Modelling the Rhizosphere Priming Effect in Combination with Soil Food Webs to Quantify Interaction between Living Plant, Soil Biota and Soil Organic Matter
Abstract
1. Introduction
2. Results
2.1. Model Description
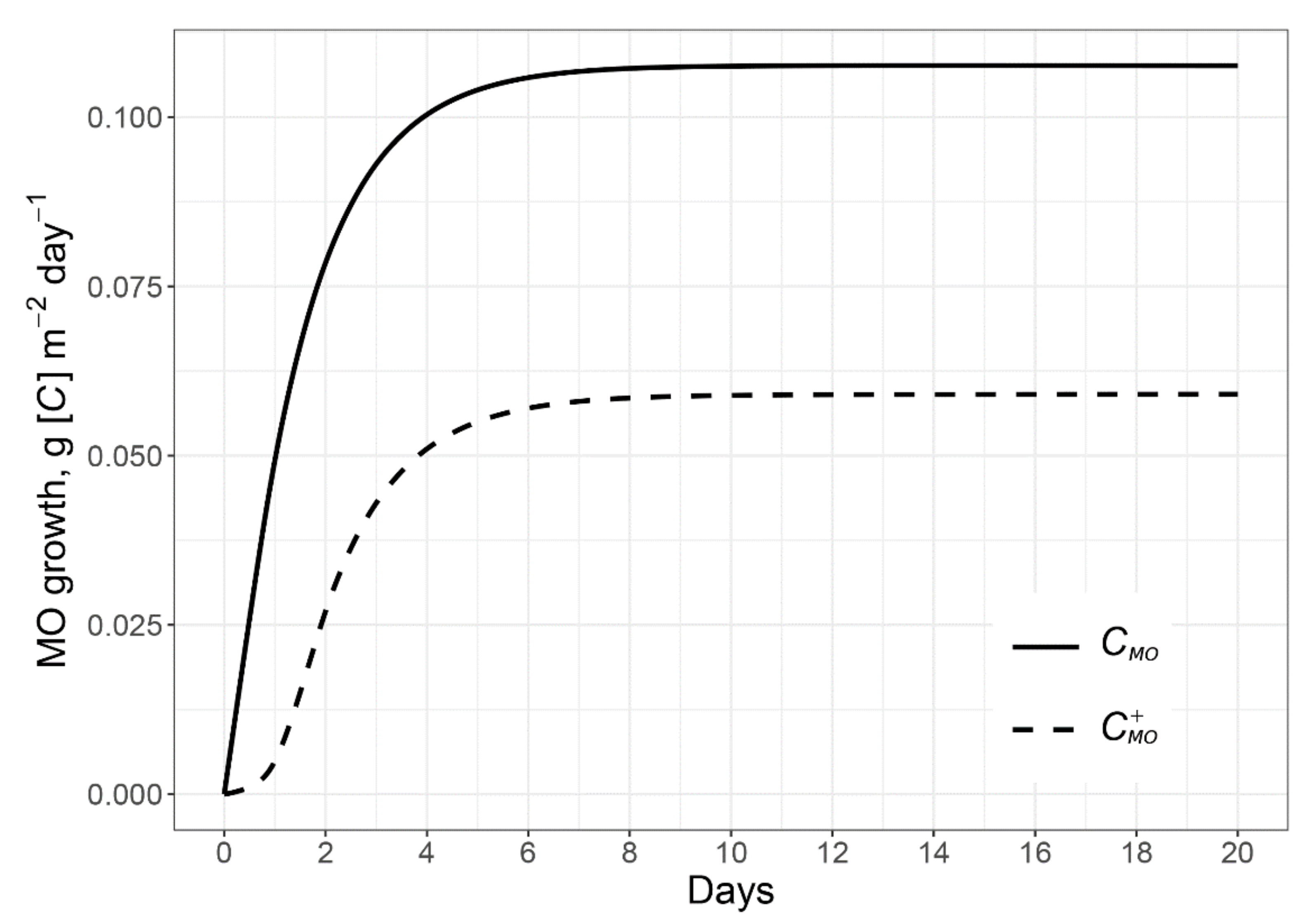
2.1.1. Microbial Growth Caused by the C and N of Root Exudates
2.1.2. Microbial Growth Due to Using Excessive C of Exudates and Mined N from the Rhizosphere SOM
2.1.3. Food Web Processes of the Soil Faunal By-Products and Available N Formation
2.2. Model Verification
2.3. Analysis of Model Sensitivity to Parameter Variations
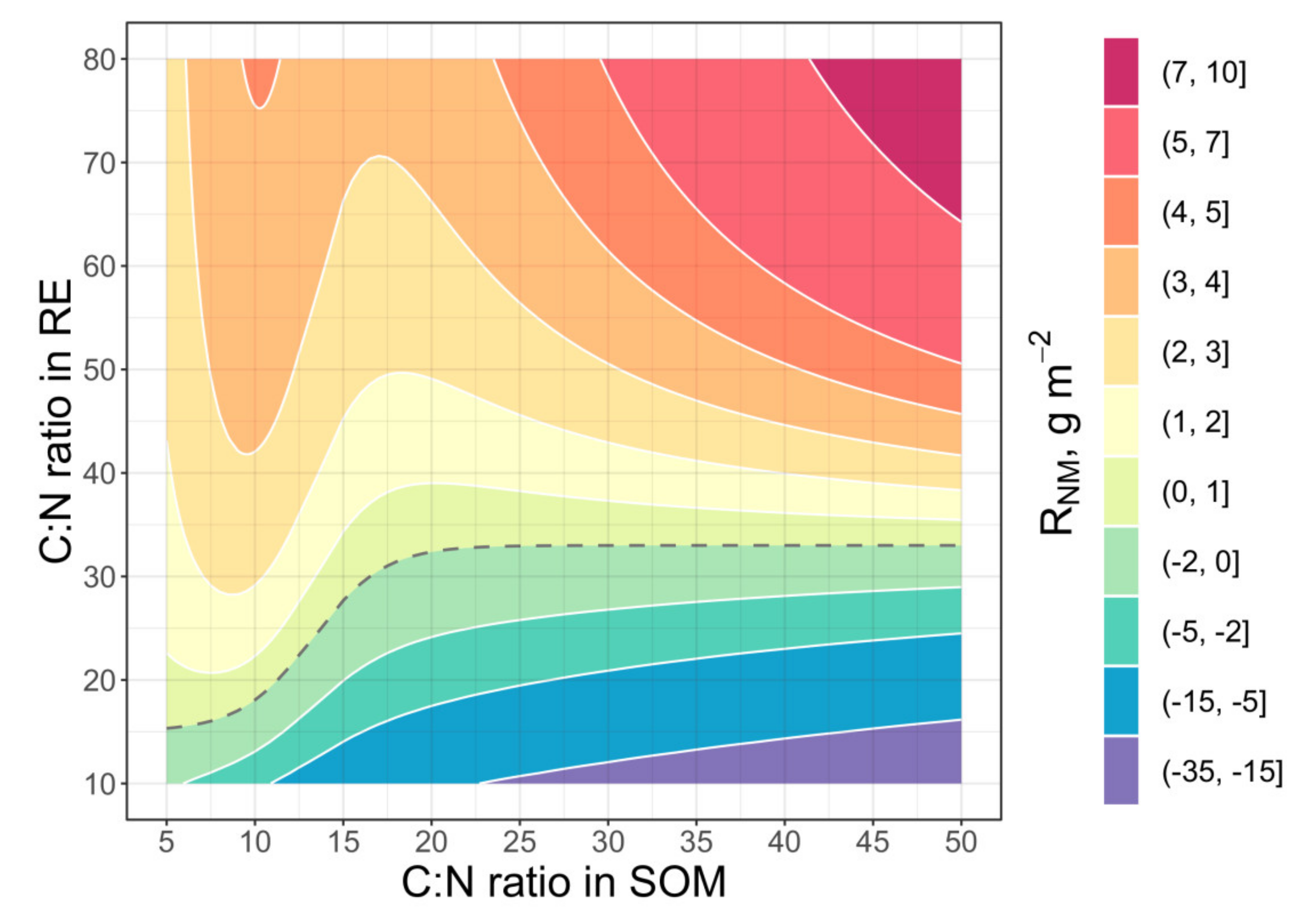
2.4. Model Testing at the Level of Rhizosphere Soil
2.5. Model Testing at the Level of Whole Soil Horizon
3. Discussion
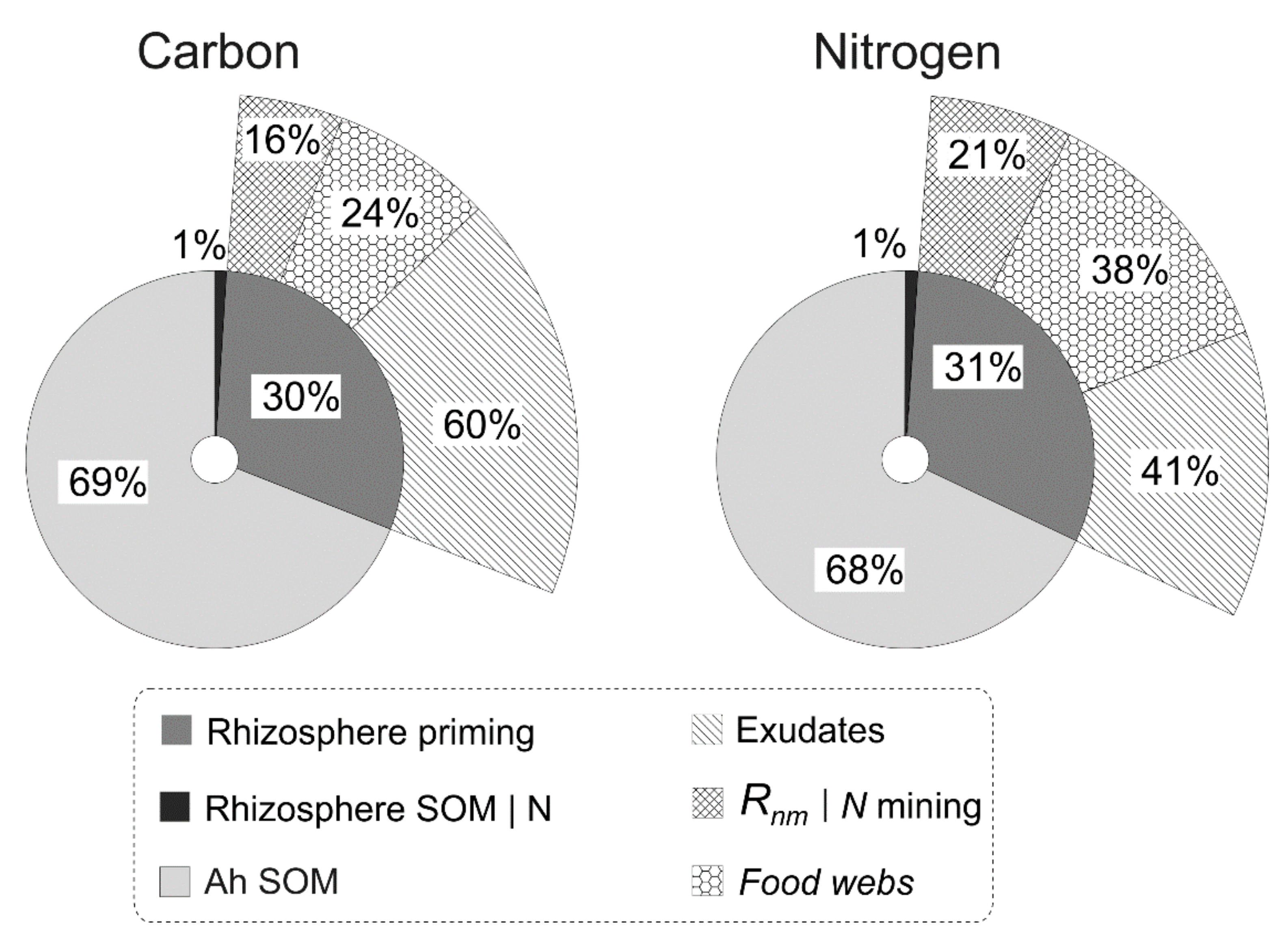
3.1. Priming Effect, N Mining and Food Webs as Processes of Fast Cycles of C and N in the Rhizosphere Soil
3.2. Experimental and Simulated Data: Plant-Microorganism-Soil Fauna Interactions in the Rhizosphere Soil
3.3. The Model Uncertainties and Future Development
4. Materials and Methods
4.1. Model Verification
4.2. Sensitivity Analysis
4.3. Model Testing
4.3.1. Rhizosphere Soil Level
4.3.2. The Level of a Whole Soil Horizon
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cheng, W.; Parton, W.J.; Gonzalez-Meler, M.A.; Phillips, R.; Asao, S.; McNickle, G.G.; Brzostek, E.; Jastrow, J.D. Synthesis and modelling perspectives of rhizosphere priming. New Phytol. 2014, 201, 31–56. [Google Scholar] [CrossRef] [PubMed]
- Huo, C.F.; Luo, Y.Q.; Cheng, W.X. Rhizosphere priming effect: A meta-analysis. Soil Biol. Biochem. 2017, 111, 78–84. [Google Scholar] [CrossRef]
- Kuzyakov, Y. Factors affecting rhizosphere priming effects. J. Plant Nutr. Soil Sci. 2002, 165, 382–396. [Google Scholar] [CrossRef]
- Qiao, N.; Xu, X.; Hu, Y.; Blagodatskaya, E.; Liu, Y.; Schaefer, D.; Kuzyakov, Y. Carbon and nitrogen additions induce distinct priming effects along an organic-matter decay continuum. Sci. Rep. 2016, 6, 19865. [Google Scholar] [CrossRef] [PubMed]
- Zhuravleva, A.I.; Alifanov, V.M.; Blagodatskaya, E.V. Effect of contrasting trophic conditions on the priming effect in gray forest soils. Eurasian Soil Sci. 2018, 51, 204–210. [Google Scholar] [CrossRef]
- Schimel, J.P.; Weintraub, M.N. The implications of exoenzyme activity on microbial carbon and nitrogen limitation in soil: A theoretical model. Soil Biol. Biochem. 2003, 35, 549–563. [Google Scholar] [CrossRef]
- Weintraub, M.N.; Scott-Denton, L.E.; Schmidt, S.K.; Monson, R.K. The effects of tree rhizodeposition on soil exoenzyme activity, dissolved organic carbon, and nutrient availability in a subalpine forest ecosystem. Oecologia 2007, 154, 327–338. [Google Scholar] [CrossRef]
- Sinsabaugh, R.L.; Belnap, J.; Findlay, S.G.; Shah, J.J.F.; Hill, B.H.; Kuehn, K.A.; Kuske, C.R.; Litvak, M.E.; Martinez, N.G.; Moorhead, D.L.; et al. Extracellular enzyme kinetics scale with resource availability. Biogeochemistry 2014, 121, 287–304. [Google Scholar] [CrossRef]
- Stock, S.C.; Köster, M.; Dippold, M.A.; Nájera, F.; Matus, F.; Merino, C.; Boyd, J.; Spielvogel, S.; Gorbushina, A.; Kuzyakov, Y. Environmental drivers and stoichiometric constraints on enzyme activities in soils from rhizosphere to continental scale. Geoderma 2019, 337, 973–982. [Google Scholar] [CrossRef]
- Blagodatsky, S.; Blagodatskaya, E.; Yuyukina, T.; Kuzyakov, Y. Model of apparent and real priming effects: Linking microbial activity with soil organic matter decomposition. Soil Biol. Biochem. 2010, 42, 1275–1283. [Google Scholar] [CrossRef]
- Bastida, F.; García, C.; Fierer, N.; Eldridge, D.J.; Bowker, M.A.; Abades, S.; Alfaro, F.D.; Berhe, A.A.; Cutler, N.A.; Gallardo, A.; et al. Global ecological predictors of the soil priming effect. Nat. Commun. 2019, 10, 3481. [Google Scholar] [CrossRef] [PubMed]
- Terrer, C.; Phillips, R.P.; Hungate, B.A.; Rosende, J.; Pett-Ridge, J.; Craig, M.E.; van Groenigen, K.J.; Keenan, T.F.; Sulman, B.N.; Stocker, B.D.; et al. A trade-off between plant and soil carbon storage under elevated CO2. Nature 2021, 591, 599–603. [Google Scholar] [CrossRef] [PubMed]
- Campbell, E.E.; Paustian, K. Current developments in soil organic matter modeling and the expansion of model applications: A review. Environ. Res. Lett. 2015, 10, 123004. [Google Scholar] [CrossRef]
- Dennis, P.G.; Miller, A.J.; Hirsch, P.R. Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol. Ecol. 2010, 72, 313–327. [Google Scholar] [CrossRef] [PubMed]
- Haichar, F.E.Z.; Marol, C.; Berge, O.; Rangel-Castro, J.I.; Prosser, J.I.; Balesdent, J.; Heulin, T.; Achouak, W. Plant host habitat and root exudates shape soil bacterial community structure. ISME J. 2008, 2, 1221–1230. [Google Scholar] [CrossRef]
- Lynch, J.M.; Whipps, J.M. Substrate flow in the rhizosphere. Plant Soil 1990, 129, 1–10. [Google Scholar] [CrossRef]
- Grayston, S.J.; Vaughan, D.; Jones, D. Rhizosphere carbon flow in trees, in comparison with annual plants: The importance of root exudation and its impact on microbial activity and nutrient availability. Appl. Soil Ecol. 1996, 5, 29–56. [Google Scholar] [CrossRef]
- Jones, D.L.; Nguyen, C.; Finlay, R.D. Carbon flow in the rhizosphere: Carbon trading at the soil–root interface. Plant Soil 2009, 321, 5–33. [Google Scholar] [CrossRef]
- Phillips, R.P.; Erlitz, Y.; Bier, R.; Bernhardt, E.S. New approach for capturing soluble root exudates in forest soils. Funct. Ecol. 2008, 22, 990–999. [Google Scholar] [CrossRef]
- Bengtson, P.; Barker, J.; Grayston, S.J. Evidence of a strong coupling between root exudation, C and N availability, and stimulated SOM decomposition caused by rhizosphere priming effects. Ecol. Evol. 2012, 2, 1843–1852. [Google Scholar] [CrossRef]
- Dijkstra, F.A.; Cheng, W. Interactions between soil and tree roots accelerate long-term soil carbon decomposition. Ecol. Lett. 2007, 10, 1046–1053. [Google Scholar] [CrossRef] [PubMed]
- Sommer, J.; Dippold, M.A.; Flessa, H.; Kuzyakov, Y. Allocation and dynamics of C and N within plant-soil system of ash and beech. J. Plant Nutr. Soil Sci. 2016, 179, 376–387. [Google Scholar] [CrossRef]
- Bais, H.P.; Weir, L.P.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant. Biol. 2006, 57, 233–266. [Google Scholar] [CrossRef] [PubMed]
- Guyonnet, J.P.; Gullemet, M.; Dubost, A.; Simon, L.; Ortet, P.; Barakat, M.; Heulin, T.; Achouk, W.; Haichar, F.E.Z. Plant nutrient resource use strategies shape active rhizosphere micribiota through root exudation. Front. Plant Sci. 2018, 9, 1662. [Google Scholar] [CrossRef] [PubMed]
- Blagodatskaya, E.; Kuzyakov, Y. Mechanisms of real and apparent priming effects and their dependence on soil microbial biomass and community structure: Critical review. Biol. Fert. Soils 2008, 45, 115–131. [Google Scholar] [CrossRef]
- Fontaine, S.; Mariotti, A.; Abbadie, L. The priming effect of organic matter: A question of microbial competition? Soil Biol. Biochem. 2003, 35, 837–843. [Google Scholar] [CrossRef]
- Mason-Jones, K.; Kuzyakov, Y. “Non-metabolizable” glucose analogue shines new light on Priming mechanisms: Triggering of microbial metabolism. Soil Biol. Biochem. 2017, 107, 68–76. [Google Scholar] [CrossRef]
- Wild, B.; Lid, J.; Pihlblad, J.; Bengtsond, P.; Rütting, T. Decoupling of priming and microbial N mining during a short-term soil incubation. Soil Biol. Biochem. 2019, 129, 71–79. [Google Scholar] [CrossRef]
- Fontaine, S.; Henault, C.; Aamor, A.; Bdioui, N.; Bloor, J.M.G.; Maire, V.; Mary, B.; Revaillot, S.; Maron, P.A. Fungi mediate long term sequestration of carbon and nitrogen in soil through their priming effect. Soil Biol. Biochem. 2011, 43, 86–96. [Google Scholar] [CrossRef]
- Guenet, B.; Danger, M.; Abbadie, L.; Lacroix, G. Priming effect: Bridging the gap between terrestrial and aquatic ecology. Ecology 2010, 91, 2850–2861. [Google Scholar] [CrossRef]
- Dijkstra, F.A.; Zhu, B.; Cheng, W. Root effects on soil organic carbon: A double-edged sword. New Phytol. 2021, 230, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Perveen, N.; Barot, S.; Alvarez, G.; Klumpp, K.; Martin, R.; Rapaport, A.; Herfurth, D.; Louault, F.; Fontaine, S. Priming effect and microbial diversity in ecosystem functioning and response to global change: A modeling approach using the SYMPHONY model. Glob. Change Biol. 2014, 20, 1174–1190. [Google Scholar] [CrossRef] [PubMed]
- Parnas, H.A. A theoretical explanation of the priming effect based on microbial growth with two limiting substrates. Soil Biol. Biochem. 1976, 8, 139–144. [Google Scholar] [CrossRef]
- Molina, J.A.E.; Hadas, A.; Clapp, C.E. Computer simulation of nitrogen turnover in soil and priming effect. Soil Biol. Biochem. 1990, 22, 349–353. [Google Scholar] [CrossRef]
- Fontaine, S.; Barot, S. Size and functional diversity of microbe populations control plant persistence and long-term soil carbon accumulation. Ecol. Lett. 2005, 8, 1075–1087. [Google Scholar] [CrossRef]
- Huang, Y.; Guenet, B.; Ciais, P.; Janssens, I.A.; Soong, J.L.; Wang, Y.; Goll, D.; Blagodatskaya, E.; Huang, Y. ORCHIMIC (v1.0), A microbe-mediated model for soil organic matter decomposition. Geosci. Model Dev. 2018, 11, 2111–2138. [Google Scholar] [CrossRef]
- Wutzler, T.; Reichstein, M. Priming and substrate quality interactions in soil organic matter models. Biogeosciences 2013, 10, 2089–2103. [Google Scholar] [CrossRef]
- Savage, K.E.; Parton, W.J.; Davidson, E.A.; Trumbore, S.E.; Frey, S.D. Long-term changes in forest carbon under temperature and nitrogen amendments in a temperate northern hardwood forest. Glob. Change Biol. 2013, 19, 2389–2400. [Google Scholar] [CrossRef]
- Raynaud, X.; Jaillard, B.; Leadley, P.W. Plants may alter competition by modifying nutrient bioavailability in rhizosphere: Amodeling approach. Am. Nat. 2008, 171, 44–58. [Google Scholar] [CrossRef]
- Valadares, R.V.; Neves, J.C.L.; Costa, M.D.; Smethurst, P.J.; Peternelli, L.A.; Jesus, G.L.; Cantarutti, R.B.; Silva, I.R. Modeling rhizosphere carbon and nitrogen cycling in Eucalyptus plantation soil. Biogeosciences 2018, 15, 4943–4954. [Google Scholar] [CrossRef]
- Finzi, A.C.; Abramoff, R.Z.; Spiller, K.S.; Brzostek, E.R.; Darby, B.A.; Kramer, M.A.; Philips, R.P. Rhizosphere processes are quantitatively important components of terrestrial carbon and nutrient cycles. Glob. Change Biol. 2015, 21, 2082–2094. [Google Scholar] [CrossRef] [PubMed]
- Allison, S.D.; Wallenstein, M.D.; Bradford, M.A. Soil-carbon response to warming dependent on microbial physiology. Nat. Geosci. 2010, 3, 336–340. [Google Scholar] [CrossRef]
- Drake, J.; Darby, B.; Giasson, M.-A.; Kramer, M.; Phillips, R.; Finzi, A. Stoichiometry constrains microbial response to root exudation-insights from a model and a field experiment in a temperate forest. Biogeosciences 2013, 10, 821–838. [Google Scholar] [CrossRef]
- Vitousek, P.M.; Howarth, R.W. Nitrogen limitation on land and in the sea: How can it occur? Biogeochemistry 1991, 13, 87–115. [Google Scholar] [CrossRef]
- Vance, E.D.; Chapin III, F.S. Substrate limitations to microbial activity in taiga forest floors. Soil Biol. Biochem. 2001, 33, 173–188. [Google Scholar] [CrossRef]
- Komarov, A.; Chertov, O.; Bykhovets, S.; Shaw, C.; Nadporozhskaya, M.; Frolov, P.; Shashkov, M.; Shanin, V.; Grabarnik, P.; Priputina, I.; et al. Romul_Hum model of soil organic matter formation coupled with soil biota activity. I. Problem formulation, model description, and testing. Ecol. Model. 2017, 345, 113–124. [Google Scholar] [CrossRef]
- De Ruiter, P.C.; Van Veen, J.A.; Moore, J.C.; Brussaard, L.; Hunt, H.W. Calculation of nitrogen mineralization in soil food webs. Plant Soil 1993, 157, 263–273. [Google Scholar] [CrossRef]
- Holtkamp, R.; van der Wal, A.; Kardol, P.; van der Putten, W.H.; de Ruiter, P.C.; Dekker, S.C. Modelling C and N mineralisation in soil food webs during secondary succession on ex-arable land. Soil Biol. Biochem. 2011, 43, 251–260. [Google Scholar] [CrossRef]
- Chertov, O.; Komarov, A.; Shaw, C.; Bykhovets, S.; Frolov, P.; Shanin, V.; Grabarnik, P.; Priputina, I.; Zubkova, E.; Shashkov, M. Romul_Hum—A model of soil organic matter formation coupling with soil biota activity. II. Parameterisation of the soil food web biota activity. Ecol. Model. 2017, 345, 125–139. [Google Scholar] [CrossRef]
- Sándor, R.; Ehrhardt, F.; Grace, P.; Recous, S.; Smith, P.; Snow, V.; Soussana, J.-F.; Basso, B.; Bhatia, A.; Brilli, L.; et al. Ensemble modelling of carbon fluxes in grasslands and croplands. Field Crop. Res. 2020, 252, 107791. [Google Scholar] [CrossRef]
- Liu, Y.; Ge, T.; Ye, J.; Liu, S.; Shibistova, O.; Wang, P.; Wang, J.; Li, Y.; Guggenberger, G.; Kuzyakov, Y.; et al. Initial utilization of rhizodeposits with rice growth in paddy soils: Rhizosphere and N fertilization effects. Geoderma 2019, 338, 30–39. [Google Scholar] [CrossRef]
- Owen, A.G.; Jones, D.L. Competition for amino acids between wheat roots and rhizosphere microorganisms and the role of amino acids in plant N acquisition. Soil Biol. Biochem. 2001, 33, 651–657. [Google Scholar] [CrossRef]
- Chen, R.; Senbayram, M.; Blagodatsky, S.; Myachina, O.; Dittert, K.; Lin, X.; Blagodatskaya, E.; Kuzyakov, Y. Soil C and N availability determine the priming effect: Microbial N mining and stoichiometry theories. Glob. Change Biol. 2014, 20, 2356–2367. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Seven, J.; Zilla, T.; Dippold, M.A.; Blagodatskaya, E.; Kuzyakov, Y. Microbial C:N:P stoichiometry and turnover depend on nutrient availability in soil: A 14C, 15N and 33P triple labelling study. Soil Biol. Biochem. 2019, 131, 206–216. [Google Scholar] [CrossRef]
- Cui, J.; Zhu, Z.; Xu, X.; Liu, S.; Jones, D.L.; Kuzyakov, Y.; Wu, J.; Ge, T. Carbon and nitrogen recycling from microbial necromass to cope with C:N stoichiometric imbalance by priming. Soil Biol. Biochem. 2020, 142, 107720. [Google Scholar] [CrossRef]
- Clarholm, M. Interactions of bacteria, protozoa and plants leading to mineralization of soil nitrogen. Soil Biol. Biochem. 1985, 17, 181–187. [Google Scholar] [CrossRef]
- Clarholm, M. Effects of plant-bacterial-amoebal interactions on plant uptake of nitrogen under field conditions. Biol. Fert. Soils 1989, 8, 373–378. [Google Scholar] [CrossRef]
- Chertov, O.G.; Gryazkin, A.V.; Smirnov, A.P.; Kovalev, N.V. Change of carbon balance and biological productivity of the forest stands at different managing regimes [Izmenenie balansa ugleroda i biologicheskoj produktivnosti lesnogo massiva pri raznyh rezhimah hozjajstvovanija. Izvestia Sankt-Peterburgskoj Lesotehniceskoj Akademii 2011, 197, 263–272. (In Russian) [Google Scholar]
- Komarov, A.; Chertov, O.; Zudin, S.; Nadporozhskaya, M.; Mikhailov, A.; Bykhovets, S.; Zudina, E.; Zoubkova, E. EFIMOD 2—A model of growth and elements cycling in boreal forest ecosystems. Ecol. Model. 2003, 170, 373–392. [Google Scholar] [CrossRef]
- Shanin, V.N.; Grabarnik, P.Y.; Bykhovets, S.S.; Chertov, O.G.; Priputina, I.V.; Shashkov, M.P.; Ivanova, N.V.; Stamenov, M.N.; Frolov, P.V.; Zubkova, E.V.; et al. Parameterization of productivity model for the most common trees species in European part of Russia for simulation of forest ecosystem dynamics [Parametrizacija modeli produkcionnogo processa dlja dominirujushhih vidov derev’ev Evropejskoj chasti RF v zadachah modelirovanija dinamiki lesnyh jekosistem. Math. Biol. Bioinformat. 2019, 14, 54–76. (In Russian) [Google Scholar] [CrossRef]
- De Vries, F.T.; Thébault, E.; Liiri, M.; Birkhofer, K.; Tsiafouli, M.A.; Bjørnlund, L.; Jørgensen, H.B.; Brady, M.V.; Christensen, S.; de Ruiter, P.C.; et al. Soil food web properties explain ecosystem services across European land use systems. Proc. Natl. Acad. Sci. USA 2013, 110, 14296–14301. [Google Scholar] [CrossRef] [PubMed]
- Kuzyakov, Y.; Hill, P.W.; Jones, D.L. Root exudate components change litter decomposition in a simulated rhizosphere depending on temperature. Plant Soil 2007, 290, 293–305. [Google Scholar] [CrossRef]
- Wichern, F.; Eberhardt, E.; Mayer, J.; Joergensen, R.G.; Muller, T. Nitrogen rhizodeposition in agricultural crops: Methods, estimates and future prospects. Soil Biol. Biochem. 2008, 40, 30–48. [Google Scholar] [CrossRef]
- Dijkstra, P.; Salpas, E.; Fairbanks, D.; Miller, E.B.; Hagerty, S.B.; van Groenigen, K.J.; Hungate, B.A.; Marks, J.C.; Koch, G.W.; Schwartz, E. High carbon use efficiency in soil microbial communities is related to balanced growth, not storage compound synthesis. Soil Biol. Biochem. 2015, 89, 35–43. [Google Scholar] [CrossRef]
- Geyer, K.M.; Dijkstra, P.; Sinsabaugh, R.; Frey, S.D. Clarifying the interpretation of carbon use efficiency in soil through methods comparison. Soil Biol. Biochem. 2019, 128, 79–88. [Google Scholar] [CrossRef]
- Manzoni, S.; Čapek, P.; Mooshammer, M.; Lindahl, B.D.; Richter, A.; Ṡantrủčková, H. Optimal metabolic regulation along resource stoichiometry gradients. Ecol. Lett. 2017, 20, 1182–1191. [Google Scholar] [CrossRef]
- Qiao, N.; Schaefer, D.; Blagodatskaya, E.; Zou, X.; Xu, X.; Kuzyakov, Y. Labile carbon retention compensates for CO2 released by priming in forest soils. Glob. Change Biol. 2014, 20, 1943–1954. [Google Scholar] [CrossRef]
- Kästner, M.; Miltner, A.; Thiele-Bruhn, S.; Liang, C. Microbial necromass in soils—Linking microbes to soil processes and carbon turnover. Front. Environ. Sci. 2021, 9, 756378. [Google Scholar] [CrossRef]
- Coleman, D.S. The microbial loop concept as used in terrestrial soil studies. Microb. Ecol. 1994, 28, 245–250. [Google Scholar] [CrossRef]
- Titlianova, A.A. Fast cycles of carbon and nitrogen turnover. In Mathematical Modelling in Ecology, Proceedings of the Second National Conference, Pushchino, Russia, 23–27 May 2011; Wiley Online Library: Hoboken, NJ, USA, 2011; pp. 269–271. (In Russian) [Google Scholar]
- Blagodatskaya, E.; Blagodatsky, S.; Dorodnikov, M.; Kuzyakov, Y. Elevated atmospheric CO2 increases microbial growth rates in soil: Results of three CO2 enrichment experiments. Glob. Change Biol. 2010, 16, 836–848. [Google Scholar] [CrossRef]
- Blagodatskaya, E.; Blagodatsky, S.; Anderson, T.H.; Kuzyakov, Y. Microbial growth and carbon use efficiency in the rhizosphere and root-free soil. PLoS ONE 2014, 9, e93282. [Google Scholar] [CrossRef] [PubMed]
- Cleveland, C.C.; Liptzin, D. C:N:P stoichiometry in soil: Is there a “Redfield ratio” for the microbial biomass? Biogeochemistry 2007, 85, 235–252. [Google Scholar] [CrossRef]
- Sterner, R.W.; Elser, J.J. Ecological Stoichiometry: The Biology of Elements from Molecules to the Biosphere; Princeton University Press: Princeton, NJ, USA, 2002; 464p. [Google Scholar]
- Clarholm, M. Soil protozoa: An under-researched microbial group gaining momentum. Soil Biol. Biochem. 2005, 37, 811–817. [Google Scholar] [CrossRef]
- Abramoff, R.; Xu, X.; Hartman, M.; O’Brien, S.; Feng, W.; Davidson, E.; Finzi, A.; Moorhead, D.; Schimel, J.; Torn, M.; et al. The Millennial model: In search of measurable pools and transformations for modeling soil carbon in the new century. Biogeochemistry 2018, 137, 51–71. [Google Scholar] [CrossRef]
- Berardi, D.; Brzostek, E.; Blanc-Betes, E.; Davison, B.; DeLucia, E.; Hartman, M.D.; Kent, J.; Parton, W.J.; Saha, D.; Hudiburg, T.W. 21st-century biogeochemical modeling: Challenges for Century-based models and where do we go from here? GCB-Bioenergy 2020, 12, 774–788. [Google Scholar] [CrossRef]
- Manzoni, S.; Taylor, P.; Richter, A.; Porporato, A.; Ågren, G. Environmental and stoichiometric controls on microbial carbon-use efficiency in soils. New Phytol. 2012, 196, 79–91. [Google Scholar] [CrossRef]
- Van Nuland, M.E.; Wooliver, R.C.; Pfennigwerth, A.A.; Read, Q.D.; Ware, I.M.; Mueller, L.; Fordyce, J.A.; Schweitzer, J.A.; Bailey, J.K. Plant–soil feedbacks: Connecting ecosystem ecology and evolution. Funct. Ecol. 2016, 30, 1032–1042. [Google Scholar] [CrossRef]
- Rutten, G.; Gómez-Aparicio, L. Plant-soil feedbacks and root responses of two Mediterranean oaks along a precipitation gradient. Plant Soil 2018, 424, 221–231. [Google Scholar] [CrossRef]
- Pena, R. Nitrogen acquisition in ectomycorrhizal symbiosis. In Molecular Mycorrhizal Symbiosis, 1st ed.; Martin, F., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2016; pp. 179–196. [Google Scholar] [CrossRef]
- Bastos, A.; Fleischer, K. Fungi are key to CO2 response of soil. Nature 2021, 591, 532–534. [Google Scholar] [CrossRef]
- Wu, L.; Xu, H.; Xiao, Q.; Huang, Y.; Suleman, M.M.; Zhu, P.; Kuzyakov, Y.; Xu, X.; Xu, M.; Zhang, W. Soil carbon balance by priming differs with single versus repeated addition of glucose and soil fertility level. Soil Biol. Biochem. 2020, 148, 107913. [Google Scholar] [CrossRef]
- He, Y.H.; Cheng, W.X.; Zhou, L.Y.; Shao, J.J.; Liu, H.Y.; Zhou, H.M.; Zhu, K.; Zhou, X.H. Soil DOC release and aggregate disruption mediate rhizosphere priming effect on soil C decomposition. Soil Biol. Biochem. 2020, 144, 107787. [Google Scholar] [CrossRef]
- Pei, J.M.; Dijkstra, F.A.; Li, J.Q.; Fang, C.M.; Su, J.H.; Zhao, J.Y.; Nie, M.; Wu, J.H. Biochar-induced reductions in the rhizosphere priming effect are weaker under elevated CO2. Soil Biol. Biochem. 2020, 142, 107700. [Google Scholar] [CrossRef]
- Zhou, J.; Zang, H.; Loeppmann, S.; Gube, M.; Kuzyakov, Y.; Pausch, J. Arbuscular mycorrhiza enhances rhizodeposition and reduces the rhizosphere priming effect on the decomposition of soil organic matter. Soil Biol. Biochem. 2020, 140, 107641. [Google Scholar] [CrossRef]
- Dijkstra, F.A.; Carrillo, Y.; Pendall, E.; Morgan, J.A. Rhizosphere priming: A nutrient perspective. Front. Microbiol. 2013, 4, 216. [Google Scholar] [CrossRef]
- Vives-Peris, V.; de Ollas, C.; Gómez-Cadenas, A.; Pérez-Clemente, R.M. Root exudates: From plant to rhizosphere and beyond. Plant Cell Rep. 2020, 39, 3–17. [Google Scholar] [CrossRef]
- Pausch, J.; Tian, J.; Riederer, M.; Kuzyakov, Y. Estimation of rhizodeposition at field scale: Upscaling of a 14C labeling study. Plant Soil 2013, 364, 273–285. [Google Scholar] [CrossRef]
- Smith, S.E.; Read, D.J. Mycorrhizal Symbiosis, 3rd ed.; Academic Press: New York, NY, USA, 2008; 800p. [Google Scholar] [CrossRef]
- Deckmyn, G.; Meyer, A.; Smits, M.M.; Ekblad, A.; Grebenc, T.; Komarov, A.; Kraigher, H. Simulating ectomycorrhizal fungi and their role in carbon and nitrogen cycling in forest ecosystems. Can. J. For. Res. 2014, 44, 535–553. [Google Scholar] [CrossRef]
- Godbold, D.L.; Hoosbeek, M.R.; Lukac, M.; Cotrufo, M.F.; Janssens, I.A.; Ceulemans, R.; Polle, A.; Velthorst, E.J.; Mugnozza, G.; De Angelis, P.; et al. Mycorrhizal hyphal turnover as a dominant process for C input into soil organic matter. Plant Soil 2006, 281, 15–24. [Google Scholar] [CrossRef]
- Murphy, C.J.; Baggs, E.M.; Morley, N.; Wall, D.P.; Paterson, E. Rhizosphere priming can promote mobilisation of N-rich compounds from soil organic matter. Soil Biol. Biochem. 2015, 81, 236–243. [Google Scholar] [CrossRef]
- Zhu, B.; Gutknecht, J.L.M.; Herman, D.J.; Keck, D.C.; Firestone, M.K.; Cheng, W.X. Rhizosphere priming effects on soil carbon and nitrogen mineralization. Soil Biol. Biochem. 2014, 76, 183–192. [Google Scholar] [CrossRef]
- Saltelli, A.; Annoni, P. How to avoid a perfunctory sensitivity analysis. Environ. Model. Softw. 2010, 25, 1508–1517. [Google Scholar] [CrossRef]




| Parameters | Measurement Units |
|---|---|
| Input variables | |
| Rhizosphere soil C | kg [C] m−2 |
| Rhizosphere soil N | kg [N] m−2 |
| Root exudate, RE, input | kg [C] m−2 day−1 |
| Root exudate C:N ratio | - |
| kg [C] m−2 | |
| Microbial grazers biomass | kg [C] m−2 |
| Arthropods biomass | kg [C] m−2 |
| Output variables | |
| Total C-CO2 emission at priming | kg [C] m−2 day−1 |
| C-CO2 emission at N mining | kg [C] m−2 day−1 |
| C-CO2 emission at rhizosphere SOM mineralisation | kg [C] m−2 day−1 |
| N available | kg [N] m−2 day−1 |
| SOC, rhizosphere soil | kg [C] m−2 |
| SON, rhizosphere soil | kg [N] m−2 |
| Parameters and Source of Data in Square Brackets | Measurement Units | Amount/Value |
|---|---|---|
| Root exudates, RE, assimilation rate by MO, KAS [3,19,51] | day−1 | 0.50 |
| Bacteria C:N ratio, CNb [48], | - | 5.0 |
| Fungi C:N ratio, CNf [48], | - | 14.0 |
| [48] | - | 10.0 |
| [48], | - | 8.0 |
| [48] | - | 0.30 * |
| Bacteria and Fungi respiration efficiency [48] | - | 0.70 |
| [3,51] | day−1 | 0.50 * |
| SOM mineralisation rate [49] | day−1 | 0.00018 |
| [36,61] | day−1 | 0.04 |
| ** | day−1 | 0.15 |
| [48] | - | 0.40 |
| [48], | - | 0.40 |
| ** | day−1 | 0.14 |
| [48] | - | 0.20 |
| [48,61] | day−1 | 0.10 |
| Parameter | Parameter Name | Standardized Coefficient of Linear Regression |
|---|---|---|
| C:N ratio of soil | CNSOM | 0.414 *** |
| C:N ratio of microorganisms (MO) | CNMO | −0.603 *** |
| C:N ratio of root exudates (RE) | CNRE | 0.181 *** |
| Efficiency of RE assimilation by microorganisms | Keff | 0.405 *** |
| Efficiency of MO assimilation by microbial grazers | KMG | −0.177 *** |
| Faunal assimilation efficiency | Kfae | 0.0534 *** |
| Intercept | ~0 | |
| R2 | 0.937 | |
| Parameters | Amount |
|---|---|
| Soil horizon Ah C pool, kg m−2 | 6.91 |
| Soil horizon Ah N pool, kg m−2 | 0.49 |
| Fine root specific length, m m−2 | 42.50 |
| Fine root diameter, mm | 1.50 |
| Fine root dry weight, kg m−2 | 0.068 |
| Diameter of rhizosphere soil tube (including root diameter), mm | 7.50 |
| Rhizosphere soil C pool, kg m−2 | 0.090 |
| Rhizosphere soil N pool, kg m−2 | 0.0064 |
| Root exudate input, kg [C] m−2 day−1 | 0.0001 … 0.0005 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chertov, O.; Kuzyakov, Y.; Priputina, I.; Frolov, P.; Shanin, V.; Grabarnik, P. Modelling the Rhizosphere Priming Effect in Combination with Soil Food Webs to Quantify Interaction between Living Plant, Soil Biota and Soil Organic Matter. Plants 2022, 11, 2605. https://doi.org/10.3390/plants11192605
Chertov O, Kuzyakov Y, Priputina I, Frolov P, Shanin V, Grabarnik P. Modelling the Rhizosphere Priming Effect in Combination with Soil Food Webs to Quantify Interaction between Living Plant, Soil Biota and Soil Organic Matter. Plants. 2022; 11(19):2605. https://doi.org/10.3390/plants11192605
Chicago/Turabian StyleChertov, Oleg, Yakov Kuzyakov, Irina Priputina, Pavel Frolov, Vladimir Shanin, and Pavel Grabarnik. 2022. "Modelling the Rhizosphere Priming Effect in Combination with Soil Food Webs to Quantify Interaction between Living Plant, Soil Biota and Soil Organic Matter" Plants 11, no. 19: 2605. https://doi.org/10.3390/plants11192605
APA StyleChertov, O., Kuzyakov, Y., Priputina, I., Frolov, P., Shanin, V., & Grabarnik, P. (2022). Modelling the Rhizosphere Priming Effect in Combination with Soil Food Webs to Quantify Interaction between Living Plant, Soil Biota and Soil Organic Matter. Plants, 11(19), 2605. https://doi.org/10.3390/plants11192605








