CRISPR-Cas9 Gene Editing of the Sal1 Gene Family in Wheat
Abstract
1. Introduction
2. Results
2.1. Sal1 Gene Family in Wheat
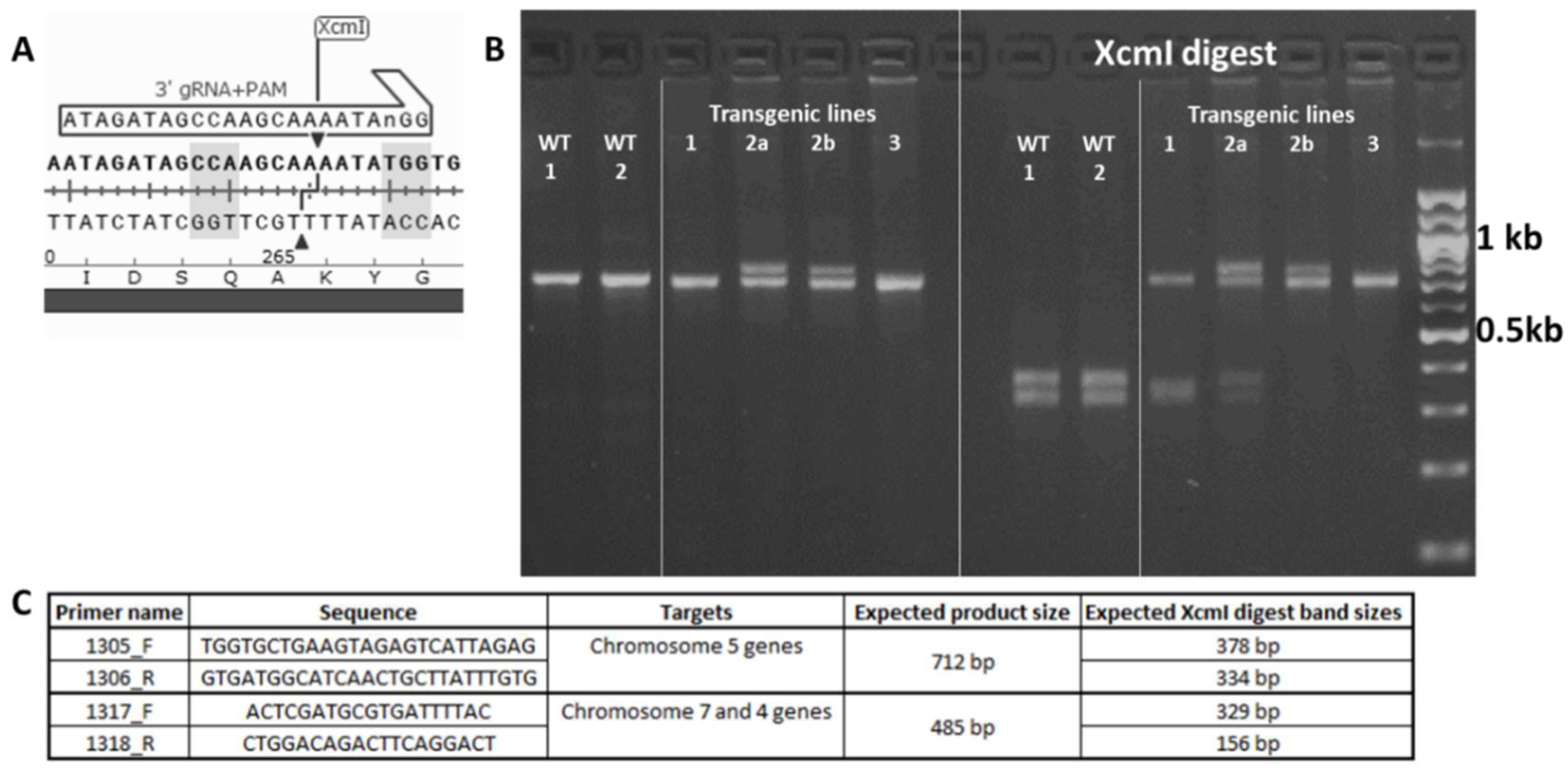
2.2. Design and Transformation of the CRISPR-Cas9 Sal1 Knockout Construct
2.3. Identification of Wheat Sal1 Mutant Lines
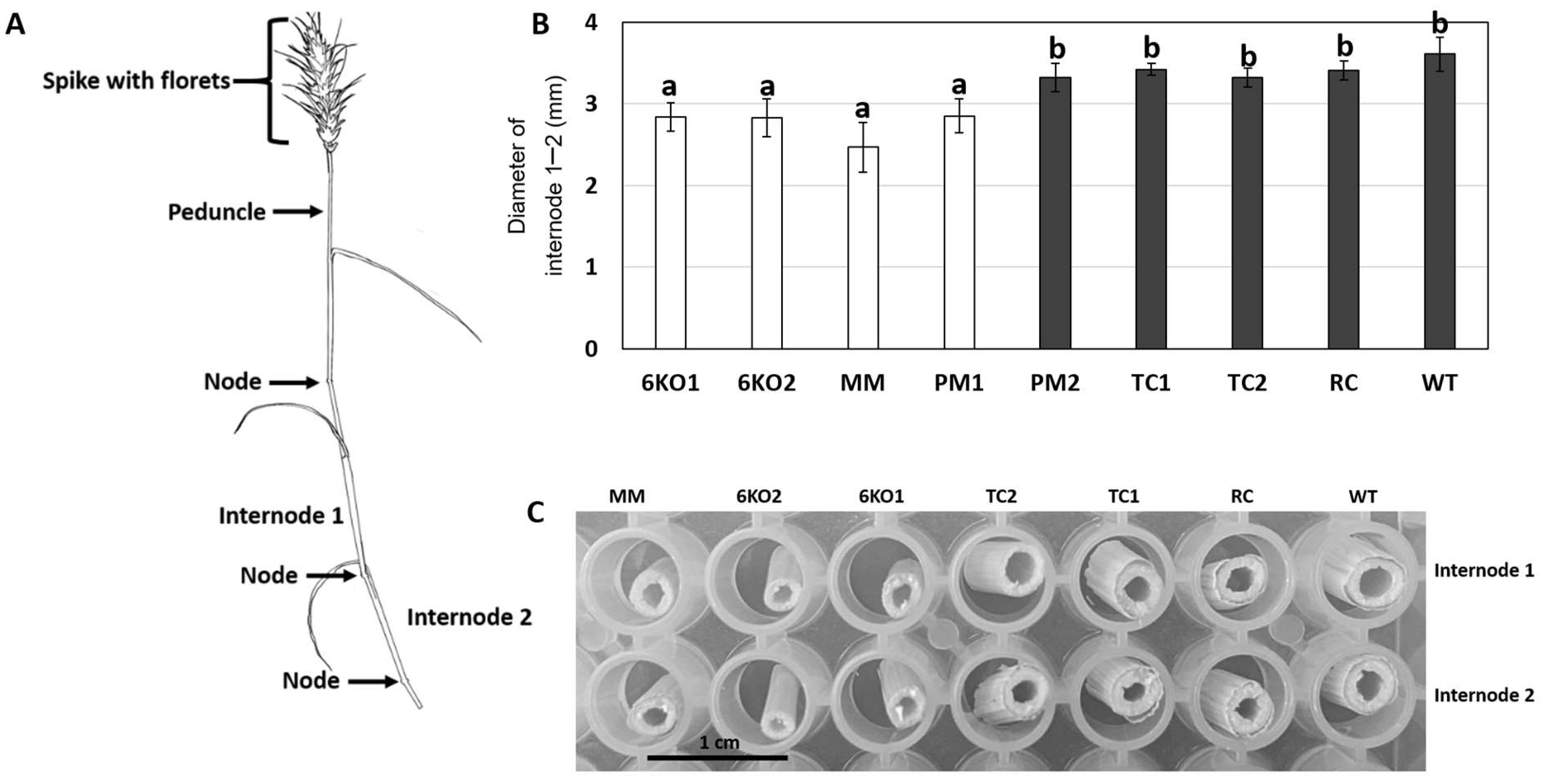
2.4. Morphological Characterization of the Sal1 Mutant Plants
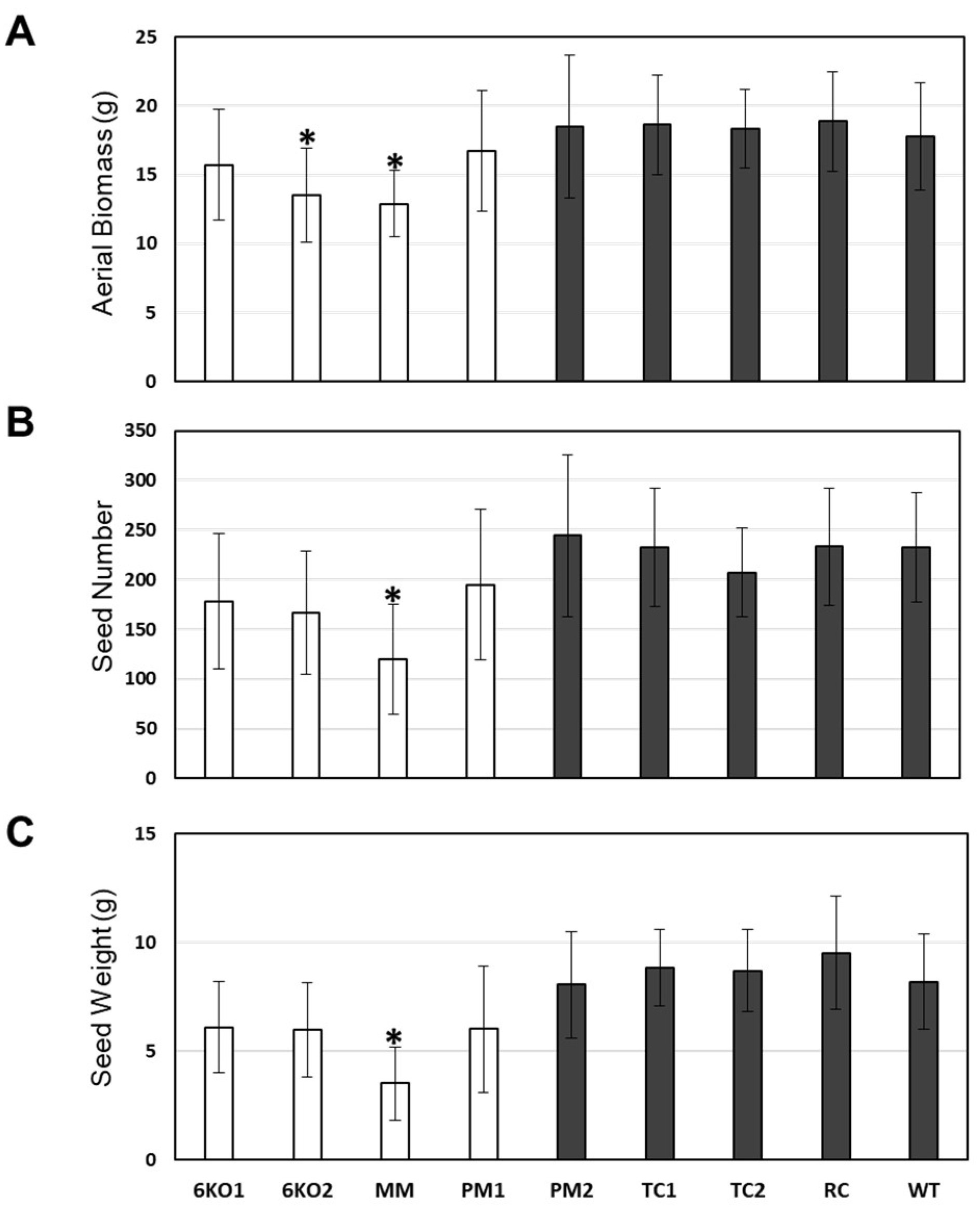
2.5. Characterization of the Stress Tolerance of the Sal1 Mutant Plants
3. Discussion
4. Materials and Methods
4.1. Identification, Cloning, and Sequencing of Bobwhite Wheat Sal1
4.2. CRISPR Sal1 Transformation Construct
4.3. Bobwhite Wheat Biolistic Transformation
4.4. Identification of Events carrying Sal1 Mutations
4.5. Quantitative Reverse Transcriptase-PCR
4.6. Growth and Characterization of Transgenic and Mutant Plants
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shewry, P.R. Wheat. J. Exp. Bot. 2009, 60, 1537–1553. [Google Scholar] [CrossRef]
- Ani Akpinar, B.; Avsar, B.; Lucas, S.J.; Budak, H. Plant Abiotic Stress Signaling. Plant Signal. Behav. 2012, 7, 1450–1455. [Google Scholar] [CrossRef]
- Reynolds, M.P.; Lewis, J.M.; Ammar, K.; Basnet, B.R.; Crespo-Herrera, L.; Crossa, J.; Dhugga, K.S.; Dreisigacker, S.; Juliana, P.; Karwat, H.; et al. Harnessing Translational Research in Wheat for Climate Resilience. J. Exp. Bot. 2021, 72, 5134–5157. [Google Scholar] [CrossRef]
- Budak, H.; Hussain, B.; Khan, Z.; Ozturk, N.Z.; Ullah, N. From Genetics to Functional Genomics: Improvement in Drought Signaling and Tolerance in Wheat. Front. Plant Sci. 2015, 6, 1012. [Google Scholar] [CrossRef]
- Pandey, D.M.; Hu, Y.; Shavrukov, Y.; Gupta, N.K. Editorial: Drought Threat: Responses and Molecular-Genetic Mechanisms of Adaptation and Tolerance in Wheat. Front. Plant Sci. 2022, 13, 3389–3391. [Google Scholar] [CrossRef]
- Bapela, T.; Shimelis, H.; Tsilo, T.J.; Mathew, I. Genetic Improvement of Wheat for Drought Tolerance: Progress, Challenges and Opportunities. Plants 2022, 11, 1331. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. The New Frontier of Genome Engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Belhaj, K.; Chaparro-Garcia, A.; Kamoun, S.; Patron, N.J.; Nekrasov, V. Editing Plant Genomes with CRISPR/Cas9. Curr. Opin. Biotechnol. 2015, 32, 76–84. [Google Scholar] [CrossRef]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef]
- Sander, J.D.; Joung, J.K. CRISPR-Cas Systems for Editing, Regulating and Targeting Genomes. Nat. Biotechnol. 2014, 32, 347–350. [Google Scholar] [CrossRef]
- Ding, Y.; Li, H.; Chen, L.L.; Xie, K. Recent Advances in Genome Editing Using CRISPR/Cas9. Front. Plant Sci. 2016, 7, 703. [Google Scholar] [CrossRef] [PubMed]
- Matres, J.M.; Hilscher, J.; Datta, A.; Armario-Nájera, V.; Baysal, C.; He, W.; Huang, X.; Zhu, C.; Valizadeh-Kamran, R.; Trijatmiko, K.R.; et al. Genome Editing in Cereal Crops: An Overview. Transgenic Res. 2021, 30. ISBN 0123456789. [Google Scholar] [CrossRef] [PubMed]
- Awan, M.J.A.; Pervaiz, K.; Rasheed, A.; Amin, I.; Saeed, N.A.; Dhugga, K.S.; Mansoor, S. Genome Edited Wheat- Current Advances for the Second Green Revolution. Biotechnol. Adv. 2022, 60, 108006. [Google Scholar] [CrossRef] [PubMed]
- Rao, Y.; Yang, X.; Pan, C.; Wang, C.; Wang, K. Advance of Clustered Regularly Interspaced Short Palindromic Repeats-Cas9 System and Its Application in Crop Improvement. Front. Plant Sci. 2022, 13, 839001. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-F.; Norville, J.E.; Aach, J.; McCormack, M.; Zhang, D.; Bush, J.; Church, G.M.; Sheen, J. Multiplex and Homologous Recombination–Mediated Genome Editing in Arabidopsis and Nicotiana Benthamiana Using Guide RNA and Cas9. Nat. Biotechnol. 2013, 31, 688–691. [Google Scholar] [CrossRef]
- Nekrasov, V.; Staskawicz, B.; Weigel, D.; Jones, J.D.G.; Kamoun, S. Targeted Mutagenesis in the Model Plant Nicotiana Benthamiana Using Cas9 RNA-Guided Endonuclease. Nat. Biotechnol. 2013, 31, 691–693. [Google Scholar] [CrossRef]
- Jiang, W.; Zhou, H.; Bi, H.; Fromm, M.; Yang, B.; Weeks, D.P. Demonstration of CRISPR/Cas9/SgRNA-Mediated Targeted Gene Modification in Arabidopsis, Tobacco, Sorghum and Rice. Nucleic Acids Res. 2013, 41, e188. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Gui, J.-L.; et al. Targeted Genome Modification of Crop Plants Using a CRISPR-Cas System. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar] [CrossRef]
- Miao, J.; Guo, D.; Zhang, J.; Huang, Q.; Qin, G.; Zhang, X.; Wan, J.; Gu, H.; Qu, L.J. Targeted Mutagenesis in Rice Using CRISPR-Cas System. Cell Res. 2013, 23, 1233–1236. [Google Scholar] [CrossRef]
- Upadhyay, S.K.; Kumar, J.; Alok, A.; Tuli, R. RNA-Guided Genome Editing for Target Gene Mutations in Wheat. G3 Genes, Genomes, Genet. 2013, 3, 2233–2238. [Google Scholar] [CrossRef]
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J.L. Simultaneous Editing of Three Homoeoalleles in Hexaploid Bread Wheat Confers Heritable Resistance to Powdery Mildew. Nat. Biotechnol. 2014, 32, 947–951. [Google Scholar] [CrossRef]
- Zhang, Y.; Liang, Z.; Zong, Y.; Wang, Y.; Liu, J.; Chen, K.; Qiu, J.L.; Gao, C. Efficient and Transgene-Free Genome Editing in Wheat through Transient Expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 2016, 7, 12617. [Google Scholar] [CrossRef]
- Li, J.; Zhang, S.; Zhang, R.; Gao, J.; Qi, Y.; Song, G.; Li, W.; Li, Y.; Li, G. Efficient Multiplex Genome Editing by CRISPR/Cas9 in Common Wheat. Plant Biotechnol. J. 2021, 19, 427–429. [Google Scholar] [CrossRef]
- Kottakota, C.; Pradhan, B.; Roychowdhury, R.; Dubey, V.K. Improvement of Wheat (Triticum spp.) Through Genetic Manipulation. In Genetically Modified Crops: Current Status, Prospects and Challenges; Kavi Kishor, P.B., Rajam, M.V., Pullaiah, T., Eds.; Springer Nature: Singapore, 2021; pp. 33–66. ISBN 9789811558979. [Google Scholar]
- Quintero, F.J.; Garcideblas, B.; Rodriguez-Navarro, A. The SAL1 Gene of Arabidopsis, Encoding an Enzyme with 3′(2′),5′-Bisphosphate Nucleotidase and Inositol Polyphosphate 1-Phosphatase Activities, Increases Salt Tolerance in Yeast. Plant Cell 1996, 8, 529–537. [Google Scholar] [CrossRef]
- Xiong, L.; Lee, H.; Huang, R.; Zhu, J.K. A Single Amino Acid Substitution in the Arabidopsis FIERY1/HOS2 Protein Confers Cold Signaling Specificity and Lithium Tolerance. Plant J. 2004, 40, 536–545. [Google Scholar] [CrossRef]
- Wilson, P.B.; Estavillo, G.M.; Field, K.J.; Pornsiriwong, W.; Carroll, A.J.; Howell, K.A.; Woo, N.S.; Lake, J.A.; Smith, S.M.; Harvey Millar, A.; et al. The Nucleotidase/Phosphatase SAL1 Is a Negative Regulator of Drought Tolerance in Arabidopsis. Plant J. 2009, 58, 299–317. [Google Scholar] [CrossRef]
- Estavillo, G.M.; Crisp, P.A.; Pornsiriwong, W.; Wirtz, M.; Collinge, D.; Carrie, C.; Giraud, E.; Whelan, J.; David, P.; Javot, H.; et al. Evidence for a SAL1-PAP Chloroplast Retrograde Pathway That Functions in Drought and High Light Signaling in Arabidopsis. Plant Cell 2011, 23, 3992–4012. [Google Scholar] [CrossRef]
- Chan, K.X.; Mabbitt, P.D.; Phua, S.Y.; Mueller, J.W.; Nisar, N.; Gigolashvili, T.; Stroeher, E.; Grassl, J.; Arlt, W.; Estavillo, G.M.; et al. Sensing and Signaling of Oxidative Stress in Chloroplasts by Inactivation of the SAL1 Phosphoadenosine Phosphatase. Proc. Natl. Acad. Sci. USA 2016, 113, E4567–E4576. [Google Scholar] [CrossRef] [PubMed]
- Shirzadian-Khorramabad, R.; Moazzenzadeh, T.; Sajedi, R.H.; Jing, H.C.; Hille, J.; Dijkwel, P.P. A Mutation in Arabidopsis SAL1 Alters Its in Vitro Activity against IP3 and Delays Developmental Leaf Senescence in Association with Lower ROS Levels. Plant Mol. Biol. 2022, 108, 549–563. [Google Scholar] [CrossRef]
- Manmathan, H.; Shaner, D.; Snelling, J.; Tisserat, N.; Lapitan, N. Virus-Induced Gene Silencing of Arabidopsis Thaliana Gene Homologues in Wheat Identifies Genes Conferring Improved Drought Tolerance. J. Exp. Bot. 2013, 64, 1381–1392. [Google Scholar] [CrossRef]
- Zhao, C.; Wang, Y.; Chan, K.X.; Marchant, D.B.; Franks, P.J.; Randall, D.; Tee, E.E.; Chen, G.; Ramesh, S.; Phua, S.Y.; et al. Evolution of Chloroplast Retrograde Signaling Facilitates Green Plant Adaptation to Land. Proc. Natl. Acad. Sci. USA 2019, 116, 5015–5020. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhu, T.; Rodriguez, J.C.; Deal, K.R.; Dubcovsky, J.; McGuire, P.E.; Lux, T.; Spannagl, M.; Mayer, K.F.X.; Baldrich, P.; et al. Aegilops Tauschii Genome Assembly Aet v5.0 Features Greater Sequence Contiguity and Improved Annotation. G3 Genes Genomes. Genet. 2021, 11, 12. [Google Scholar] [CrossRef]
- Zhou, Y.; Bai, S.; Li, H.; Sun, G.; Zhang, D.; Ma, F.; Zhao, X.; Nie, F.; Li, J.; Chen, L.; et al. Introgressing the Aegilops Tauschii Genome into Wheat as a Basis for Cereal Improvement. Nat. Plants 2021, 7, 774–786. [Google Scholar] [CrossRef]
- Zhu, T.; Wang, L.; Rodriguez, J.C.; Deal, K.R.; Avni, R.; Distelfeld, A.; McGuire, P.E.; Dvorak, J.; Luo, M.C. Improved Genome Sequence of Wild Emmer Wheat Zavitan with the Aid of Optical Maps. G3 Genes Genomes Genet. 2019, 9, 619–624. [Google Scholar] [CrossRef]
- Scott, M.F.; Botigué, L.R.; Brace, S.; Stevens, C.J.; Mullin, V.E.; Stevenson, A.; Thomas, M.G.; Fuller, D.Q.; Mott, R. A 3,000-Year-Old Egyptian Emmer Wheat Genome Reveals Dispersal and Domestication History. Nat. Plants 2019, 5, 1120–1128. [Google Scholar] [CrossRef]
- Maccaferri, M.; Harris, N.S.; Twardziok, S.O.; Pasam, R.K.; Gundlach, H.; Spannagl, M.; Ormanbekova, D.; Lux, T.; Prade, V.M.; Milner, S.G.; et al. Durum Wheat Genome Highlights Past Domestication Signatures and Future Improvement Targets. Nat. Genet. 2019, 51, 885–895. [Google Scholar] [CrossRef]
- Walkowiak, S.; Gao, L.; Monat, C.; Haberer, G.; Kassa, M.T.; Brinton, J.; Ramirez-Gonzalez, R.H.; Kolodziej, M.C.; Delorean, E.; Thambugala, D.; et al. Multiple Wheat Genomes Reveal Global Variation in Modern Breeding. Nature 2020, 588, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Wang, L.; Rimbert, H.; Rodriguez, J.C.; Deal, K.R.; De Oliveira, R.; Choulet, F.; Keeble-Gagnère, G.; Tibbits, J.; Rogers, J.; et al. Optical Maps Refine the Bread Wheat Triticum Aestivum Cv. Chinese Spring Genome Assembly. Plant J. 2021, 107, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Abe, F.; Mascher, M.; Haberer, G.; Gundlach, H.; Spannagl, M.; Shirasawa, K.; Isobe, S. Chromosome-Scale Genome Assembly of the Transformation-Amenable Common Wheat Cultivar ’Fielder. DNA Res. 2021, 28, 1–7. [Google Scholar] [CrossRef]
- Prykhozhij, S.V.; Rajan, V.; Gaston, D.; Berman, J.N. CRISPR Multitargeter: A Web Tool to Find Common and Unique CRISPR Single Guide RNA Targets in a Set of Similar Sequences. PLoS ONE 2015, 10, e0119372. [Google Scholar] [CrossRef]
- Čermák, T.; Curtin, S.J.; Gil-Humanes, J.; Čegan, R.; Kono, T.J.Y.Y.; Konečná, E.; Belanto, J.J.; Starker, C.G.; Mathre, J.W.; Greenstein, R.L.; et al. A Multipurpose Toolkit to Enable Advanced Genome Engineering in Plants. Plant Cell 2017, 29, 1196–1217. [Google Scholar] [CrossRef]
- Hahn, F.; Sanjurjo Loures, L.; Sparks, C.A.; Kanyuka, K.; Nekrasov, V. Efficient Crispr/Cas-mediated Targeted Mutagenesis in Spring and Winter Wheat Varieties. Plants 2021, 10, 1481. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.; Turesson, H.; Nicolia, A.; Fält, A.S.; Samuelsson, M.; Hofvander, P. Efficient Targeted Multiallelic Mutagenesis in Tetraploid Potato (Solanum Tuberosum) by Transient CRISPR-Cas9 Expression in Protoplasts. Plant Cell Rep. 2017, 36, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Banakar, R.; Eggenberger, A.L.; Lee, K.; Wright, D.A.; Murugan, K.; Zarecor, S.; Lawrence-Dill, C.J.; Sashital, D.G.; Wang, K. High-Frequency Random DNA Insertions upon Co-Delivery of CRISPR-Cas9 Ribonucleoprotein and Selectable Marker Plasmid in Rice. Sci. Rep. 2019, 9, 19902. [Google Scholar] [CrossRef] [PubMed]
- Lawlor, D.W. Genetic Engineering to Improve Plant Performance under Drought: Physiological Evaluation of Achievements, Limitations, and Possibilities. J. Exp. Bot. 2013, 64, 83–108. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-New Capabilities and Interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Christensen, A.H.; Quail, P.H. Ubiquitin Promoter-Based Vectors for High-Level Expression of Selectable and/or Screenable Marker Genes in Monocotyledonous Plants. Transgenic Res. 1996, 5, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Weeks, J.T.; Anderson, O.D.; Blechl, A.E. Rapid Production of Multiple Independent Lines of Fertile Transgenic Wheat (Triticum aestivum). Plant Physiol. 1993, 102, 1077–1084. [Google Scholar] [CrossRef]
- Okubara, P.A.; Blechl, A.E.; McCormick, S.P.; Alexander, N.J.; Dill-Macky, R.; Hohn, T.M. Engineering Deoxynivalenol Metabolism in Wheat through the Expression of a Fungal Trichothecene Acetyltransferase Gene. Theor. Appl. Genet. 2002, 106, 74–83. [Google Scholar] [CrossRef]
- Thilmony, R.; Guttman, M.E.; Lin, J.W.; Blechl, A.E. The Wheat HMW-Glutenin 1Dy10 Gene Promoter Controls Endosperm Expression in Brachypodium Distachyon. GM Crop. Food Biotechnol. Agric. Food Chain 2014, 5, 36–43. [Google Scholar] [CrossRef][Green Version]
- Rajasekaran, K.; Majumdar, R.; Sickler, C.; Wei, Q.; Cary, J.; Bhatnagar, D. Fidelity of a Simple Liberty Leaf-Painting Assay to Validate Transgenic Maize Plants Expressing the Selectable Marker Gene, Bar. J. Crop Improv. 2017, 31, 628–636. [Google Scholar] [CrossRef]
- Dudziak, K.; Sozoniuk, M.; Szczerba, H.; Kuzdraliński, A.; Kowalczyk, K.; Börner, A.; Nowak, M. Identification of Stable Reference Genes for QPCR Studies in Common Wheat (Triticum aestivum L.) Seedlings under Short-Term Drought Stress. Plant Methods 2020, 16, 58. [Google Scholar] [CrossRef] [PubMed]



| Species | Genomes | Accession Name (Year Sequence Released) [Citation] | Sal1 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 5A | 5B | 5D | 4A-1 | 4A-2 | 7A | 7D | |||
| Aegilops tauschii | DD | AL8/78, AY61, AY17, XJ02, T093 (2021) [33,34] | + | + | |||||
| Triticum turgidum subsp. dicoccoides | AABB | Zavitan (2019) [35,36] | + | + | + | ||||
| Triticum turgidum subsp. durum | AABB | Svevo (2019) [37] | + | + | + | + | + | ||
| Triticum turgidum subsp. Durum | AABB | Kronos (2017) | + | + | + | + | + | ||
| Triticum spelta | AABBDD | PI190962 (2020) [38] | + | + | + | + | + | + | + |
| Triticum aestivum | AABBDD | Chinese Spring (2021) [39] | + | + | + | + | + | + | + |
| Triticum aestivum | AABBDD | Norin 61 (2020) [38] | + | + | + | + | + | + | + |
| Triticum aestivum | AABBDD | ArinaLrFor (2020) [38] | + | + | + | + | + | + | |
| Triticum aestivum | AABBDD | Jagger (2020) [38] | + | + | + | + | + | + | |
| Triticum aestivum | AABBDD | Julius (2020) [38] | + | + | + | + | + | + | |
| Triticum aestivum | AABBDD | LongReach Lancer (2020) [38] | + | + | + | + | + | + | |
| Triticum aestivum | AABBDD | Mace (2020) [38] | + | + | + | + | + | + | |
| Triticum aestivum | AABBDD | SY Mattis (2020) [38] | + | + | + | + | + | + | |
| Triticum aestivum | AABBDD | Fielder (2021) [40] | + | + | + | + | + | + | |
| Triticum aestivum | AABBDD | Bobwhite (this study) | + | + | + | + | + | + | |
| Triticum aestivum | AABBDD | CDC Landmark (2020) [38] | + | + | + | + | + | ||
| Triticum aestivum | AABBDD | CDC Stanley (2020) [38] | + | + | + | + | + |
| Name of Line | Line Description | Sal1 | Genotype Method | Phenotype | Transgenes Present | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 5B | 5D | 4A-1 | 4A-2 | 7A | 7D | |||||
| WT | wildtype Bobwhite | WT | WT | WT | WT | WT | WT | CAPS and sequence | WT | no |
| RC | regeneration control | WT | WT | WT | WT | WT | WT | CAPS | WT | no |
| TC1 | transgenic control 1 | WT | WT | WT | WT | WT | WT | CAPS | WT | yes |
| TC2 | transgenic control 2 | WT | WT | WT | WT | WT | WT | CAPS | WT | yes |
| PM1 | partial mutant 1 | WT | WT | not amplified | WT | mutant * | WT | CAPS and sequence | mutant stems | no |
| PM2 | partial mutant 2 | WT | mutant | not amplified | not amplified | WT | not amplified | CAPS and sequence | WT | no |
| MM | multiple mutant | mutant | mutant | mutant* | mutant | mutant | mutant | CAPS and sequence | mutant stems | yes |
| 6KO1 | 6 gene knockout 1 | mutant | mutant | mutant | mutant | mutant | mutant | CAPS and sequence | mutant stems | yes |
| 6KO2 | 6 gene knockout 2 | mutant | mutant | mutant | mutant | mutant | mutant | CAPS and sequence | mutant stems | yes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohr, T.; Horstman, J.; Gu, Y.Q.; Elarabi, N.I.; Abdallah, N.A.; Thilmony, R. CRISPR-Cas9 Gene Editing of the Sal1 Gene Family in Wheat. Plants 2022, 11, 2259. https://doi.org/10.3390/plants11172259
Mohr T, Horstman J, Gu YQ, Elarabi NI, Abdallah NA, Thilmony R. CRISPR-Cas9 Gene Editing of the Sal1 Gene Family in Wheat. Plants. 2022; 11(17):2259. https://doi.org/10.3390/plants11172259
Chicago/Turabian StyleMohr, Toni, James Horstman, Yong Q. Gu, Nagwa I. Elarabi, Naglaa A. Abdallah, and Roger Thilmony. 2022. "CRISPR-Cas9 Gene Editing of the Sal1 Gene Family in Wheat" Plants 11, no. 17: 2259. https://doi.org/10.3390/plants11172259
APA StyleMohr, T., Horstman, J., Gu, Y. Q., Elarabi, N. I., Abdallah, N. A., & Thilmony, R. (2022). CRISPR-Cas9 Gene Editing of the Sal1 Gene Family in Wheat. Plants, 11(17), 2259. https://doi.org/10.3390/plants11172259






