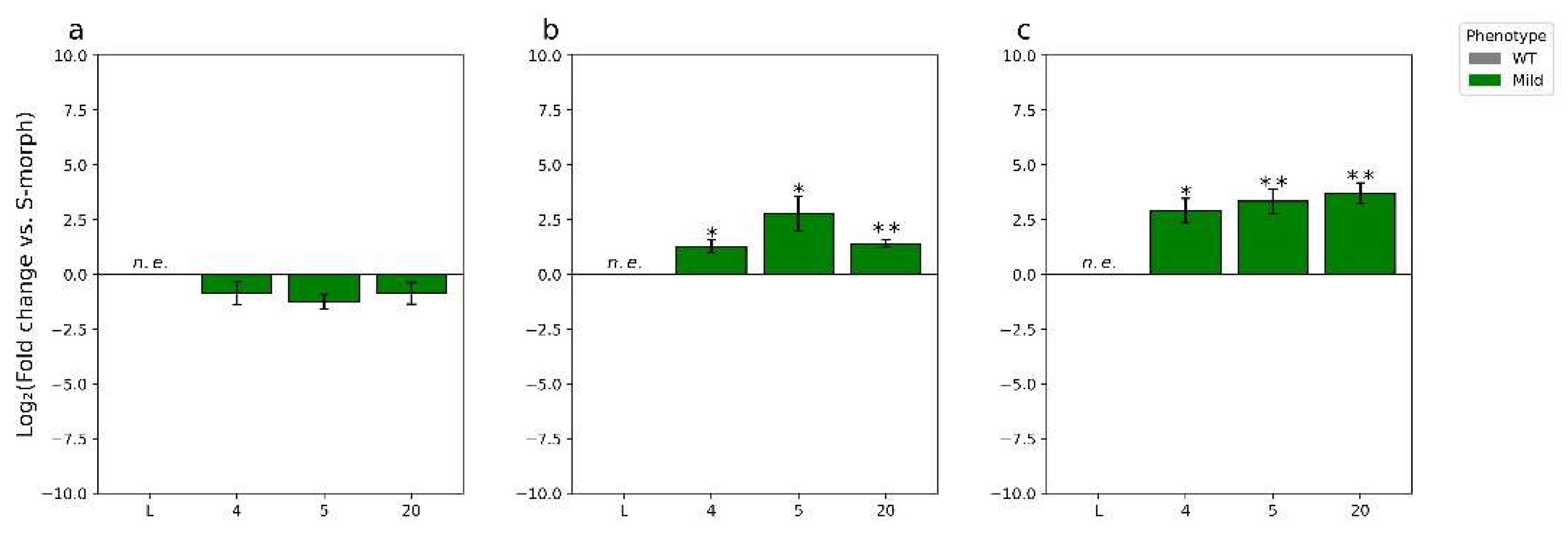
2.2. RT-qPCR Analysis of YUC6 Expression in Knockdown Lines
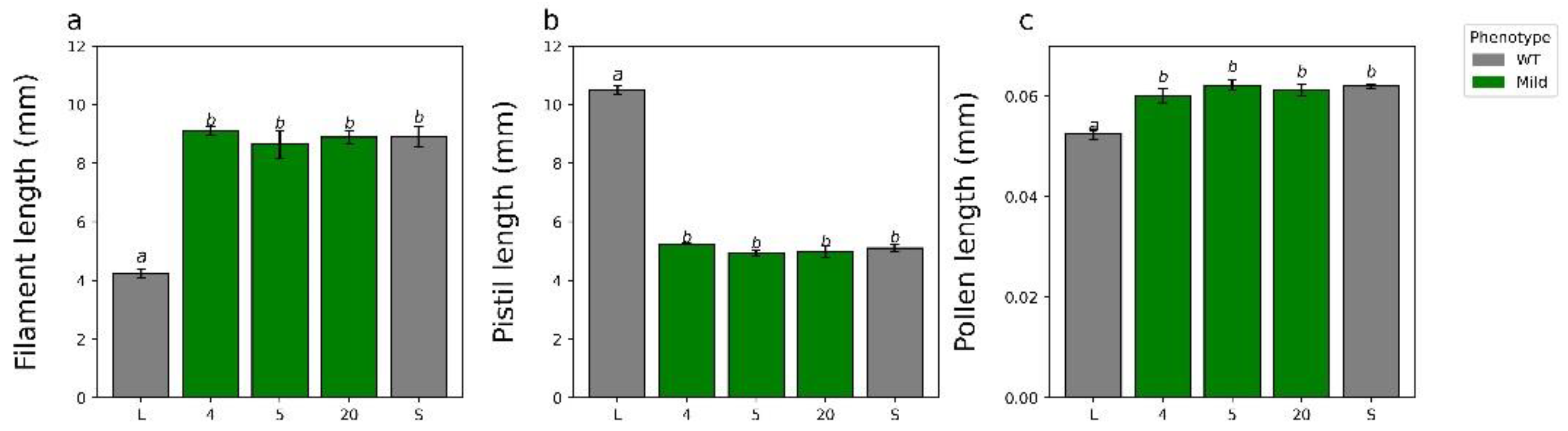
2.3. Additional Characterization of YUC6 Knockdown Lines
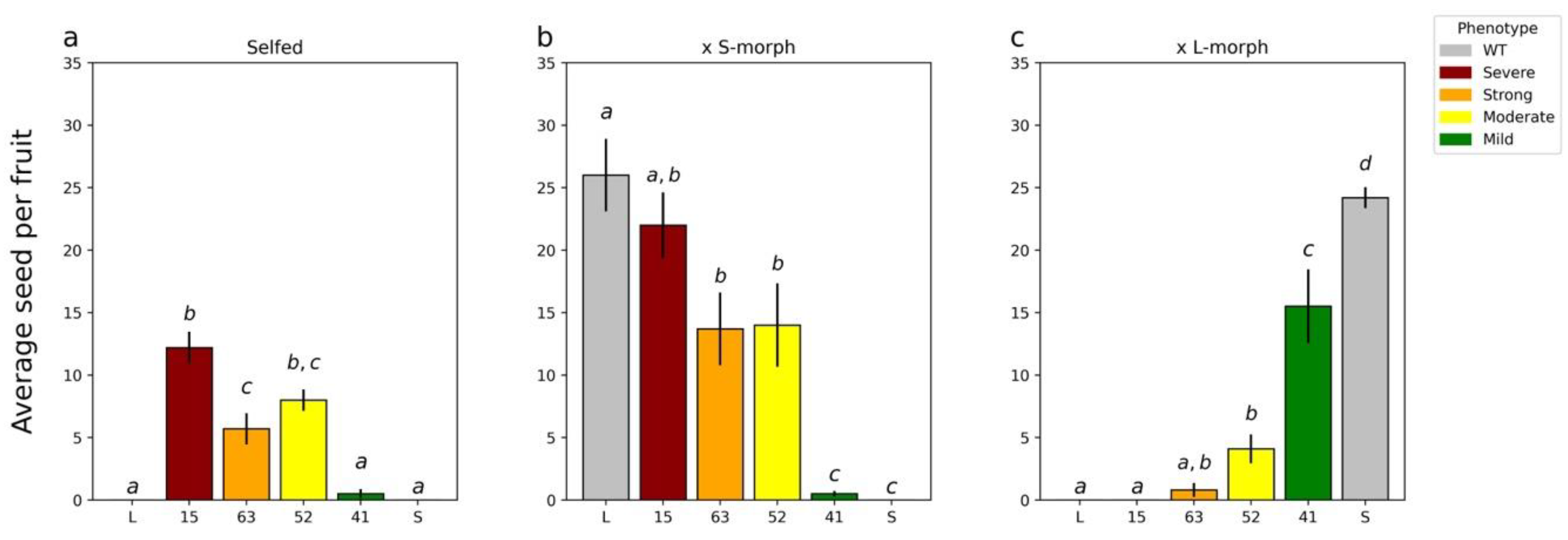
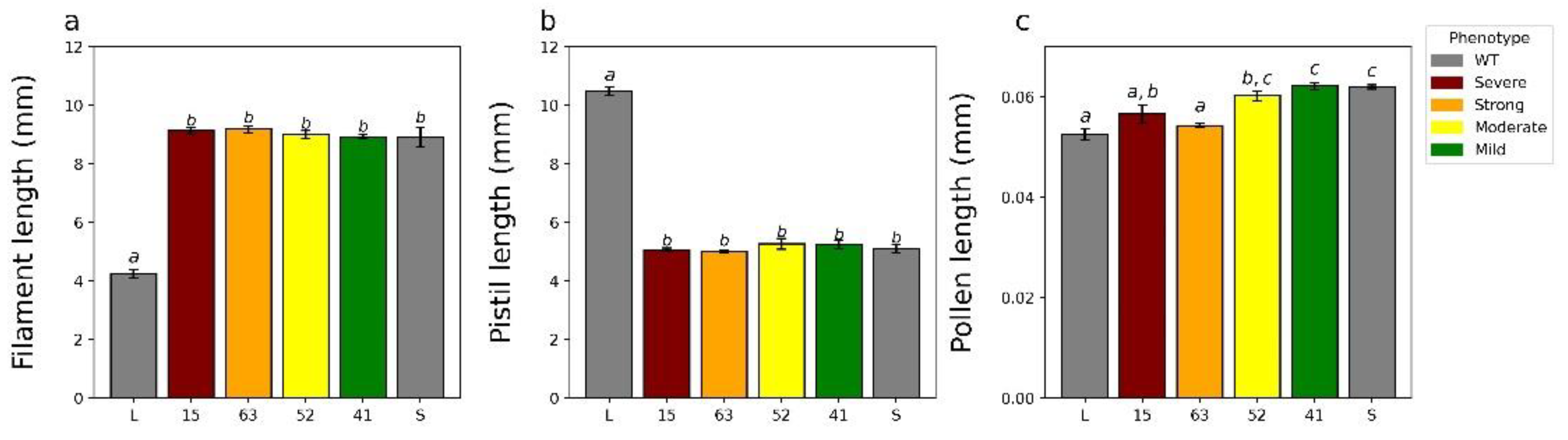
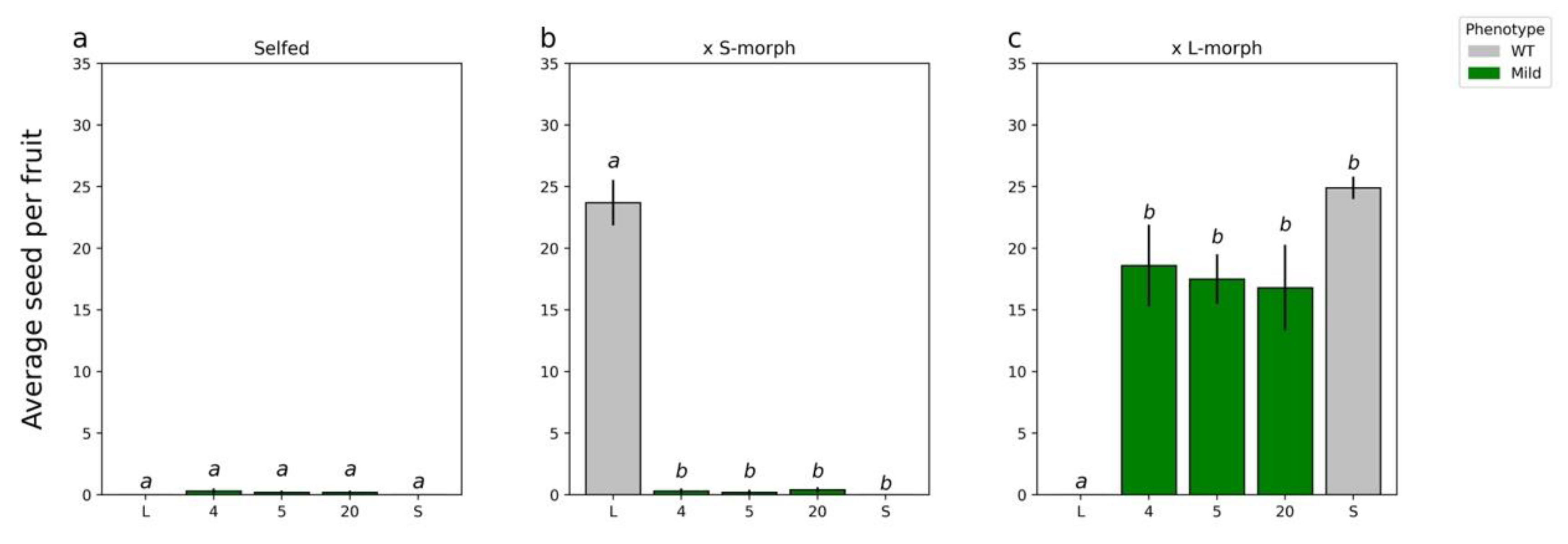
2.4. Characterizing the T1 Generation
3. Discussion
3.1. TjYUC6 Is the Male Determinant Factor in Distylous Turnera
3.2. Is Neofunctionalization of YUC Homologs a Common Mechanism in Breeding Barrier Evolution?
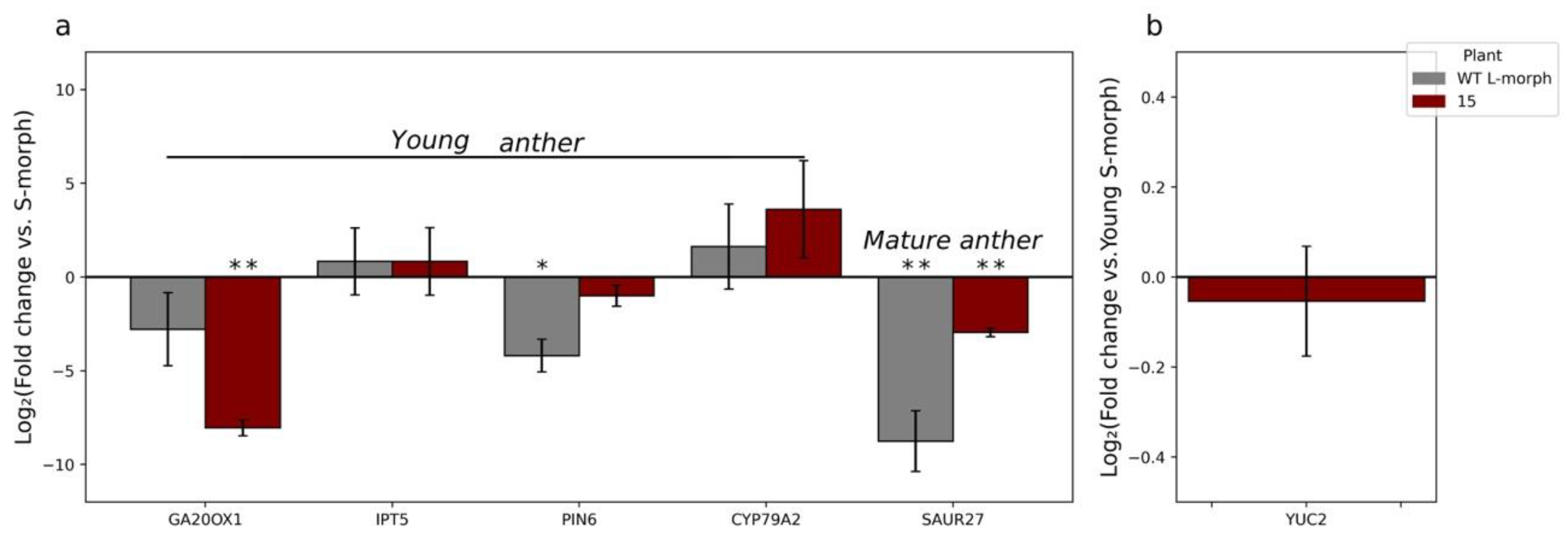
3.3. RT-qPCR Analysis Reveals Expression of TjYUC6 in Mature Pollen
3.4. YUC6 Acts Pleiotropically, Establishing Pollen Size Dimorphisms in Addition to Mating Type
4. Methods
4.1. Plasmid Construction and Transformation
4.2. Genotyping
4.3. Quantification of Expression
4.4. Phenotyping
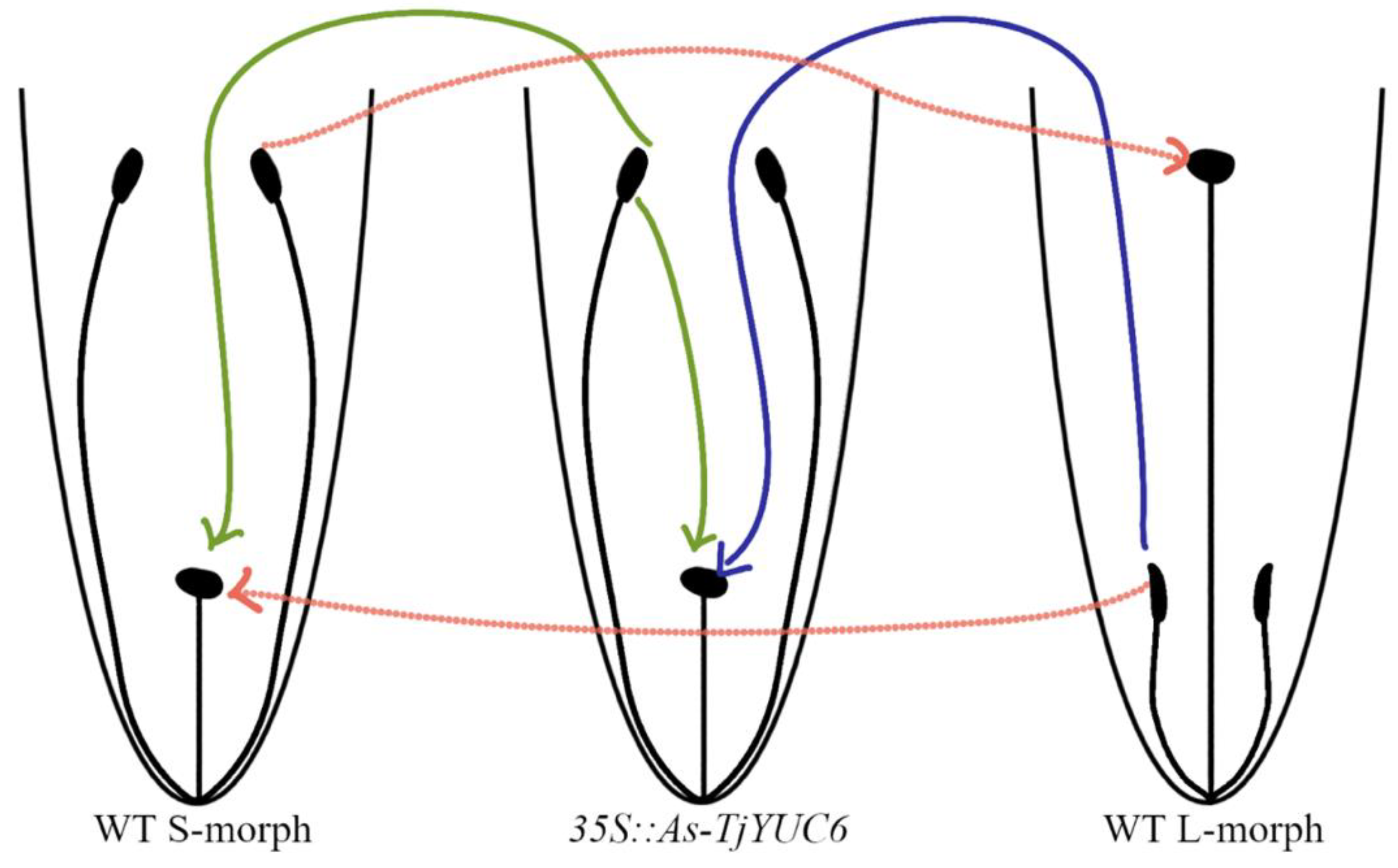
- Transformants were initially self-pollinated and those that set seed considered self-compatible.
- To confirm the retention of female mating type, transformants were crossed as pistil parents with the WT S- and L-morphs. Lines retaining the S-morph female mating type would only set seed with the L-morph.
- To confirm the change in male mating type, the transformants were crossed as pollen parents with WT S- and L-morphs. Lines showing a change in male mating type would only set seed with the S-morph.
5. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barrett, S.C.H. A most complex marriage arrangement, recent advances on heterostyly and unresolved questions. New Phytol. 2019, 224, 1051–1067. [Google Scholar] [CrossRef] [PubMed]
- Bateson, W.; Gregory, R.P. On the inheritance of heterostylism in Primula. Proc. R. Soc. Lond. Ser. B 1905, 76, 581–586. [Google Scholar] [CrossRef]
- Dowrick, V.P. Heterostyly and homostyly in Primula obconica. Heredity 1956, 10, 219–236. [Google Scholar] [CrossRef]
- Schwander, T.; Libbrecht, R.; Keller, L. Supergenes and complex phenotypes. Curr. Biol. 2014, 24, R288–R294. [Google Scholar] [CrossRef]
- Gutiérrez-Valencia, J.; Hughes, P.W.; Berdan, E.L.; Slotte, T. The genomic architecture and evolutionary fates of supergenes. Genome Biol. Evol. 2021, 13, evab057. [Google Scholar] [CrossRef]
- Ushijima, K.; Ikeda, K.; Nakano, R.; Matsubara, M.; Tsuda, Y.; Kubo, Y. Genetic control of floral morph and petal pigmentation in Linum grandiflorum Desf.; a heterostylous flax. Hort. J. 2015, 84, 261–268. [Google Scholar] [CrossRef]
- Nowak, M.D.; Russo, G.; Schlapbach, R.; Huu, C.N.; Lenhard, M.; Conti, E. The draft genome of Primula veris yields insights into the molecular basis of heterostyly. Genome Biol. 2015, 16, 12. [Google Scholar] [CrossRef]
- Yasui, Y.; Hirakawa, H.; Ueno, M.; Matsui, K.; Katsube-Tanaka, T.; Yang, S.J.; Aii, J.; Sato, S.; Mori, M. Assembly of the draft genome of buckwheat and its applications in identifying agronomically useful genes. DNA Res. 2016, 23, 215–224. [Google Scholar] [CrossRef]
- Shore, J.S.; Hamam, H.J.; Chafe, P.D.J.; Labonne, J.D.J.; Henning, P.M.; McCubbin, A.G. The long and short of the S-locus in Turnera (Passifloraceae). New Phytol. 2019, 224, 1316–1329. [Google Scholar] [CrossRef]
- Franklin-Tong, V.E. Self-Incompatibility in Flowering Plants; Springer: Berlin/Heidelberg, Germany, 2008. [Google Scholar] [CrossRef]
- Yasui, Y.; Mori, M.; Aii, J.; Abe, T.; Masumoto, D.; Sato, S.; Hayashi, Y.; Ohnishi, O.; Ota, T. S-Locus Early Flowering 3 is exclusively present in the genomes of short-styled buckwheat plants that exhibit heteromorphic self-incompatibility. PLoS ONE 2012, 7, e31264. [Google Scholar] [CrossRef]
- Zhao, H.; Xu, D.; Tian, T.; Kong, F.; Lin, K.; Gan, S.; Zhang, H.; Li, G. Molecular and functional dissection of EARLY-FLOWERING 3 (ELF3) and ELF4 in Arabidopsis. Plant Sci. 2021, 303, 110786. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cocker, J.M.; Wright, J.; Webster, M.A.; McMullan, M.; Dyer, S.; Swarbreck, D.; Caccamo, M.; Oosterhout, C.V.; Gilmartin, P.M. Genetic architecture and evolution of the S locus supergene in Primula vulgaris. Nat. Plants 2016, 2, 16188. [Google Scholar] [CrossRef]
- Potente, G.; Léveillé-Bourret, É.; Yousefi, N.; Choudhury, R.R.; Keller, B.; Diop, S.I.; Duijsings, D.; Pirovano, W.; Lenhard, M.; Szövényi, P.; et al. Comparative genomics elucidates the origin of a supergene controlling floral heteromorphism. Mol. Biol. Evol. 2022, 39, msac035. [Google Scholar] [CrossRef]
- Burrows, B.A.; McCubbin, A.G. Sequencing the genomic regions flanking S-linked PvGLO sequences confirm the presence of two GLO loci, one of which lies adjacent to the style-length determinant gene CYP734A50. Plant Reprod. 2017, 30, 53–67. [Google Scholar] [CrossRef]
- Huu, C.N.; Kappel, C.; Keller, B.; Sicard, A.; Takebayashi, Y.; Breuninger, H.; Nowak, M.D.; Bäurle, I.; Himmelbach, A.; Burkart, M.; et al. Presence versus absence of CYP734A50 underlies the style-length dimorphism in primroses. eLife 2016, 5, e17956. [Google Scholar] [CrossRef]
- Huu, C.N.; Plaschil, S.; Himmelbach, A.; Kappel, C.; Lenhard, M. Female self-incompatibility type in heterostylous Primula is determined by the brassinosteroid-inactivating cytochrome P450 CYP734A50. Curr. Biol. 2021, 32, 671–676. [Google Scholar] [CrossRef]
- Huu, C.N.; Keller, B.; Conti, E.; Kappel, C.; Lenhard, M. Supergene evolution via stepwise duplications and neofunctionalization of a floral-organ identity gene. Proc. Natl. Acad. Sci. USA 2020, 117, 23149–23157. [Google Scholar] [CrossRef]
- Matzke, C.M.; Shore, J.S.; Neff, M.M.; McCubbin, A.G. The Turnera style S-locus gene TsBAHD possesses brassinosteroid-inactivating activity when expressed in Arabidopsis thaliana. Plants 2020, 9, 1566. [Google Scholar] [CrossRef]
- Matzke, C.M.; Hamam, H.J.; Henning, P.M.; Dougherty, K.; Shore, J.S.; Neff, M.M.; McCubbin, A.G. Pistil mating type and morphology are mediated by the brassinosteroid inactivating activity of the S-locus gene BAHD in heterostylous Turnera species. Int. J. Mol. Sci. 2021, 22, 10603. [Google Scholar] [CrossRef]
- Henning, P.M.; Shore, J.S.; McCubbin, A.G. Transcriptome and network analyses of heterostyly in Turnera subulata provide mechanistic insights: Are S-loci a red-light for pistil elongation? Plants 2020, 9, 713. [Google Scholar] [CrossRef]
- Foote, H.C.; Ride, J.P.; Franklin-Tong, V.E.; Walker, E.A.; Lawrence, M.J.; Franklin, F.C. Cloning and expression of a distinctive class of self-incompatibility (S) gene from Papaver rhoeas L. Proc. Natl. Acad. Sci. USA 1994, 91, 2265–2269. [Google Scholar] [CrossRef] [PubMed]
- Kakeda, K.; Jordan, N.D.; Conner, A.; Ride, J.P.; Franklin-Tong, V.E.; Franklin, F.C.H. Identification of residues in a hydrophilic loop of the Papaver rhoeas S protein that play a crucial role in recognition of incompatible pollen. Plant Cell 1998, 10, 1723–1731. [Google Scholar] [CrossRef]
- de Graaf, B.H.; Vatovec, S.; Juárez-Díaz, J.A.; Chai, L.; Kooblall, K.; Wilkins, K.A.; Zou, H.; Forbes, T.; Franklin, F.C.H.; Franklin-Tong, V.E. The Papaver self-incompatibility pollen S-determinant, PrpS, functions in Arabidopsis thaliana. Curr. Biol. 2012, 22, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, K.A.; Bosch, M.; Haque, T.; Teng, N.; Poulter, N.S.; Franklin-Tong, V.E. Self-incompatibility-induced programmed cell death in field poppy pollen involves dramatic acidification of the incompatible pollen tube cytosol. Plant Physiol. 2015, 167, 766–779. [Google Scholar] [CrossRef] [PubMed]
- Takeshima, R.; Nishio, T.; Komatsu, S.; Kurauchi, N.; Matsui, K. Identification of a gene encoding polygalacturonase expressed specifically in short styles in distylous common buckwheat (Fagopyrum esculentum). Heredity 2019, 123, 492–502. [Google Scholar] [CrossRef] [PubMed]
- McCubbin, A.G.; Lee, C.; Hetrick, A. Identification of genes showing differential expression between morphs in developing flowers of Primula vulgaris. Sex. Plant Reprod. 2006, 19, 63–72. [Google Scholar] [CrossRef]
- Burrows, B.; McCubbin, A. Examination of S-locus regulated differential expression in Primula vulgaris floral development. Plants 2018, 7, 38. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Luo, Z.; Yuan, S.; Mei, L.; Zhang, D. Global transcriptome and gene co-expression network analyses on the development of distyly in Primula oreodoxa. Heredity 2019, 123, 784–794. [Google Scholar] [CrossRef] [PubMed]
- Khosravi, D.; Joulaie, R.; Shore, J.S. Immunocytochemical distribution of polygalacturonase and pectins in styles of distylous and homostylous Turneraceae. Sex. Plant Reprod. 2003, 16, 179–190. [Google Scholar] [CrossRef]
- Khosravi, D.; Yang, E.C.C.; Siu, K.W.M.; Shore, J.S. High level of α-dioxygenase in short styles of distylous Turnera species. Int. J. Plant Sci. 2004, 165, 995–1006. [Google Scholar] [CrossRef]
- Cohen, J.I. De novo Sequencing and comparative transcriptomics of floral development of the distylous species Lithospermum multiflorum. Front. Plant Sci. 2016, 7, 1934. [Google Scholar] [CrossRef] [PubMed]
- Stern, D.L. The genetic causes of convergent evolution. Nat. Rev. Genet. 2013, 14, 751–764. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Qu, Y.; Song, G.; Lei, F. Genomic insights into the adaptive convergent evolution. Curr. Genom. 2019, 20, 81–89. [Google Scholar] [CrossRef]
- Gomes, G.L.B.; Scortecci, K.C. Auxin and its role in plant development: Structure, signaling, regulation and response mechanisms. Plant Biol. 2021, 23, 894–904. [Google Scholar] [CrossRef] [PubMed]
- Mashiguchi, K.; Tanaka, K.; Sakai, T.; Sugawara, S.; Kawaide, H.; Natsume, M.; Hanada, A.; Yaeno, T.; Shirasu, K.; Yao, H.; et al. The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18512–18517. [Google Scholar] [CrossRef] [PubMed]
- Morffy, N.; Strader, L.C. Old Town Roads: Routes of auxin biosynthesis across kingdoms. Curr. Opin. Plant Biol. 2020, 55, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Vosolsobě, S.; Skokan, R.; Petrášek, J. The evolutionary origins of auxin transport: What we know and what we need to know. J. Exp. Bot. 2020, 71, 3287–3295. [Google Scholar] [CrossRef]
- Wang, C.; Liu, Y.; Li, S.-S.; Han, G.-Z. Insights into the origin and evolution of the plant hormone signaling machinery. Plant Physiol. 2015, 167, 872–886. [Google Scholar] [CrossRef]
- Mutte, S.K.; Kato, H.; Rothfels, C.; Melkonian, M.; Wong, G.K.-S.; Weijers, D. Origin and evolution of the nuclear auxin response system. eLife 2018, 7, e33399. [Google Scholar] [CrossRef]
- Zhao, Y. Essential roles of local auxin biosynthesis in plant development and in adaptation to environmental changes. Annu. Rev. Plant Biol. 2018, 69, 417–435. [Google Scholar] [CrossRef]
- Du, M.; Spalding, E.P.; Gray, W.M. Rapid auxin-mediated cell expansion. Annu. Rev. Plant Biol. 2020, 71, 379–402. [Google Scholar] [CrossRef] [PubMed]
- Gasser, C.S.; Skinner, D.J. Development and evolution of the unique ovules of flowering plants. Curr. Top. Dev. Biol. 2019, 131, 373–399. [Google Scholar] [CrossRef] [PubMed]
- Cucinotta, M.; Cavalleri, A.; Chandler, J.W.; Colombo, L. Auxin and Flower Development: A Blossoming Field. Cold Spring Harb. Persp. Biol. 2021, 13, a039974. [Google Scholar] [CrossRef] [PubMed]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Blakeslee, J.J.; Spatola Rossi, T.; Kriechbaumer, V. Auxin biosynthesis: Spatial regulation and adaptation to stress. J. Exp. Bot. 2019, 70, 5041–5049. [Google Scholar] [CrossRef] [PubMed]
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007, 31, 425–448. [Google Scholar] [CrossRef]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006, 20, 1790–1799. [Google Scholar] [CrossRef]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 2007, 19, 2430–2439. [Google Scholar] [CrossRef]
- Chen, Q.; Dai, X.; De Paoli, H.; Cheng, Y.; Takebayashi, Y.; Kasahara, H.; Kamiya, Y.; Zhao, Y. Auxin Overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol. 2014, 55, 1072–1079. [Google Scholar] [CrossRef]
- Hou, X.; Liu, S.; Pierri, F.; Dai, X.; Qu, L.-J.; Zhao, Y. Allelic analyses of the Arabidopsis YUC1 locus reveal residues and domains essential for the functions of YUC family of flavin monooxygenases. J. Integr. Plant Biol. 2011, 53, 54–62. [Google Scholar] [CrossRef]
- Yamaguchi, N.; Huang, J.; Tatsumi, Y.; Abe, M.; Sugano, S.S.; Kojima, M.; Takebayashi, Y.; Kiba, T.; Yokoyama, R.; Nishitani, K.; et al. Chromatin-mediated feed-forward auxin biosynthesis in floral meristem determinacy. Nat. Commun. 2018, 9, 5290. [Google Scholar] [CrossRef]
- Kriechbaumer, V.; Wang, P.; Hawes, C.; Abell, B.M. Alternative splicing of the auxin biosynthesis gene YUCCA4 determines its subcellular compartmentation: YUCCA4 and auxin biosynthesis. Plant J. 2012, 70, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Cecchetti, V.; Altamura, M.M.; Falasca, G.; Costantino, P.; Cardarelli, M. Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. Plant Cell 2008, 20, 1760–1774. [Google Scholar] [CrossRef] [PubMed]
- Cecchetti, V.; Celebrin, D.; Napoli, N.; Ghelli, R.; Brunetti, P.; Costantino, P.; Cardarelli, M. An auxin maximum in the middle layer controls stamen development and pollen maturation in Arabidopsis. New Phytol. 2017, 213, 1194–1207. [Google Scholar] [CrossRef] [PubMed]
- Cabot, C.; Sibole, J.V.; Barceló, J.; Poschenrieder, C. Luxury zinc supply acts as antiaging agent and enhances reproductive fitness in Arabidopsis thaliana. Plant Sci. 2021, 304, 110805. [Google Scholar] [CrossRef] [PubMed]
- Medberry, S.L.; Lockhart, B.E.; Olszewski, N.A. The Commelina yellow mottle virus promoter is a strong promoter in vascular and reproductive tissues. Plant Cell 1992, 4, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Mascarenhas, J.; Hamilton, D.A. Artifacts in the localization of GUS activity in anthers of petuniatransformedwithaCaMV35s-GUS construct. Plant J. 1992, 2, n405–n408. [Google Scholar] [CrossRef]
- Mesa, M.C.D.; Santiago-Domenech, N.; Pliego-Alfaro, F.; Quesada, M.A.; Mecado, J.A. The CaMV 35S promoter is highly active on floral organs and pollen of transgenic strawberry plants. Plant Cell Rep. 2004, 23, 32–38. [Google Scholar] [CrossRef]
- Wilkinson, J.E.; Twell, D.; Lindsey, K. Activities of CaMV 35S and nos promoters in pollen: Implications for field release of transgenic plants. J. Exp. Bot. 1997, 48, 265–275. [Google Scholar] [CrossRef]
- Sunilkumar, G.; Mohr, L.; Lopata-Finch, E.; Emani, C.; Rathore, K.S. Developmental and tissue-specific expression of CaMV 35S promoter in cotton as revealed by GFP. Plant Mol. Biol. 2002, 50, 463–479. [Google Scholar] [CrossRef] [PubMed]
- Iwano, M.; Shiba, H.; Funato, M.; Shimosato, H.; Takayama, S.; Isogai, A. Immunohistochemical studies on translocation of pollen S-haplotype determinant in self-incompatibility of Brassica rapa. Plant Cell Physiol. 2003, 44, 428–436. [Google Scholar] [CrossRef]
- Squires, J.; Stephens, J.; Shoelz, J.E.; Palukaitis, P. Assessment of CaMV-mediated gene silencing and integration of CaMV into GM plants with a 35S RNA promoter. Environ. Biosafety Res. 2007, 6, 259–270. [Google Scholar] [CrossRef]
- Suzuki, M.; Yamazaki, C.; Mitsui, M.; Kakei, Y.; Mitani, Y.; Nakamura, A.; Ishii, T.; Soeno, K.; Shimada, Y. Transcriptional feedback regulation of YUCCA genes in response to auxin levels in Arabidopsis. Plant Cell Rep. 2015, 34, 1343–1352. [Google Scholar] [CrossRef]
- Yao, X.; Tian, L.; Yang, J.; Zhao, Y.-N.; Zhu, Y.-X.; Dai, X.; Zhao, Y.; Yang, Z.-N. Auxin production in diploid microsporocytes is necessary and sufficient for early stages of pollen development. PLoS Genet. 2018, 14, e1007397. [Google Scholar] [CrossRef]
- Cao, X.; Yang, H.; Shang, C.; Ma, S.; Liu, L.; Cheng, J. The Roles of Auxin Biosynthesis YUCCA Gene Family in Plants. Int. J. Mol. Sci. 2019, 20, 6343. [Google Scholar] [CrossRef]
- Sakata, T.; Oshino, T.; Miura, S.; Tomabechi, M.; Tsunaga, Y.; Higashitani, N.; Miyazawa, Y.; Takahashi, H.; Watanabe, M.; Higashitani, A. Auxins reverse plant male sterility caused by high temperatures. Proc. Natl. Acad. Sci. USA 2010, 107, 8569–8574. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xie, W.F.; Zhang, L.; Valpuesta, V.; Ye, Z.W.; Gao, Q.H.; Duan, K. Auxin biosynthesis by the YUCCA6 flavin monooxygenase gene in Woodland Strawberry. J. Integr. Plant Biol. 2014, 56, 350–363. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Ma, Y.; Liu, N.; Xu, J.; Hu, Q.; Li, Y.; Wu, Y.; Xie, S.; Zhu, L.; Min, L.; et al. microRNAs involved in auxin signaling modulate male sterility under high-temperature stress in cotton (Gossypium hirsutum). Plant J. 2017, 91, 977–994. [Google Scholar] [CrossRef] [PubMed]
- Gallavotti, A.; Barazesh, S.; Malcomber, S.; Hall, D.; Jackson, D.; Schmidt, R.J.; McSteen, P. sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc. Natl. Acad. Sci. USA 2008, 105, 15196–15201. [Google Scholar] [CrossRef] [PubMed]
- Cardarelli, M.; Costantino, P. An auxin switch for male fertility. Nat. Plants 2018, 4, 408–409. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Chen, Y.; Liu, L.; See, Y.H.B.; Mao, C.; Gan, Y.; Yu, H. OsFTIP7 determines auxin-mediated anther dehiscence in rice. Nat. Plants 2018, 4, 495–504. [Google Scholar] [CrossRef]
- Smyth, D.R.; Bowman, J.L.; Meyerowitz, E.M. Early flower development in Arabidopsis. Plant Cell 1990, 2, 755–767. [Google Scholar] [CrossRef] [PubMed]
- Gómez, J.F.; Talle, B.; Wilson, Z.A. Anther and pollen development: A conserved developmental pathway: Conservation of anther and pollen development. J. Integr. Plant Biol. 2015, 57, 876–891. [Google Scholar] [CrossRef]
- Yuan, S.; Barrett, S.C.H.; Daun, T.; Qian, X.; Shi, M.; Zhang, D. Ecological correlates and genetic consequences of evolutionary transitions from distyly to homostyly. Ann. Bot. 2017, 120, 775–789. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Li, A.; Zhang, D. Cryptic self-incompatibility and distyly in Hedyotis acutangula Champ. (Rubiaceae). Plant Biol. 2010, 12, 484–494. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Barrales, R.; Vargas, P.; Arroyo, J. New evidence for the Darwinian hypothesis of heterostyly: Breeding systems and pollinators in Narcissus sect. Apodanthi. New Phytol. 2006, 171, 553–567. [Google Scholar] [CrossRef]
- Nakamura, K.; Denda, T.; Kameshima, O.; Yokota, M. Breakdown of distyly in a tetraploid variety of Ophiorrhiza japonica (Rubiaceae) and its phylogenetic analysis. J. Plant Res. 2007, 120, 501–509. [Google Scholar] [CrossRef]
- Hu, X.; Liao, Z.; Zhang, B.; Yue, J.; Wang, Z.; Jie, X.; Liu, J. Transcriptome sequencing and screening of genes related to sex determination of Trichosanthes kirilowii Maxim. PLoS ONE 2020, 15, e0239230. [Google Scholar] [CrossRef]
- Devani, R.S.; Sinha, S.; Banerjee, J.; Sinha, R.K.; Bendahmane, A.; Banerjee, A.K. De novo transcriptome assembly from flower buds of dioecious, gynomonoecious and chemically masculinized female Coccinia grandis reveals genes associated with sex expression and modification. BMC Plant Biol. 2017, 17, 241. [Google Scholar] [CrossRef] [PubMed]
- West, N. A Mechanism for Sex Determination in Dioecious Cultivated Spinach. Ph.D. Thesis, Wayne State University, Detroit, MI, USA, 2020. [Google Scholar]
- Harkess, A.; Mercati, F.; Shan, H.; Sunseri, F.; Falavigna, A.; Leebens-Mack, J. Sex-biased gene expression in dioecious garden asparagus ( Asparagus officinalis). New Phytol. 2015, 207, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Golenberg, E.M.; West, N.W. Hormonal interactions and gene regulation can link monoecy and environmental plasticity to the evolution of dioecy in plants. Am. J. Bot. 2013, 100, 1022–1037. [Google Scholar] [CrossRef]
- Zhang, X.; Jia, Y.; Liu, Y.; Chen, D.; Luo, Y.; Niu, S. Challenges and perspectives in the study of self-incompatibility in Orchids. Int. J. Mol. Sci. 2021, 22, 12901. [Google Scholar] [CrossRef]
- Hasenstein, K.H.; Zavada, M.S. Auxin modification of the incompatibility response in Theobroma cacao. Physiol. Plant. 2001, 112, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Eklund, D.M.; Ishizaki, K.; Flores-Sandoval, E.; Kikuchi, S.; Takebayashi, Y.; Tsukamoto, S.; Hirakawa, Y.; Nonomura, M.; Kato, H.; Kouno, M.; et al. Auxin Produced by the indole-3-pyruvic acid pathway regulates development and gemmae dormancy in the liverwort Marchantia polymorpha. Plant Cell 2015, 27, 1650–1669. [Google Scholar] [CrossRef] [PubMed]
- Ganders, F.R. The biology of heterostyly. N. Z. J. Bot. 1979, 17, 607–635. [Google Scholar] [CrossRef]
- Chafe, P.D.J.; Lee, T.; Shore, J.S. Development of a genetic transformation system for distylous Turnera joelii (Passifloraceae) and characterization of a self-compatible mutant. Plant Cell Tissue Organ Cult. 2015, 120, 507–517. [Google Scholar] [CrossRef]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; Dezonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Henning, P.M.; Shore, J.S.; McCubbin, A.G. The S-Gene YUC6 Pleiotropically Determines Male Mating Type and Pollen Size in Heterostylous Turnera (Passifloraceae): A Novel Neofunctionalization of the YUCCA Gene Family. Plants 2022, 11, 2640. https://doi.org/10.3390/plants11192640
Henning PM, Shore JS, McCubbin AG. The S-Gene YUC6 Pleiotropically Determines Male Mating Type and Pollen Size in Heterostylous Turnera (Passifloraceae): A Novel Neofunctionalization of the YUCCA Gene Family. Plants. 2022; 11(19):2640. https://doi.org/10.3390/plants11192640
Chicago/Turabian StyleHenning, Paige M., Joel S. Shore, and Andrew G. McCubbin. 2022. "The S-Gene YUC6 Pleiotropically Determines Male Mating Type and Pollen Size in Heterostylous Turnera (Passifloraceae): A Novel Neofunctionalization of the YUCCA Gene Family" Plants 11, no. 19: 2640. https://doi.org/10.3390/plants11192640
APA StyleHenning, P. M., Shore, J. S., & McCubbin, A. G. (2022). The S-Gene YUC6 Pleiotropically Determines Male Mating Type and Pollen Size in Heterostylous Turnera (Passifloraceae): A Novel Neofunctionalization of the YUCCA Gene Family. Plants, 11(19), 2640. https://doi.org/10.3390/plants11192640







