Quantitative Proteomics and Functional Characterization Reveal That Glutathione Peroxidases Act as Important Antioxidant Regulators in Mulberry Response to Drought Stress
Abstract
:1. Introduction
2. Results
2.1. Physiological and Biochemical Characterization of the Mulberry under Drought Stress
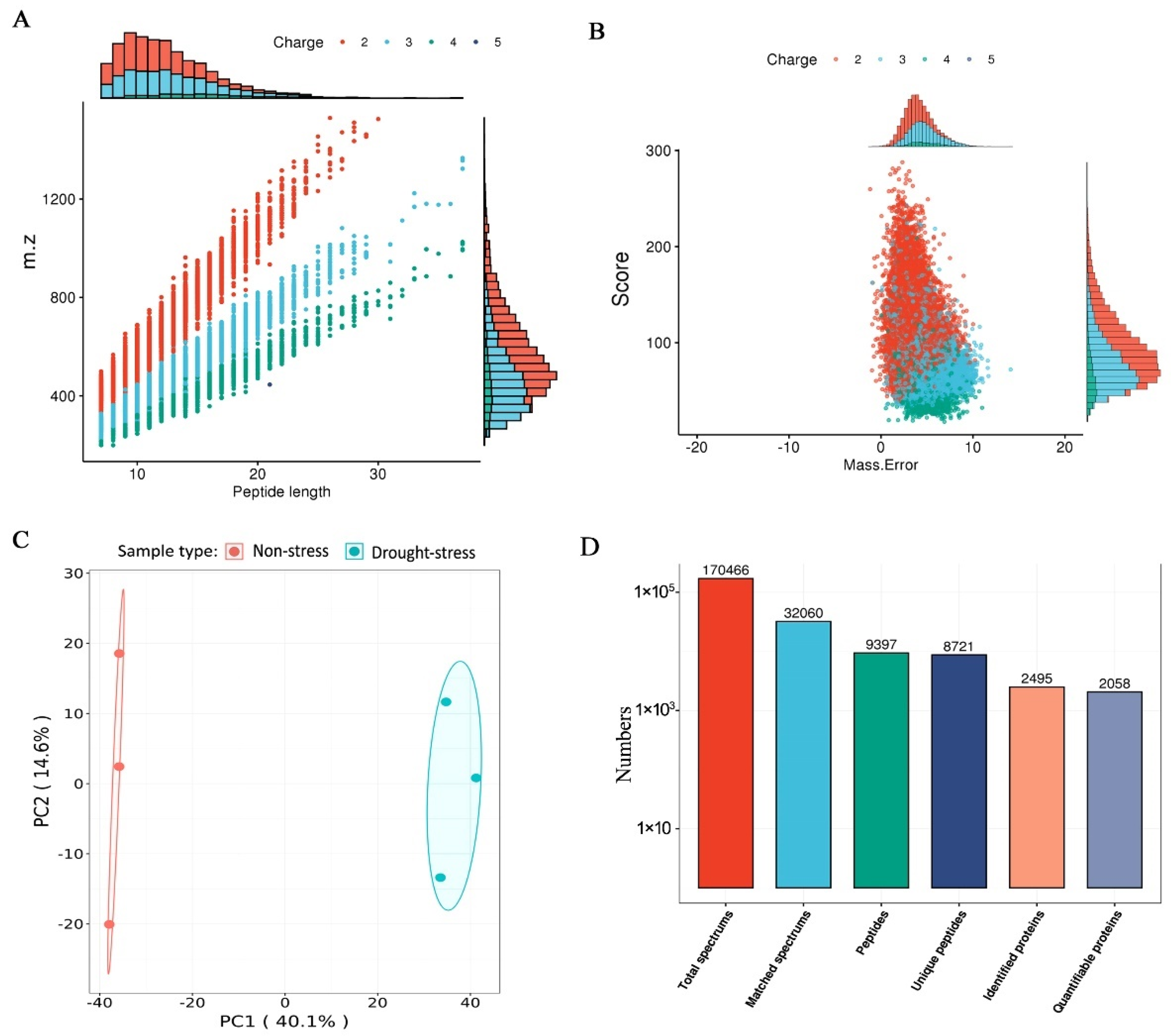
2.2. Quantitative Proteomic Analysis of the Mulberry under Drought Stress
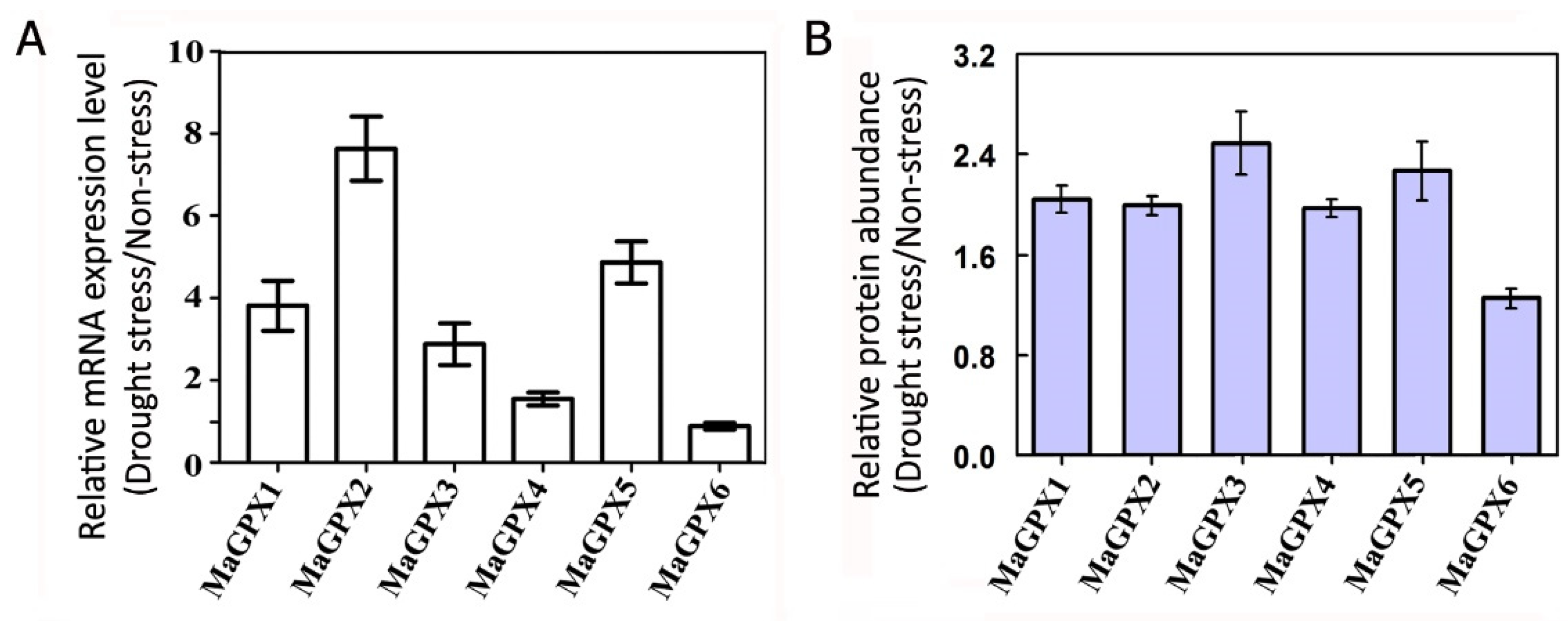
2.3. Abundance of Antioxidant Enzymes Especially GPX Isoforms Were Systematically Increased in the Mulberry under Drought Stress
2.4. Enzymatic Activity of GPX Was Increased in the Mulberry under Drought Stress
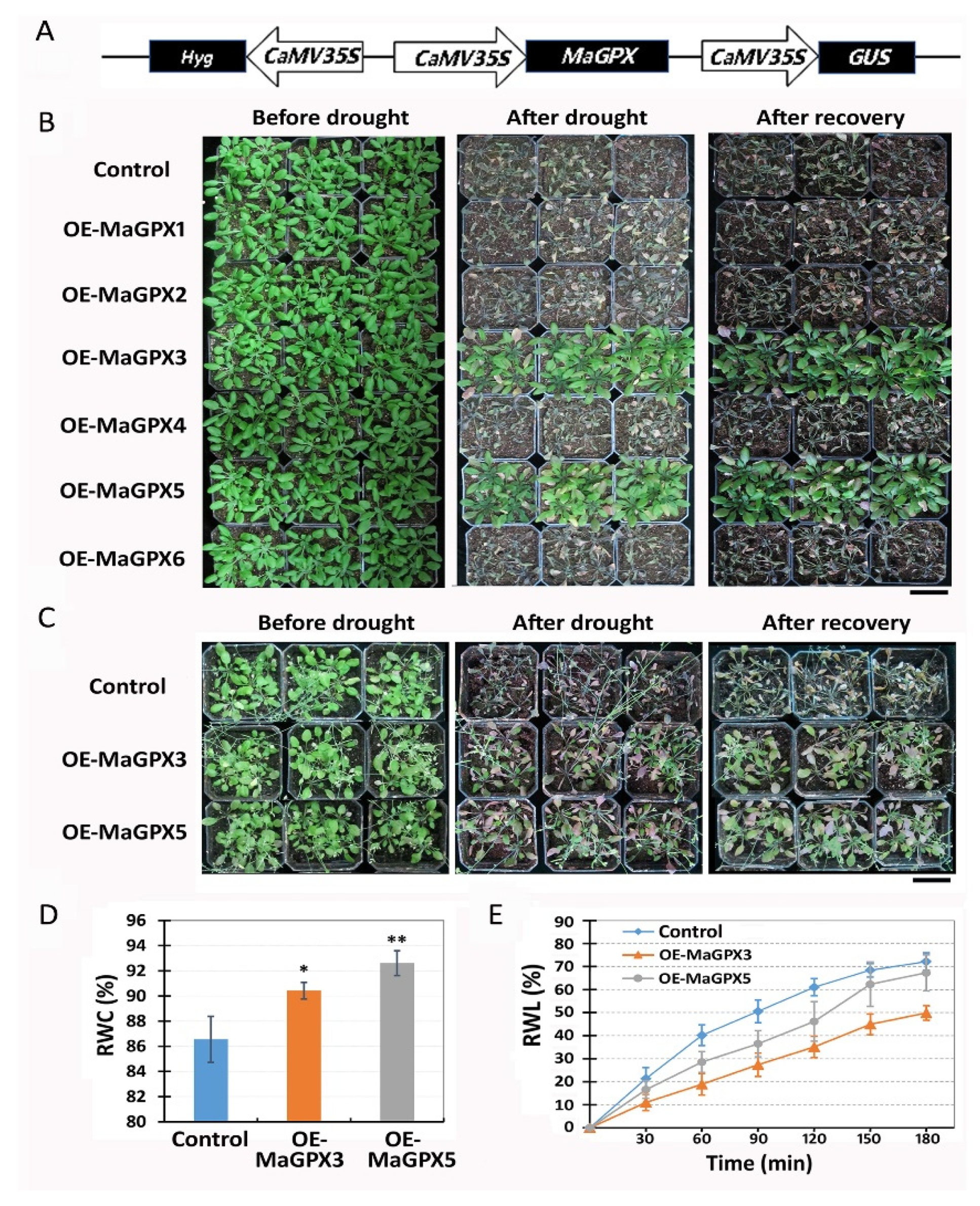
2.5. Ectopic Overexpression of Mulberry GPX Isoforms Confered Drought Resistance in Transgenic Arabidopsis
2.6. Ectopic Overexpression of Mulberry GPX Isoforms Improve Both Antioxidant Enzymatic Activities and ROS Scavenging in Transgenic Arabidopsis under Drought Stress
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Drought Treatments
4.3. Biochemical Measurements
4.4. iTRAQ-Based Quantitative Proteomics and Bioinformatic Analysis
4.5. Ectopic Overexpression of Mulberry GPX Genes in Arabidopsis
4.6. RNA Extraction and Quantitative RT-PCR
4.7. ROS Measurement and Histochemical Staining
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- He, X. Progress of comprehensive utilization of mulberry resources and development countermeasure. Acta Sericologica Sin. 2005, 31, 4–7. [Google Scholar]
- Qin, J.; He, N.J.; Wang, Y.; Xiang, Z.H. Ecological issues of mulberry and sustainable development. J. Resour. Ecol. 2012, 3, 330–339. [Google Scholar]
- Yin, H.; Wang, L.; Liu, G.; Huang, G.; Wei, L.; Wu, J. Effect of mulberry (Morus alba Linn) on soil and water conservation. Agric. Sci. Technol. 2016, 17, 2308–2311. [Google Scholar]
- Wang, J.; Ma, S.; Cao, Y.; Li, X.; Song, Y. The advantage of the mulberry using in the forestry’s sustainable development. Chin. Agric. Sci. Bull. 2004, 20, 72–73. [Google Scholar]
- Su, C.; Xue, Z.M.; Jiao, F. Cultivation features and ecological protective functions of adaptive mulberry germplasm resources in Loess Plateau of Northern Shaanxi. Acta Sericologica Sin. 2012, 38, 146–151. [Google Scholar]
- Lu, H.; Ding, T.Y.; Zuo, S.C.; Wu, L.L.; Ji, D.F.; Qin, J. Mulberry resource utilization and development strategy of “Ecologically and Economically Integrated” mulberry industry in Xinjiang. Acta Sericologica Sin. 2013, 39, 166–170. [Google Scholar]
- Bhusal, N.; Han, S.-G.; Yoon, T.-M. Impact of drought stress on photosynthetic response, leaf water potential, and stem sap flow in two cultivars of bi-leader apple trees (Malus × domestica Borkh.). Sci. Hortic. 2019, 246, 535–543. [Google Scholar] [CrossRef]
- Huang, G.T.; Ma, S.L.; Bai, L.P.; Zhang, L.; Ma, H.; Jia, P.; Liu, J.; Zhong, M.; Guo, Z.F. Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 2012, 39, 969–987. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Raghavendra, A.S.; Gonugunta, V.K.; Christmann, A.; Grill, E. ABA perception and signalling. Trends Plant Sci. 2010, 15, 395–401. [Google Scholar] [CrossRef]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, Y.; Zhu, J.K. Thriving under stress: How plants balance growth and the stress response. Dev. Cell 2020, 55, 529–543. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Koussevitzky, S.; Mittler, R.; Miller, G. ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 2012, 35, 259–270. [Google Scholar] [CrossRef]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [Green Version]
- Waszczak, C.; Carmody, M.; Kangasjarvi, J. Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef]
- Halliwell, B. Reactive species and antioxidants. Redox biology is a fundamental theme of aerobic life. Plant Physiol. 2006, 141, 312–322. [Google Scholar] [CrossRef]
- Foyer, C.H.; Shigeoka, S. Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiol. 2011, 155, 93–100. [Google Scholar] [CrossRef]
- Ozyigit, I.I.; Filiz, E.; Vatansever, R.; Kurtoglu, K.Y.; Koc, I.; Ozturk, M.X.; Anjum, N.A. Identification and comparative analysis of H2O2-scavenging enzymes (ascorbate peroxidase and glutathione peroxidase) in selected plants employing bioinformatics approaches. Front. Plant Sci. 2016, 7, 301. [Google Scholar] [CrossRef]
- Gaber, A.; Ogata, T.; Maruta, T.; Yoshimura, K.; Tamoi, M.; Shigeoka, S. The involvement of Arabidopsis glutathione peroxidase 8 in the suppression of oxidative damage in the nucleus and cytosol. Plant Cell Physiol. 2012, 53, 1596–1606. [Google Scholar] [CrossRef] [PubMed]
- Islam, T.; Manna, M.; Reddy, M.K. Glutathione peroxidase of Pennisetum glaucum (PgGPx) is a functional Cd2+ dependent peroxiredoxin that enhances tolerance against salinity and drought stress. PLoS ONE 2015, 10, e0143344. [Google Scholar] [CrossRef] [PubMed]
- Navrot, N.; Collin, V.; Gualberto, J.; Gelhaye, E.; Hirasawa, M.; Rey, P.; Knaff, D.B.; Issakidis, E.; Jacquot, J.P.; Rouhier, N. Plant glutathione peroxidases are functional peroxiredoxins distributed in several subcellular compartments and regulated during biotic and abiotic stresses. Plant Physiol. 2006, 142, 1364–1379. [Google Scholar] [CrossRef] [PubMed]
- Passaia, G.; Margis-Pinheiro, M. Glutathione peroxidases as redox sensor proteins in plant cells. Plant Sci. 2015, 234, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Paiva, A.; Passaia, G.; Lobo, A.; Jardim-Messeder, D.; Silveira, J.; Margis-Pinheiro, M. Mitochondrial glutathione peroxidase (OsGPX3) has a crucial role in rice protection against salt stress. Environ. Exp. Bot. 2019, 158, 12–21. [Google Scholar] [CrossRef]
- Iqbal, A.; Yabuta, Y.; Takeda, T.; Nakano, Y.; Shigeoka, S. Hydroperoxide reduction by thioredoxin-specific glutathione peroxidase isoenzymes of Arabidopsis thaliana. FEBS J. 2006, 273, 5589–5597. [Google Scholar] [CrossRef] [PubMed]
- Bela, K.; Horvath, E.; Galle, A.; Szabados, L.; Tari, I.; Csiszar, J. Plant glutathione peroxidases: Emerging role of the antioxidant enzymes in plant development and stress responses. J. Plant Physiol. 2015, 176, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Zhang, F.; Zhai, F.; Su, Y.; Zhou, Y.; Ge, Z.; Tilak, P.; Eirich, J.; Finkemeier, I.; Fu, L.; et al. Rice GLUTATHIONE PEROXIDASE1-mediated oxidation of bZIP68 positively regulates ABA-independent osmotic stress signaling. Mol. Plant 2022, 15, 651–670. [Google Scholar] [CrossRef]
- Gong, Z.Z.; Xiong, L.M.; Shi, H.Z.; Yang, S.H.; Herrera-Estrella, L.R.; Xu, G.H.; Chao, D.Y.; Li, J.R.; Wang, P.Y.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef]
- Wang, X.L.; Cai, X.F.; Xu, C.X.; Wang, Q.H.; Dai, S.J. Drought-responsive mechanisms in plant leaves revealed by proteomics. Int. J. Mol. Sci. 2016, 17, 1706. [Google Scholar] [CrossRef]
- Krol, A.; Weidner, S. Changes in the proteome of grapevine leaves (Vitis vinifera L.) during long-term drought stress. J. Plant Physiol. 2017, 211, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.J.; Chen, J.; Ghoto, K.; Hu, W.J.; Gao, G.F.; Luo, M.R.; Li, Z.; Simon, M.; Zhu, X.Y.; Zheng, H.L. Proteomic analysis on mangrove plant Avicennia marina leaves reveals nitric oxide enhances the salt tolerance by up-regulating photosynthetic and energy metabolic protein expression. Tree Physiol. 2018, 38, 1605–1622. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Esteso, M.J.; Casado-Vela, J.; Selles-Marchart, S.; Pedreno, M.A.; Bru-Martinez, R. Differential plant proteome analysis by isobaric tags for relative and absolute quantitation (iTRAQ). Methods Mol. Biol. 2014, 1072, 155–169. [Google Scholar] [CrossRef] [PubMed]
- Li, S.J.; Zhang, M.J.; Meng, C.T.; Su, C.; Dong, X.L.; Chen, K.M.; Li, W.Q. Gene cloning and expression analysis of the MaGPX family in Morus alba L. Acta Bot. Boreali-Occident. Sin. 2019, 39, 991–1000. [Google Scholar]
- Khaleghi, A.; Naderi, R.; Brunetti, C.; Maserti, B.E.; Salami, S.A.; Babalar, M. Morphological, physiochemical and antioxidant responses of Maclura pomifera to drought stress. Sci. Rep. 2019, 9, 19250. [Google Scholar] [CrossRef]
- Bhusal, N.; Lee, M.; Reum Han, A.; Han, A.; Kim, H.S. Responses to drought stress in Prunus sargentii and Larix kaempferi seedlings using morphological and physiological parameters. For. Ecol. Manag. 2020, 465, 118099. [Google Scholar] [CrossRef]
- Sofo, A.; Scopa, A.; Nuzzaci, M.; Vitti, A. Ascorbate peroxidase and catalase activities and their genetic regulation in plants subjected to drought and salinity stresses. Int. J. Mol. Sci. 2015, 16, 13561–13578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caverzan, A.; Passaia, G.; Rosa, S.B.; Ribeiro, C.W.; Lazzarotto, F.; Margis-Pinheiro, M. Plant responses to stresses: Role of ascorbate peroxidase in the antioxidant protection. Genet. Mol. Biol. 2012, 35, 1011–1019. [Google Scholar] [CrossRef]
- Chen, M.; Li, K.; Li, H.; Song, C.P.; Miao, Y. The glutathione peroxidase gene family in Gossypium hirsutum: Genome-wide identification, classification, gene expression and functional analysis. Sci. Rep. 2017, 7, 44743. [Google Scholar] [CrossRef]
- Attacha, S.; Solbach, D.; Bela, K.; Moseler, A.; Wagner, S.; Schwarzlander, M.; Aller, I.; Muller, S.J.; Meyer, A.J. Glutathione peroxidase-like enzymes cover five distinct cell compartments and membrane surfaces in Arabidopsis thaliana. Plant Cell Environ. 2017, 40, 1281–1295. [Google Scholar] [CrossRef]
- Pandey, S.; Fartyal, D.; Agarwal, A.; Shukla, T.; James, D.; Kaul, T.; Negi, Y.K.; Arora, S.; Reddy, M.K. Abiotic stress tolerance in plants: Myriad roles of ascorbate peroxidase. Front. Plant Sci. 2017, 8, 581. [Google Scholar] [CrossRef] [PubMed]
- Bobrovskikh, A.; Zubairova, U.; Kolodkin, A.; Doroshkov, A. Subcellular compartmentalization of the plant antioxidant system: An integrated overview. PeerJ 2020, 8, e9451. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Jang, M.G.; Noh, H.Y.; Lee, H.J.; Sukweenadhi, J.; Kim, J.H.; Kim, S.Y.; Kwon, W.S.; Yang, D.C. Molecular characterization of two glutathione peroxidase genes of Panax ginseng and their expression analysis against environmental stresses. Gene 2014, 535, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Diao, Y.; Xu, H.; Li, G.; Yu, A.; Yu, X.; Hu, W.; Zheng, X.; Li, S.; Wang, Y.; Hu, Z. Cloning a glutathione peroxidase gene from Nelumbo nucifera and enhanced salt tolerance by overexpressing in rice. Mol. Biol. Rep. 2014, 41, 4919–4927. [Google Scholar] [CrossRef] [PubMed]
- Vieira Dos Santos, C.; Rey, P. Plant thioredoxins are key actors in the oxidative stress response. Trends Plant Sci. 2006, 11, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Geigenberger, P.; Thormahlen, I.; Daloso, D.M.; Fernie, A.R. The unprecedented versatility of the plant thioredoxin system. Trends Plant Sci. 2017, 22, 249–262. [Google Scholar] [CrossRef]
- Milla, M.A.R.; Maurer, A.; Huete, A.R.; Gustafson, J.P. Glutathione peroxidase genes in Arabidopsis are ubiquitous and regulated by abiotic stresses through diverse signaling pathways. Plant J. 2003, 36, 602–615. [Google Scholar] [CrossRef]
- Miao, Y.; Lv, D.; Wang, P.; Wang, X.C.; Chen, J.; Miao, C.; Song, C.P. An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 2006, 18, 2749–2766. [Google Scholar] [CrossRef]
- Gaber, A.; Yoshimura, K.; Yamamoto, T.; Yabuta, Y.; Takeda, T.; Miyasaka, H.; Nakano, Y.; Shigeoka, S. Glutathione peroxidase-like protein of Synechocystis PCC 6803 confers tolerance to oxidative and environmental stresses in transgenic Arabidopsis. Physiol. Plant 2006, 128, 251–262. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, M.; Teng, Y.; Jia, S.; Yu, D.; Wei, T.; Chen, C.; Song, W. Overexpression of the Glutathione Peroxidase 5 (RcGPX5) gene from Rhodiola crenulata increases drought tolerance in Salvia miltiorrhiza. Front. Plant Sci. 2018, 9, 1950. [Google Scholar] [CrossRef]
- Toscano, S.; Farieri, E.; Ferrante, A.; Romano, D. Physiological and biochemical responses in two ornamental shrubs to drought stress. Front. Plant Sci. 2016, 7, 645. [Google Scholar] [CrossRef] [PubMed]
- Zhong, T.; Zhang, L.; Sun, S.; Zeng, H.; Han, L. Effect of localized reduction of gibberellins in different tobacco organs on drought stress tolerance and recovery. Plant Biotechnol. Rep. 2014, 8, 399–408. [Google Scholar] [CrossRef]
- Li, W.Q.; Zhang, M.J.; Gan, P.F.; Qiao, L.; Yang, S.Q.; Miao, H.; Wang, G.F.; Zhang, M.M.; Liu, W.T.; Li, H.F.; et al. CLD1/SRL1 modulates leaf rolling by affecting cell wall formation, epidermis integrity and water homeostasis in rice. Plant J. 2017, 92, 904–923. [Google Scholar] [CrossRef] [PubMed]
- Ramel, F.; Sulmon, C.; Bogard, M.; Couee, I.; Gouesbet, G. Differential patterns of reactive oxygen species and antioxidative mechanisms during atrazine injury and sucrose-induced tolerance in Arabidopsis thaliana plantlets. BMC Plant Biol. 2009, 9, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Protein ID | Protein Description | Fold Change | p Value | Regulation | MW [kDa] | Coverage [%] | Unique Peptides | a PSMs | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|
| W9S7L1 | Glutaredoxin domain-containing protein | 2.96 | 0.000474 | Up | 13.3890 | 14.4 | 2 | 2 | Chloroplast |
| W9SE23 | Peroxidase | 2.85 | 0.000120 | Up | 36.4880 | 13.6 | 4 | 27 | Chloroplast |
| W9QHE0 | Glutathione peroxidase | 2.48 | 0.000983 | Up | 18.4420 | 13.4 | 1 | 21 | Cytoplasm |
| W9QT41 | Glutathione peroxidase | 2.27 | 0.003078 | Up | 18.9620 | 38.2 | 8 | 20 | Mitochondria |
| W9QH65 | Glutathione peroxidase | 2.04 | 0.000421 | Up | 26.5190 | 18.6 | 5 | 45 | Chloroplast |
| W9RCX8 | Glutathione S-transferase | 2.02 | 0.001778 | Up | 26.7850 | 48.3 | 9 | 74 | Cytoplasm |
| W9RT74 | Glutathione peroxidase | 1.99 | 0.000374 | Up | 20.5060 | 32.1 | 5 | 21 | Cytoplasm |
| W9SDB3 | Glutathione peroxidase | 1.97 | 0.000211 | Up | 26.0130 | 29.2 | 5 | 37 | Chloroplast |
| W9SDD7 | Thioredoxin | 1.85 | 0.000336 | Up | 13.2220 | 22.7 | 3 | 9 | Chloroplast |
| W9QZP8 | Glutaredoxin-C5 | 1.77 | 0.013193 | Up | 18.9360 | 26.1 | 3 | 9 | Chloroplast |
| W9SBA6 | Glutaredoxin domain-containing protein | 1.69 | 0.001711 | Up | 15.0310 | 30.8 | 3 | 34 | Chloroplast |
| W9S168 | Glutathione S-transferase | 1.68 | 0.000215 | Up | 39.9490 | 30.5 | 11 | 69 | Chloroplast |
| W9SW93 | Thioredoxin-like 3-1 | 1.67 | 0.000097 | Up | 21.2490 | 11.5 | 2 | 3 | Cytoplasm |
| W9RMD9 | Thioredoxin-like fold containing protein | 1.66 | 0.000454 | Up | 35.4970 | 13 | 5 | 17 | Vacuolar membrane |
| W9SCK7 | Monothiol glutaredoxin-S16 | 1.57 | 0.001539 | Up | 32.5700 | 14.4 | 3 | 10 | Chloroplast |
| W9QVC2 | Peroxiredoxin | 1.56 | 0.004432 | Up | 22.4790 | 42.1 | 7 | 52 | Chloroplast, mitochondria |
| W9SEV0 | Peroxiredoxin | 1.54 | 0.000664 | Up | 17.2870 | 36.4 | 6 | 44 | Cytoplasm |
| W9R4T1 | Thioredoxin O1 | 1.53 | 0.002092 | Up | 28.3650 | 12.3 | 4 | 13 | Chloroplast |
| W9SCK7 | Monothiol glutaredoxin-S16 | 1.57 | 0.001539 | Up | 32.5700 | 14.4 | 3 | 10 | Chloroplast |
| W9QVC2 | Peroxiredoxin | 1.56 | 0.004432 | Up | 22.4790 | 42.1 | 7 | 52 | Chloroplast, mitochondria |
| W9SEM5 | Ferredoxin-thioredoxin reductase, catalytic chain | 1.56 | 0.002258 | Up | 16.2540 | 40 | 6 | 14 | Chloroplast |
| W9SEV0 | Peroxiredoxin | 1.54 | 0.000664 | Up | 17.2870 | 36.4 | 6 | 44 | Cytoplasm |
| W9R4T1 | Thioredoxin O1 | 1.53 | 0.002092 | Up | 28.3650 | 12.3 | 4 | 13 | Chloroplast |
| W9SVF9 | Monothiol glutaredoxin-S7 | 0.64 | 0.006054 | Down | 20.0390 | 9.8 | 1 | 1 | Chloroplast |
| W9RQI7 | L-ascorbate oxidase-like protein | 0.64 | 0.007718 | Down | 60.3300 | 4.6 | 2 | 3 | Chloroplast |
| W9SBU2 | SodC protein | 0.62 | 0.005244 | Down | 29.3360 | 34.2 | 6 | 89 | Chloroplast |
| W9RYX2 | Thioredoxin reductase | 0.53 | 0.004546 | Down | 56.9680 | 8.1 | 3 | 4 | Chloroplast |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Li, W.; Li, S.; Gao, J.; Gan, T.; Li, Q.; Bao, L.; Jiao, F.; Su, C.; Qian, Y. Quantitative Proteomics and Functional Characterization Reveal That Glutathione Peroxidases Act as Important Antioxidant Regulators in Mulberry Response to Drought Stress. Plants 2022, 11, 2350. https://doi.org/10.3390/plants11182350
Zhang M, Li W, Li S, Gao J, Gan T, Li Q, Bao L, Jiao F, Su C, Qian Y. Quantitative Proteomics and Functional Characterization Reveal That Glutathione Peroxidases Act as Important Antioxidant Regulators in Mulberry Response to Drought Stress. Plants. 2022; 11(18):2350. https://doi.org/10.3390/plants11182350
Chicago/Turabian StyleZhang, Minjuan, Wenqiang Li, Shuaijun Li, Junru Gao, Tiantian Gan, Qinying Li, Lijun Bao, Feng Jiao, Chao Su, and Yonghua Qian. 2022. "Quantitative Proteomics and Functional Characterization Reveal That Glutathione Peroxidases Act as Important Antioxidant Regulators in Mulberry Response to Drought Stress" Plants 11, no. 18: 2350. https://doi.org/10.3390/plants11182350
APA StyleZhang, M., Li, W., Li, S., Gao, J., Gan, T., Li, Q., Bao, L., Jiao, F., Su, C., & Qian, Y. (2022). Quantitative Proteomics and Functional Characterization Reveal That Glutathione Peroxidases Act as Important Antioxidant Regulators in Mulberry Response to Drought Stress. Plants, 11(18), 2350. https://doi.org/10.3390/plants11182350







