Phosphoprotein Profile of Rice (Oryza sativa L.) Seedlings under Osmotic Stress after Pretreatment with Chitosan
Abstract
1. Introduction
2. Results
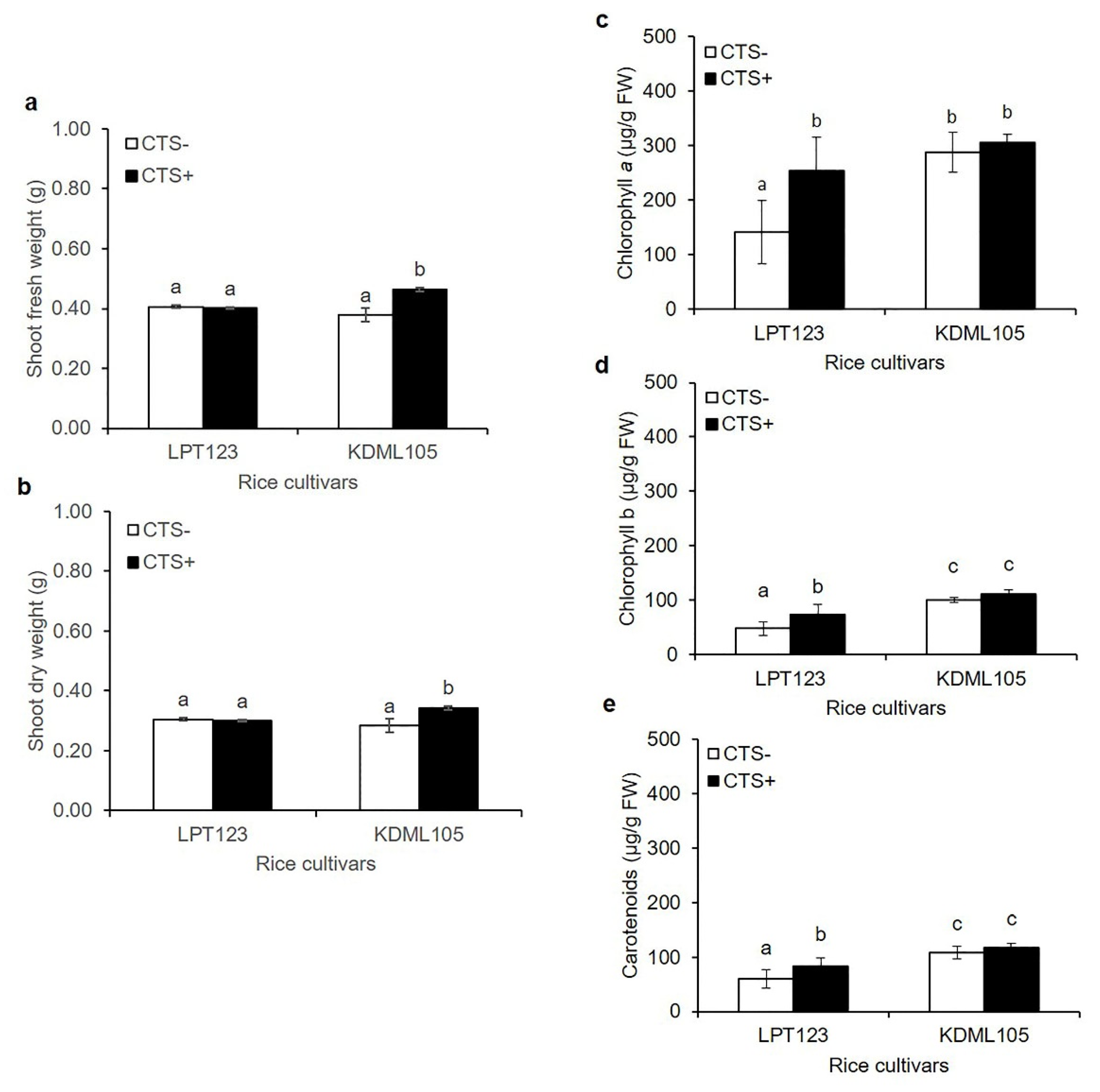
2.1. Growth and Photosynthetic Pigments Induced by CTS Treatment and Osmotic Stress
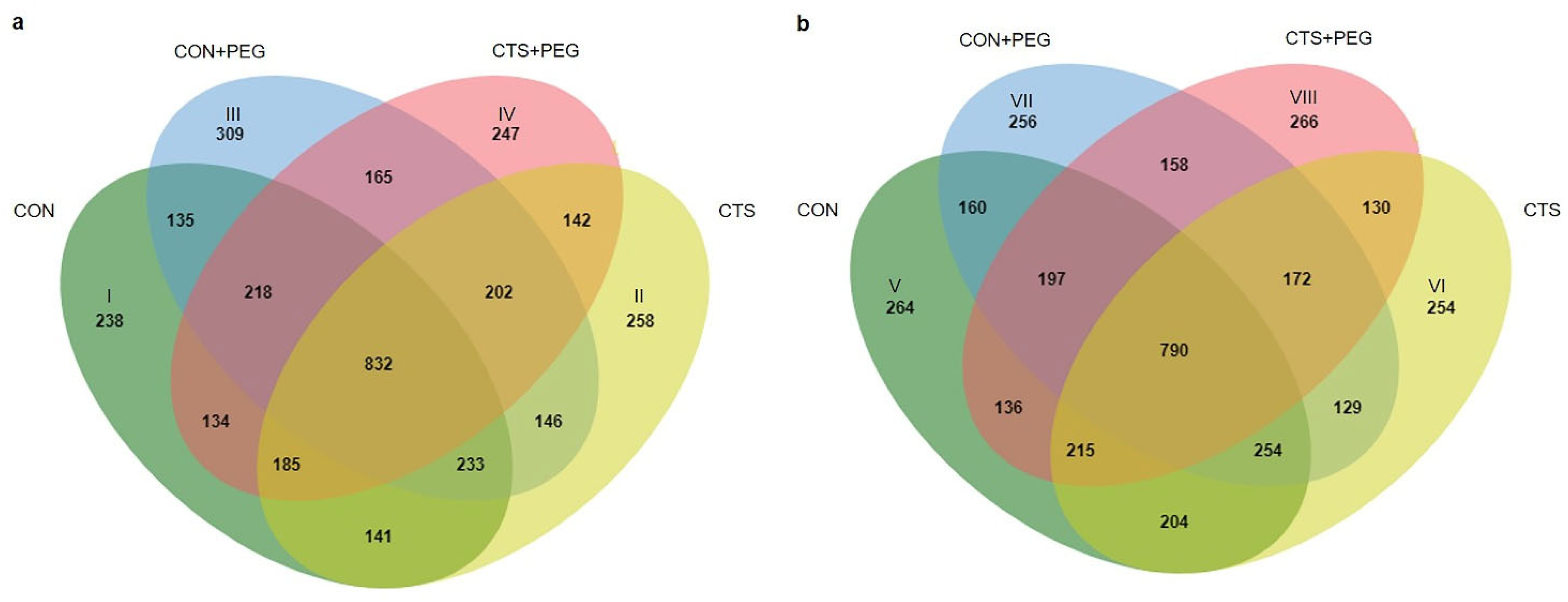
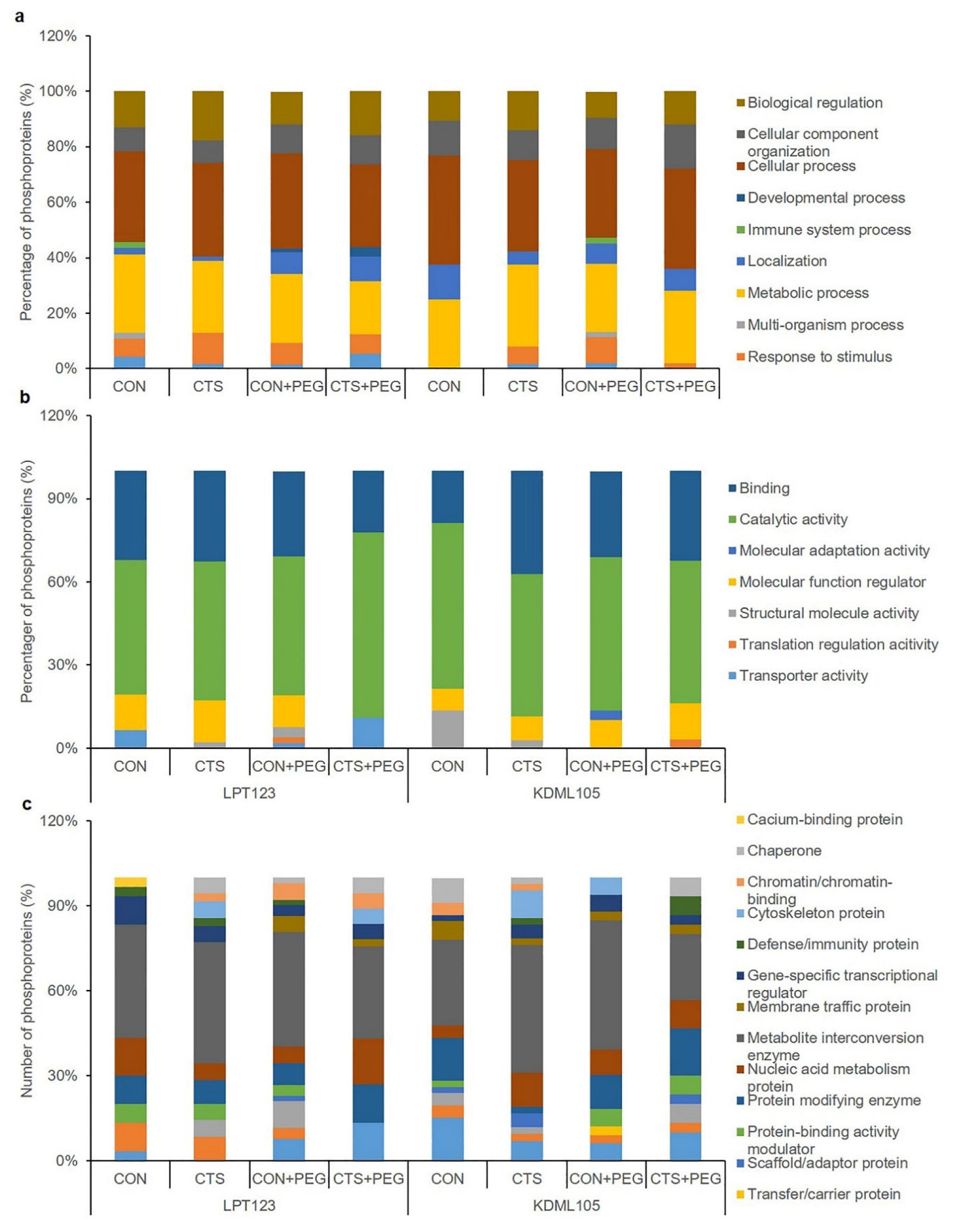
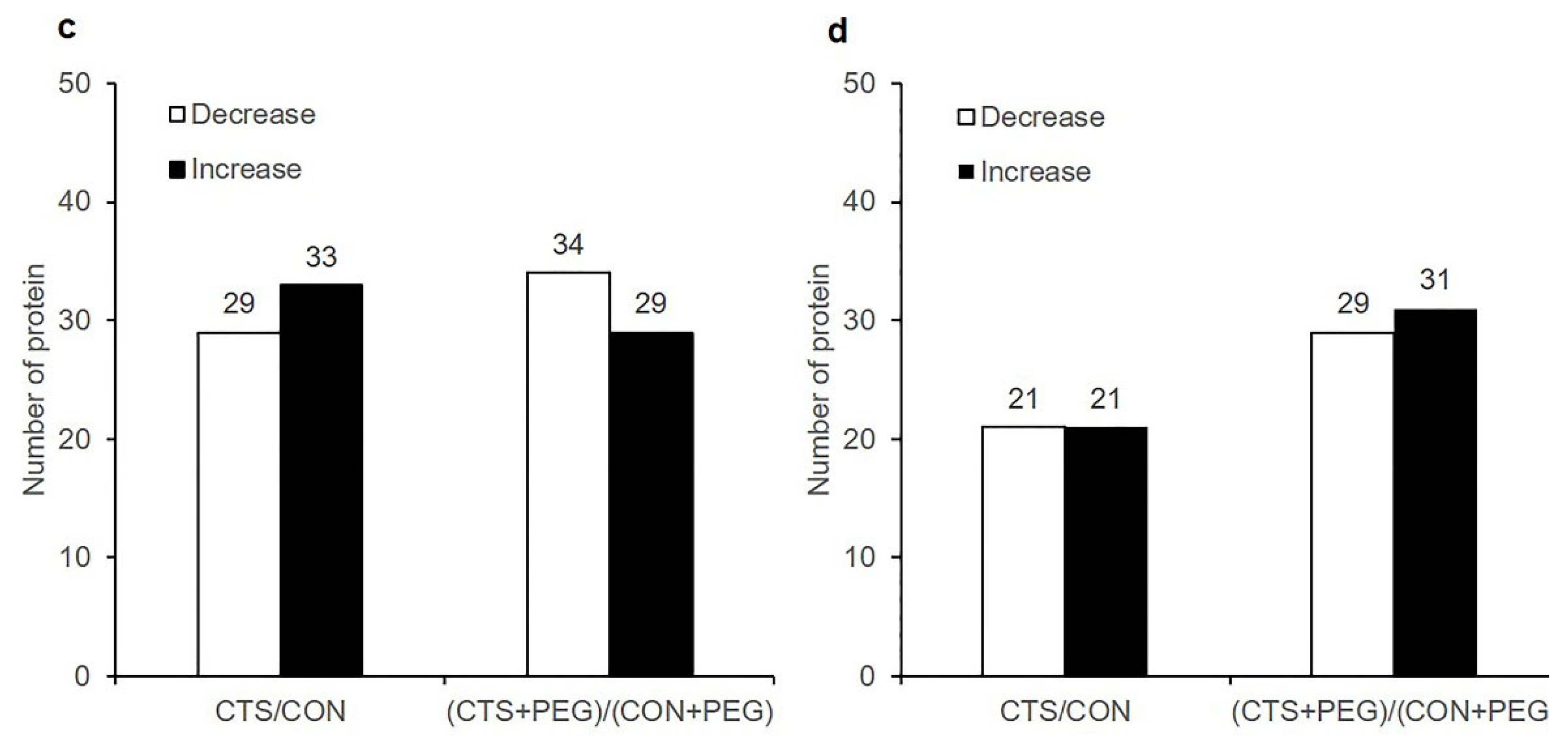
2.2. Differentially Expressed Proteins (DEPs) and Functional Classification
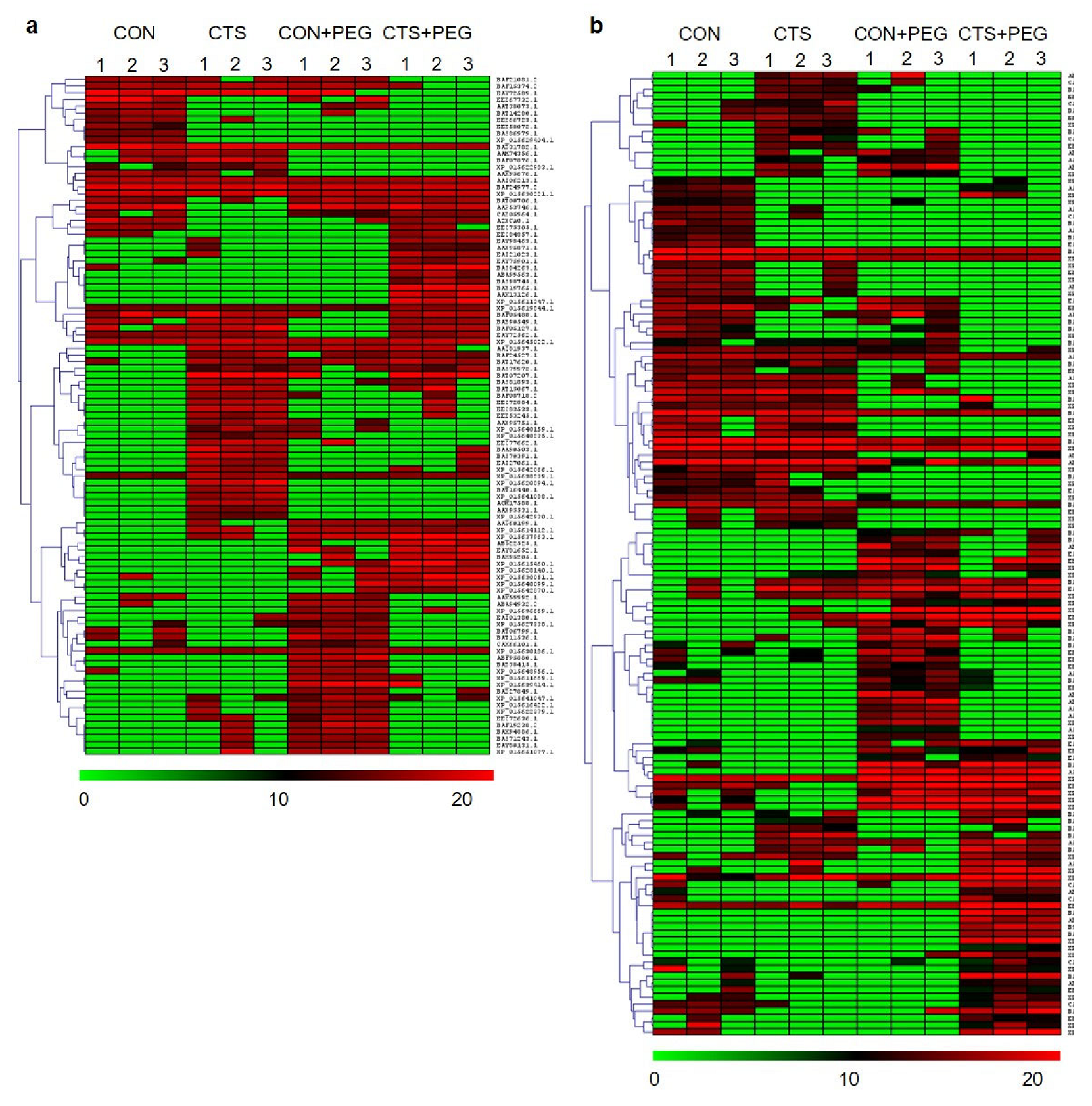
2.3. Clustering Analysis and Identification of CTS-Responsive Proteins
2.4. Identification of Phosphorylated Proteins and Phosphorylation Motifs Induced by CTS Treatment
3. Discussion
3.1. CTS Affected Growth Enhancement and Photosynthetic Pigments in the Osmotic Stress Condition
3.2. Quantitative Phosphoproteomics Analysis as a Powerful Tool for Analysis of Leaf Phosphoproteins
3.3. Protein Kinases Associated with Signal Transduction Induced by CTS and the Stress Response
3.4. Phosphoproteins Involved in the Defense Response
3.5. Phosphoproteins Involved in Transmembrane Transport
3.6. Phosphoproteins Involved in Transcription
3.7. Phosphoproteins Involved in Protein Folding and Degradation
3.8. Phosphoproteins Relative to Plant Metabolism
3.9. Predicted Phosphorylation Sites of the Peptides
4. Materials and Methods
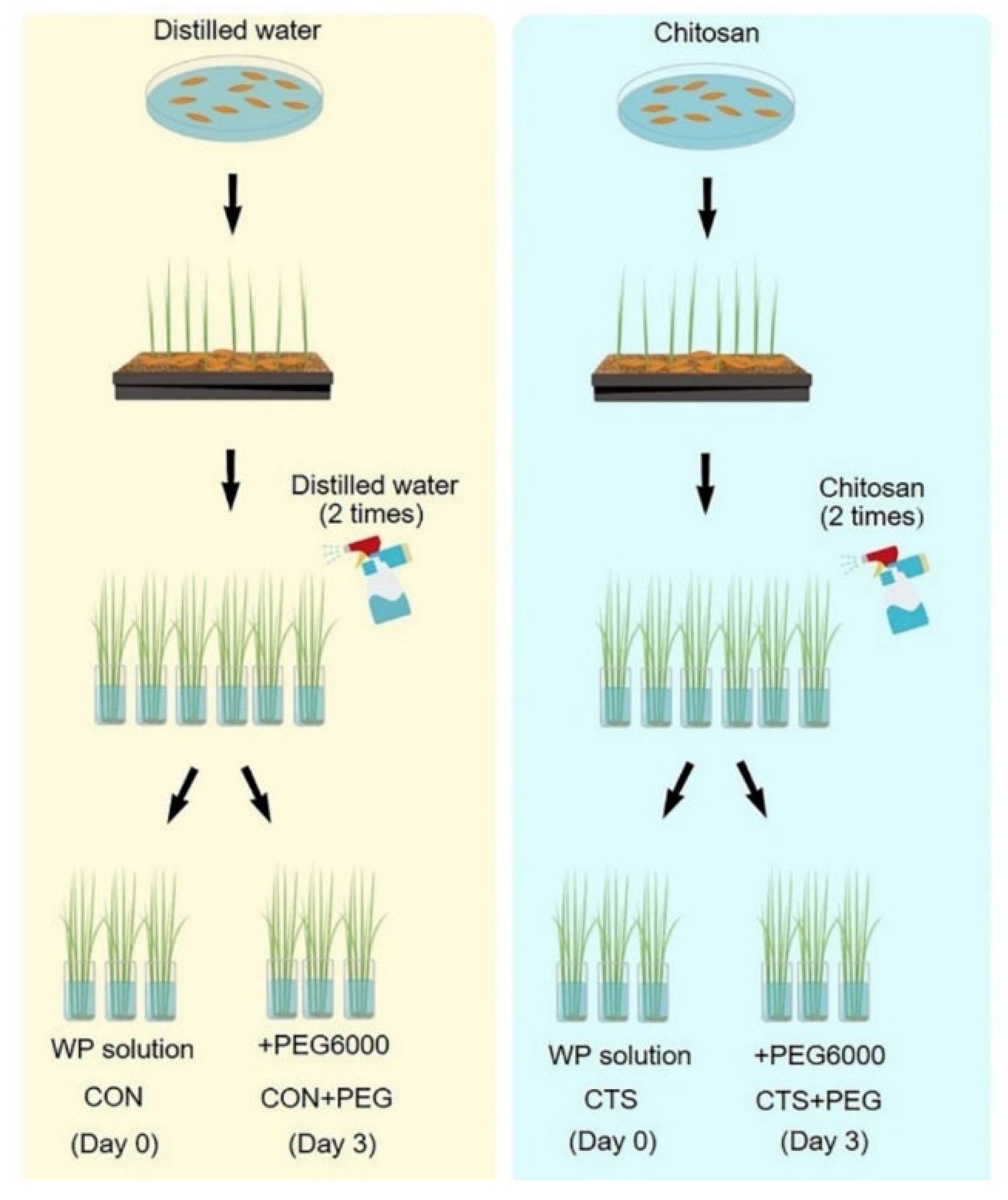
4.1. Plant Material, Growth Conditions, CTS, and Osmotic Stress Treatments
4.2. Photosynthetic Pigments Determination
4.3. Total Protein Extraction
4.4. Phosphoprotein Enrichment and Digestion
4.5. Liquid Chromatography–Mass Spectrophotometry (LC-MS/MS)
4.6. Data Analysis and Bioinformatics
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, M.; Lv, D.; Ge, P.; Bian, Y.; Chen, G.; Zhu, G.; Li, X.; Yan, Y. Phosphoproteome analysis reveals new drought response and defense mechanisms of seedling leaves in bread wheat (Triticum aestivum L.). J. Proteome. Res. 2014, 109, 290–308. [Google Scholar] [CrossRef] [PubMed]
- Kean, T.; Thanou, M. Chitin and chitosan-sources, production and medical applications. In Rsc Polymer Chemistry Series Renewable Resources for Functional Polymers And Biomaterials, 1st ed.; Peter, A.W., Ed.; Royal Society of Chemistry: London, UK, 2011; pp. 327–361. [Google Scholar] [CrossRef]
- Jones, M.; Kujundzic, M.; John, S.; Bismarck, A. Crab vs. Mushroom: A review of crustacean and fungal chitin in wound treatment. Mar. Drugs 2020, 18, 64. [Google Scholar] [CrossRef]
- Bittelli, M.; Flury, M.; Campbell, G.; Nichols, E. Reduction of transpiration through foliar application of chitosan. Agric. For. Meteorol. 2001, 107, 167–175. [Google Scholar] [CrossRef]
- Górnik, K.; Mieczyslaw, G.; Duda, B.R. The effect of chitosan on rooting of grapevine cuttings and on subsequent plant growth under drought and temperature stress. J. Fruit. Ornam. Plant. Res. 2008, 16, 333–343. [Google Scholar]
- Gadalla, S.; Amany, A.R. Improving growth and yield of cowpea by foliar application of chitosan under water stress. Egypt J. Biol. 2012, 14, 14–16. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Z.; Li, Y.P.; Zhang, X.Q.; Ma, X.; Huang, L.K.; Yan, Y.; Peng, Y. Chitosan and spermine enhance drought resistance in white clover, associated with changes in endogenous phytohormones and polyamines, and antioxidant metabolism. Funct. Plant Biol. 2018, 45, 1205–1222. [Google Scholar] [CrossRef]
- Emami Bistgani, Z.; Siadat, S.A.; Bakhshandeh, A.; Ghasemi Pirbalouti, A.; Hashemi, M. Interactive effects of drought stress and chitosan application on physiological characteristics and essential oil yield of Thymus daenensis Celak. Crop. J. 2017, 5, 407–415. [Google Scholar] [CrossRef]
- Ghasemi Pirbalouti, A.; Malekpoor, F.; Salimi, A.; Golparvar, A. Exogenous application of chitosan on biochemical and physiological characteristics, phenolic content and antioxidant activity of two species of basil (Ocimum ciliatum and Ocimum basilicum) under reduced irrigation. Sci. Hortic. 2017, 217, 114–122. [Google Scholar] [CrossRef]
- Boonlertnirun, S.; Sarobol, E.; Meechoui, S.; Sooksathan, I. Drought recovery and grain yield potential of rice after chitosan application. Witthayasan Kasetsat 2007, 41, 1–6. [Google Scholar]
- Vasconsuelo, A.; Giulietti, A.M.; Boland, R. Signal transduction events mediating chitosan stimulation of anthraquinone synthesis in Rubia tinctorum. Plant Sci. 2004, 166, 405–413. [Google Scholar] [CrossRef]
- Hidangmayum, A.; Dwivedi, P.; Katiyar, D.; Hemantaranjan, A. Application of chitosan on plant responses with special reference to abiotic stress. Physiol. Mol. Biol. Plants 2019, 25, 313–326. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Y.; Zhang, X.; Merewitz, E.; Peng, Y.; Ma, X.; Huang, L.; Yan, Y. Metabolic pathways regulated by chitosan contributing to drought resistance in white clover. J. Proteome Res. 2017, 16, 3039–3052. [Google Scholar] [CrossRef]
- Zhao, J.; Pan, L.; Zhou, M.; Yang, Z.; Meng, Y.; Zhang, X. Comparative physiological and transcriptomic analyses reveal mechanisms of improved osmotic stress tolerance in Annual ryegrass by exogenous chitosan. Genes 2019, 10, 853. [Google Scholar] [CrossRef]
- Chamnanmanoontham, N.; Pongprayoon, W.; Pichayangkura, R.; Roytrakul, S.; Chadchawan, S. Chitosan enhances rice seedling growth via gene expression network between nucleus and chloroplast. Plant Growth Regul. 2015, 75, 101–114. [Google Scholar] [CrossRef]
- Thingholm, T.E.; Jensen, O.N.; Larsen, M.R. Analytical strategies for phosphoproteomics. Proteomics 2009, 9, 1451–1468. [Google Scholar] [CrossRef]
- Fíla, J.; Honys, D. Enrichment techniques employed in phosphoproteomics. Amino Acids 2012, 43, 1025–1047. [Google Scholar] [CrossRef]
- Pongprayoon, W.; Roytraku, S.; Pichyangkur, R.; Chadchawan, S. The role of hydrogen peroxide in chitosan-induced resistance to osmotic stress in rice (Oryza sativa L.). Plant Growth Regul. 2013, 70, 159–173. [Google Scholar] [CrossRef]
- Pongprayoon, W.; Maksup, S.; Phaonakrop, N.; Jaresitthikunchai, J.; Uawisetwathana, U.; Panya, A.; Roytrakul, S. Phosphoproteome analysis reveals chitosan-induced resistance to osmotic stress in rice (Oryza sativa L.) seedlings. J. Plant Interact. 2022, 17, 894–910. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, T.; Liang, L.; Li, Z.; Hassan, M.; Peng, Y.; Wang, D. Enhanced photosynthesis, carbohydrates, and energy metabolism associated with chitosan-induced drought tolerance in creeping bentgrass. Crop Sci. 2020, 60, 1064–1076. [Google Scholar] [CrossRef]
- Limpanavech, P.; Chaiyasuta, S.; Vongpromek, R.; Pichyangkura, R.; Khunwasi, C.; Chadchawan, S.; Lotrakul, P.; Bunjongrat, R.; Chaidee, A.; Bangyeekhun, T. Chitosan effects on floral production, gene expression, and anatomical changes in the Dendrobium orchid. Sci. Hortic. 2008, 116, 65–72. [Google Scholar] [CrossRef]
- Mayya, V.; Han, D.K. Phosphoproteomics by mass spectrometry: Insights, implications, applications and limitations. Expert Rev. Proteom. 2009, 6, 605–618. [Google Scholar] [CrossRef][Green Version]
- Qeli, E.; Ahrens, C.H. PeptideClassifier for protein inference and targeted quantitative proteomics. Nat. Biotechnol. 2010, 28, 647–650. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Silva-Sanchez, C.; Zhang, T.; Chen, S.; Li, H. Phosphoproteomics technologies and applications in plant biology research. Front. Plant Sci. 2015, 6, 430. [Google Scholar] [CrossRef] [PubMed]
- Wolanin, P.M.; Thomason, P.A.; Stock, J.B. Histidine protein kinases: Key signal transducers outside the animal kingdom. Genome Biol. 2002, 3, 1–8. [Google Scholar] [CrossRef]
- Nongpiur, R.; Soni, P.; Karan, R.; Singla-Pareek, S.L.; Pareek, A. Histidine kinases in plants: Cross talk between hormone and stress responses. Plant Signal. Behav. 2012, 7, 1230–1237. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Wu, L.; Zhao, F.; Zhang, D.; Li, N.; Zhu, G.; Li, C.; Wang, W. Phosphoproteomic analysis of the response of maize leaves to drought, heat and their combination stress. Front. Plant Sci. 2015, 6, 298. [Google Scholar] [CrossRef] [PubMed]
- Petutschnig, E.K.; Jones, A.M.; Serazetdinova, L.; Lipka, U.; Lipka, V. The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J. Biol. Chem. 2010, 285, 28902–28911. [Google Scholar] [CrossRef] [PubMed]
- Povero, G.; Loreti, E.; Pucciariello, C.; Santaniello, A.; Di Tommaso, D.; Tommaso, G.D.; Kapetis, D.; Zolezzi, F.; Piaggesi, A.; Perata, P. Transcript profiling of chitosan-treated Arabidopsis seedlings. J. Plant Res. 2011, 124, 619–629. [Google Scholar] [CrossRef]
- Dao, T.T.H.; Linthorst, H.J.M.; Verpoorte, R. Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 2011, 10, 397–412. [Google Scholar] [CrossRef]
- Faoro, F.; Maffi, D.; Cantu, D.; Iriti, M. Chemical-induced resistance against powdery mildew in barley: The effects of chitosan and benzothiadiazole. BioControl 2008, 53, 387–401. [Google Scholar] [CrossRef]
- Pichyangkura, R.; Chadchawan, S. Biostimulant activity of chitosan in horticulture. Sci. Hortic. 2015, 196, 49–65. [Google Scholar] [CrossRef]
- McHale, L.; Tan, X.; Koehl, P.; Michelmore, R.W. Plant NBS-LRR proteins: Adaptable guards. Genome Biol. 2006, 7, 212. [Google Scholar] [CrossRef]
- Boyes, D.C.; Nam, J.; Dangl, J.L. The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc. Natl. Acad. Sci. USA 1998, 95, 15849–15854. [Google Scholar] [CrossRef]
- Kang, J.; Park, J.; Choi, H.; Burla, B.; Kretzschmar, T.; Lee, Y.; Martinoia, E. Plant ABC transporters. Arabidopsis Book 2011, 9, e0153. [Google Scholar] [CrossRef]
- Srivastava, N.; Gonugunta, V.K.; Puli, M.R.; Raghavendra, A.S. Nitric oxide production occurs downstream of reactive oxygen species in guard cells during stomatal closure induced by chitosan in abaxial epidermis of Pisum sativum. Planta 2009, 229, 757–765. [Google Scholar] [CrossRef]
- Lopez-Moya, F.; Escudero, N.; Zavala-Gonzalez, E.A.; Esteve-Bruna, D.; Blázquez, M.A.; Alabadí, D.; Lopez-Llorca, L.V. Induction of auxin biosynthesis and WOX5 repression mediate changes in root development in Arabidopsis exposed to chitosan. Sci. Rep. 2017, 7, 16813. [Google Scholar] [CrossRef]
- Möller, B.; Hove, C.; Xiang, D.; Williams, N.; González López, L.; Yoshida, S.; Smit, M.; Datla, R.; Weijers, D. Auxin response cell-autonomously controls ground tissue initiation in the early Arabidopsis embryo. Proc. Natl. Acad. Sci. USA 2017, 114, E2533–E2539. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, J.; Zhang, W.; Yan, S.; Wang, R.; Zhao, J.; Li, Y.; Qi, Z.; Sun, Z.; Zhu, Z. The putative auxin efflux carrier OsPIN3t is involved in the drought stress response and drought tolerance. Plant J. 2012, 72, 805–816. [Google Scholar] [CrossRef]
- Geng, W.; Li, Z.; Hassan, M.J.; Peng, Y. Chitosan regulates metabolic balance, polyamine accumulation, and Na+ transport contributing to salt tolerance in creeping bentgrass. BMC Plant Biol. 2020, 20, 506. [Google Scholar] [CrossRef]
- Lahr, R.M.; Mack, S.M.; Héroux, A.; Blagden, S.P.; Bousquet-Antonelli, C.; Deragon, J.M.; Berman, A.J. The La-related protein 1-specific domain repurposes HEAT-like repeats to directly bind a 5’TOP sequence. Nucleic Acids Res. 2015, 43, 8077–8088. [Google Scholar] [CrossRef]
- Gu, L.; Xu, T.; Lee, K.; Lee, K.H.; Kang, H. A chloroplast-localized DEAD-box RNA helicase AtRH3 is essential for intron splicing and plays an important role in the growth and stress response in Arabidopsis thaliana. Plant Physiol. Biochem. 2014, 82, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Dinkins, R.; Reddy, M.S.; Leng, M.; Collins, G.B. Overexpression of the Arabidopsis thaliana MinD1 gene alters chloroplast size and number in transgenic tobacco plants. Planta 2001, 214, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.; Zhang, X.; Li, J.; Su, X.; Liu, J. Overexpression of a tobacco J-domain protein enhances drought tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2014, 83, 100–106. [Google Scholar] [CrossRef]
- Hu, T.; Hu, Z.; Zeng, H.; Qv, X.; Chen, G. Tomato lipoxygenase D involved in the biosynthesis of jasmonic acid and tolerance to abiotic and biotic stress in tomato. Plant Biotechnol. Rep. 2015, 9, 37–45. [Google Scholar] [CrossRef]
- Callis, J.; Vierstra, R.D. Protein degradation in signaling. Curr. Opin. Plant Biol. 2000, 3, 381–386. [Google Scholar] [CrossRef]
- Yang, P.; Smalle, J.; Lee, S.; Yan, N.; Emborg, T.J.; Vierstra, R.D. Ubiquitin C-terminal hydrolases 1 and 2 affect shoot architecture in Arabidopsis. Plant J. 2007, 51, 441–457. [Google Scholar] [CrossRef]
- Azevedo, C.; Santos-Rosa, M.J.; Shirasu, K. The U-box protein family in plants. Trends Plant Sci. 2001, 6, 354–358. [Google Scholar] [CrossRef]
- Zanella, M.; Borghi, G.L.; Pirone, C.; Thalmann, M.; Pazmino Costa, D.A.; Santelia, D.; Trost, P.; Sparla, F. β-amylase 1 (BAM1) degrades transitory starch to sustain proline biosynthesis during drought stress. J. Exp. Bot. 2016, 67, 1819–1826. [Google Scholar] [CrossRef]
- Ali, Q.; Athar, H.; Haider, M.; Shahid, S.; Aslam, N.; Shehzad, F.; Naseem, J.; Ashraf, R.; Ali, A.; Hussain, S.M. Role of amino acids in improving abiotic stress tolerance to plants. In Plant Tolerance to Environmental Stress, 1st ed.; Hasanuzzaman, M., Fujita, M., Oku, H., Tofazzal, I.M., Eds.; CRC Press: Boca Raton, FL, USA, 2019; pp. 175–204. [Google Scholar] [CrossRef]
- Binder, S. Branched-chain amino acid metabolism in Arabidopsis thaliana. Arabidopsis Book 2010, 8, e0137. [Google Scholar] [CrossRef]
- Young, D.H.; Koöhle, H.; Kauss, H. Effect of Chitosan on membrane permeability of suspension-cultured Glycine max and Phaseolus vulgaris cells. Plant Physiol. 1982, 70, 1449–1454. [Google Scholar] [CrossRef]
- Trotel-Aziz, P.; Couderchet, M.; Vernet, G.; Aziz, A. Chitosan stimulates defense reactions in grapevine leaves and inhibits development of Botrytis cinerea. Eur. J. Plant Pathol. 2006, 114, 405–413. [Google Scholar] [CrossRef]
- Rakwal, R.; Tamogami, S.; Agrawal, G.K.; Iwahashi, H. Octadecanoid signaling component “burst” in rice (Oryza sativa L) seedling leaves upon wounding by cut and treatment with fungal elicitor chitosan. Biochem. Biophys. 2002, 295, 1041–1045. [Google Scholar] [CrossRef]
- Hanano, A.; Burcklen, M.; Flenet, M.; Ivancich, A.; Louwagie, M.; Garin, J.; Blée, E. Plant seed peroxygenase is an original heme-oxygenase with an EF-hand calcium binding motif. J. Biol. Chem. 2006, 281, 33140–33151. [Google Scholar] [CrossRef]
- Meesapyodsuk, D.; Qiu, X. A peroxygenase pathway involved in the biosynthesis of epoxy fatty acids in oat. Plant Physiol. 2011, 157, 454–463. [Google Scholar] [CrossRef][Green Version]
- Hanano, A.; Almousally, I.; Shaban, M.; Rahman, F.; Hassan, M.; Murphy, D.J. Specific caleosin/peroxygenase and lipoxygenase activities are tissue-differentially expressed in date palm (Phoenix dactylifera L.) seedlings and are further induced following exposure to the toxin 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin. Front. Plant Sci. 2017, 7, 2025. [Google Scholar] [CrossRef]
- Su, H.G.; Zhang, X.H.; Wang, T.; Wei, W.L.; Wang, Y.X.; Chen, J.; Zhou, Y.B.; Chen, M.; Ma, Y.Z.; Xu, Z.S.; et al. Genome-wide identification, evolution, and expression of GDSL-type esterase/lipase gene family in soybean. Front. Plant Sci. 2020, 11, 726. [Google Scholar] [CrossRef]
- Bekker-Jensen, D.B.; Bernhardt, O.M.; Hogrebe, A.; Martinez-Val, A.; Verbeke, L.; Gandhi, T.; Olsen, J.V. Rapid and site-specific deep phosphoproteome profiling by data-independent acquisition without the need for spectral libraries. Nat. Commun. 2020, 11, 787. [Google Scholar] [CrossRef]
- Rodríguez, E.M.; Svensson, J.T.; Malatrasi, M.; Choi, D.W.; Close, T.J. Barley Dhn13 encodes a KS-type dehydrin with constitutive and stress responsive expression. Theor. Appl. Genet. 2005, 110, 852–858. [Google Scholar] [CrossRef]
- Vajrabhaya, M.; Vajrabhaya, T. Somaclonal variation for salt tolerance in rice. In Rice; Bajaj, Y.P.S., Ed.; Springer: Berlin/Heidelberg, Germany, 1991; pp. 368–382. [Google Scholar]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar]
- Nakagami, H.; Sugiyama, N.; Ishihama, Y.; Shirasu, K. Shotguns in the front line: Phosphoproteomics in plants. Plant Cell Physiol. 2012, 53, 118–124. [Google Scholar] [CrossRef]
- Thomas, P.D.; Kejariwal, A.; Campbell, M.J.; Mi, H.; Diemer, K.; Guo, N.; Ladunga, I.; Ulitsky-Lazareva, B.; Muruganujan, A.; Rabkin, S.; et al. PANTHER: A browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. Spec. Publ. 2003, 31, 334–341. [Google Scholar] [CrossRef]
- Saeed, A.I.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M.; et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques 2003, 34, 378. [Google Scholar] [CrossRef]
| Cultivar | Accession Number | Protein Name | Peptide | Peptide Sequence and Predicted Phosphorylation Site (*) | Score | Intensity | Functional Category | |
|---|---|---|---|---|---|---|---|---|
| CON | CTS | |||||||
| LPT123 | BAS81451.1 | Adenylyltransferase and sulfurtransferase | AINSSIK | AINS*SIK | 0.598 | 0.0000 | 16.7085 | Signal transduction |
| BAS81893.1 | Os03g0108200 | FAMASRPR | FAMASRPR | 0.0000 | 16.9662 | Signal transduction | ||
| BAT07207.1 | Receptor-like protein kinase | VLLELVHGRK | VLLELVHGRK | 0.0000 | 22.9446 | Signal transduction | ||
| ACM17588.1 | NBS-LRR disease resistance protein | NLRYLNLAR | NLRY*LNLAR | 0.879 | 0.0000 | 17.6093 | Defense response | |
| AAX95751.1 | WAT1-related protein | MTSSSLK | MTSSSLK | 0.0000 | 18.0284 | Transport | ||
| AAX95531.1 | Swi1 | VRYIGR | VRYIGR | 0.0000 | 15.5903 | Transcription | ||
| BAT16440.1 | KAT8 regulatory NSL complex | GLLQLHSLYR | GLLQLHS*LYR | 0.537 | 0.0000 | 17.5308 | Transcription | |
| XP_015620894.1 | DEAD-box ATP-dependent RNA helicase | QRQAVQTK | QRQAVQT*K | 0.693 | 0.0000 | 21.8363 | Transcription | |
| XP_015621184.1 | La-related protein 1A isoform X1 | VPDSQR | VPDS*QR | 0.754 | 0.0000 | 13.6500 | Transcription | |
| XP_015642930.1 | Septum site-determining protein | AGFFSFFGG | AGFFS*FFGG | 0.556 | 0.0000 | 14.8483 | Transcription | |
| BAF08718.2 | ATP-dependent DNA helicase | KFEHEPK | KFEHEPK | 0.0000 | 16.1209 | Translation | ||
| BAS70391.1 | E3 ubiquitin-protein ligase | DLLNATKR | DLLNATKR | 0.0000 | 18.5204 | Protein degradation | ||
| XP_015640159.1 | Arginyl-tRNA--protein transferase 2 | QSSVNKNTVR | QSSVNKNTVR | 0.0000 | 17.3091 | Protein degradation | ||
| XP_015641088.1 | Peroxygenase 4 | MASKPADVTATGGGGVAVVR | MAS*KPADVTATGGGGVAVVR | 0.520 | 0.0000 | 17.6856 | Metabolic process | |
| KDML105 | BAD53112.1 | Chalcone synthase | QIGDSIGR | QIGDS*IGR | 0.762 | 0.0000 | 20.6759 | Defense response |
| BAF29919.2 | Phenylalanine ammonia-lyase | VDAAEAFR | VDAAEAFR | 0.0000 | 17.6121 | Defense response | ||
| CAE03366.1 | Osjnbb0065l13.9 | AGMAVWMRR | AGMAVWMRR | 0.0000 | 18.5368 | Transcription | ||
| CAE04537.2 | Osjnba0040d17.5 | YILSAPILKGR | YILSAPILKGR | 0.0000 | 16.7900 | Transcription | ||
| CAE05823.1 | Osjnba0028m15.15 | VHKDYK | VHKDYK | 0.0000 | 17.3996 | Transcription | ||
| ABF99104.1 | Ribosomal protein s14p/S29e | NLSFFMADK | NLSFFMADK | 0.0000 | 17.5785 | Translation | ||
| XP_015622394.1 | UDP-N-acetylglucosamine O-acyltransferase | RLLIASR | RLLIASR | 0.0000 | 18.1314 | Metabolic process | ||
| Cultivar | Accession Number | Protein Name | Peptide | Peptide Sequence and Predicted Phosphorylation Site (*) | Score | Intensity | Functional Category | |
|---|---|---|---|---|---|---|---|---|
| CON+PEG | CTS+PEG | |||||||
| LPT123 | AAK13126.1 | Histidine-kinase-like protein | AEVTMYHLR | AEVTMYHLR | 0.0000 | 19.2691 | Signal transduction | |
| AAX95871.1 | Protein kinase domain | KVVEHNGK | KVVEHNGK | 0.0000 | 13.8992 | Signal transduction | ||
| BAS86337.1 | WD-domain-containing protein | LVIFDG | LVIFDG | 0.0000 | 16.1930 | Signal transduction | ||
| BAF05127.1 | Disease resistance protein RPM1 | IGGMR | IGGMR | 0.0000 | 17.5347 | Defense response | ||
| XP_015611347.1 | ABC transporter C family member 4 | SSLLGCILGEMR | SS*LLGCILGEMR | 0.532 | 0.0000 | 20.2736 | Transport | |
| XP_015612553.1 | Cytochrome b561 | SGDTSSR | SGDT*S*S*R | 0.667, 0.984, 0.670 | 0.0000 | 17.7423 | Transport | |
| BAF08853.2 | DNA polymerase epsilon catalytic subunit | EEGVLLK | EEGVLLK | 0.0000 | 16.4910 | Replication | ||
| AFI71858.1 | Beta-amylase 1, chloroplastic | MSESGSPR | MS*ESGSPR | 0.566 | 0.0000 | 16.3572 | Metabolic process | |
| BAS93628.1 | Chaperone protein dnaJ 49 | LTKGMDGNK | LT*KGMDGNK | 0.629 | 0.0000 | 16.7405 | Protein folding | |
| BAS95271.1 | T-complex protein | DPPVFLRI | DPPVFLRI | 0.0000 | 16.4615 | Protein folding | ||
| KDML105 | XP_015617069.1 | Receptor-like serine/threonine-protein kinase | TAQAK | T*AQAK | 0.621 | 0.0000 | 16.2963 | Signal transduction |
| XP_015624189.1 | B3 domain-containing protein Os02g0598200 | TSNQNGEKNMK | T*SNQNGEKNMK | 0.660 | 0.0000 | 18.3402 | Signal transduction | |
| AAQ74383.1 | Na/H antiporter | SLHSPLLTR | S*LHS*PLLT*R | 0.567, 0.596, 0.524 | 0.0000 | 19.2654 | Transport | |
| B9G2A8.1 | Auxin transport protein BIG | KLGSSILSSR | KLGS*SILS*S*R | 0.731, 0.853, 0.899 | 0.0000 | 19.2937 | Transport | |
| XP_015642960.1 | Membrane protein of ER body-like protein | AGLKVITIIDK | AGLKVITIIDK | 0.0000 | 21.3910 | Transport | ||
| XP_015626888.1 | Serine/arginine-rich SC35-like splicing factor SCL30 | EHEVDK | EHEVDK | 0.0000 | 20.8443 | Transcription | ||
| AAT77858.1 | Translational activator | AILGGSEGK | AILGGSEGK | 0.0000 | 18.8387 | Translation | ||
| ABA95184.1 | Lipase family protein | DVLTLVTK | DVLT*LVTK | 0.511 | 0.0000 | 16.2153 | Metabolic process | |
| KDML105 | ABF96062.1 | Branched-chain amino acid aminotransferase | RNSPNSIDSK | RNS*PNS*IDS*K | 0.734, 0.985, 0.993 | 0.0000 | 19.0565 | Metabolic process |
| XP_015613842.1 | U-box domain-containing protein 45 | GSSCK | GSS*CK | 0.810 | 0.0000 | 19.7904 | Protein degradation | |
| XP_015627057.1 | Ubiquitin carboxyl-terminal hydrolase 15 | VEALKKPSK | VEALKKPS*K | 0.980 | 0.0000 | 20.0381 | Protein degradation | |
| XP_015651426.1 | Polyadenylation specificity factor | HLGAAQIDR | HLGAAQIDR | 0.0000 | 20.2841 | Metabolic process | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pongprayoon, W.; Panya, A.; Jaresitthikunchai, J.; Phaonakrop, N.; Roytrakul, S. Phosphoprotein Profile of Rice (Oryza sativa L.) Seedlings under Osmotic Stress after Pretreatment with Chitosan. Plants 2022, 11, 2729. https://doi.org/10.3390/plants11202729
Pongprayoon W, Panya A, Jaresitthikunchai J, Phaonakrop N, Roytrakul S. Phosphoprotein Profile of Rice (Oryza sativa L.) Seedlings under Osmotic Stress after Pretreatment with Chitosan. Plants. 2022; 11(20):2729. https://doi.org/10.3390/plants11202729
Chicago/Turabian StylePongprayoon, Wasinee, Atikorn Panya, Janthima Jaresitthikunchai, Narumon Phaonakrop, and Sittiruk Roytrakul. 2022. "Phosphoprotein Profile of Rice (Oryza sativa L.) Seedlings under Osmotic Stress after Pretreatment with Chitosan" Plants 11, no. 20: 2729. https://doi.org/10.3390/plants11202729
APA StylePongprayoon, W., Panya, A., Jaresitthikunchai, J., Phaonakrop, N., & Roytrakul, S. (2022). Phosphoprotein Profile of Rice (Oryza sativa L.) Seedlings under Osmotic Stress after Pretreatment with Chitosan. Plants, 11(20), 2729. https://doi.org/10.3390/plants11202729







