Transcriptome Analysis of Eggplant under Salt Stress: AP2/ERF Transcription Factor SmERF1 Acts as a Positive Regulator of Salt Stress
Abstract
:1. Introduction
2. Results
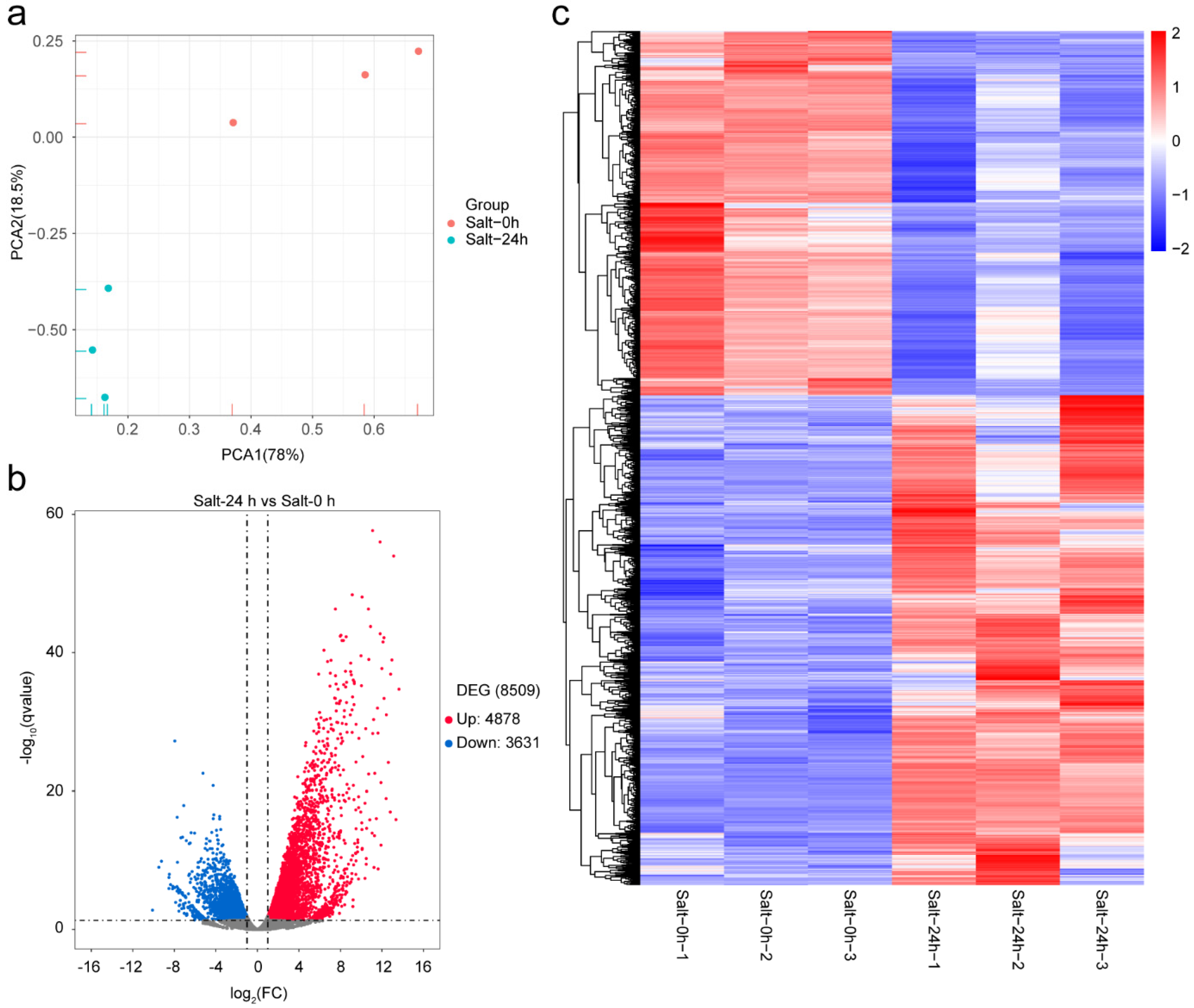
2.1. Sequencing Data Summary and DEGs Analysis
2.2. GO Analysis of DEGs
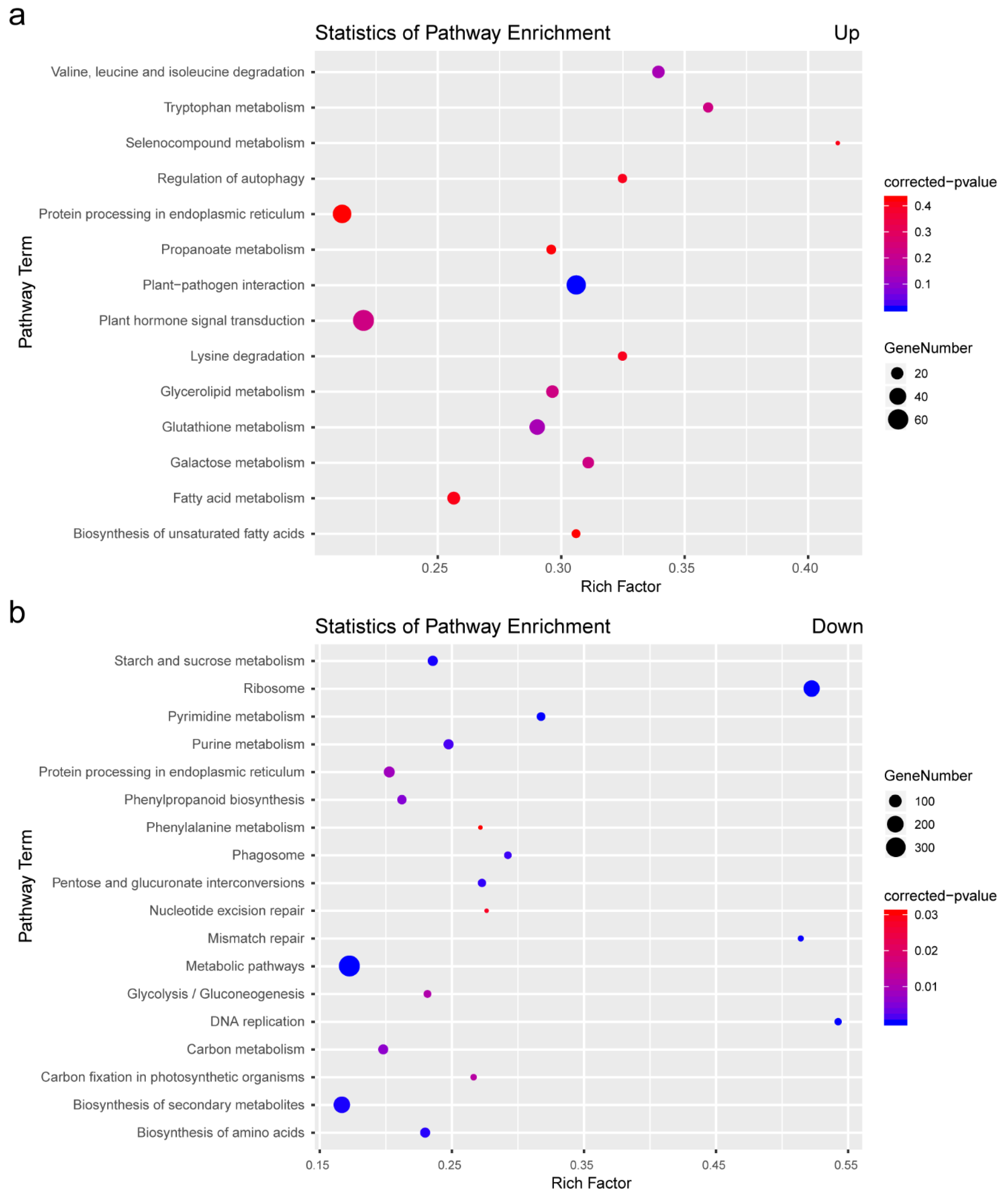
2.3. KEGG Analysis of DEGs
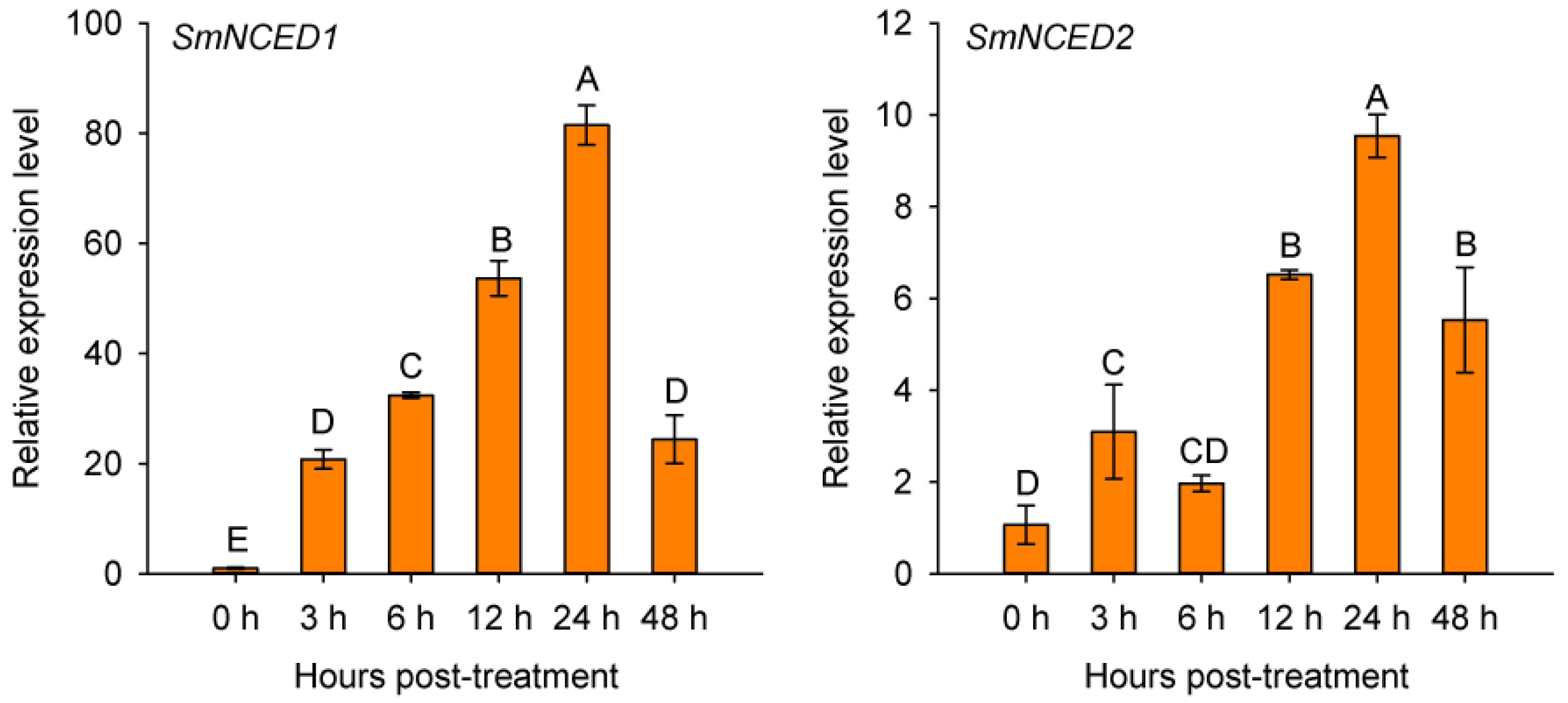
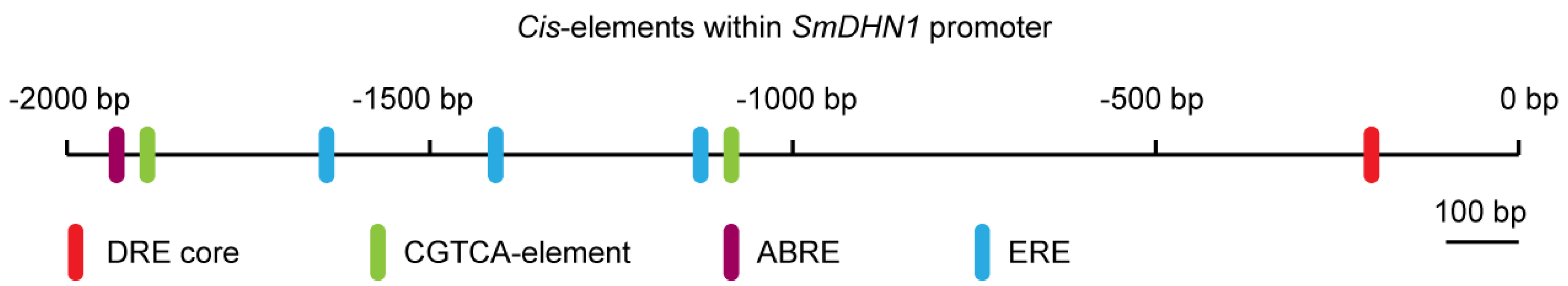
2.4. Sequence Analysis of SmERF1
2.5. Subcellular Localization of SmERF1
2.6. Transcriptional Activation Activity Analysis
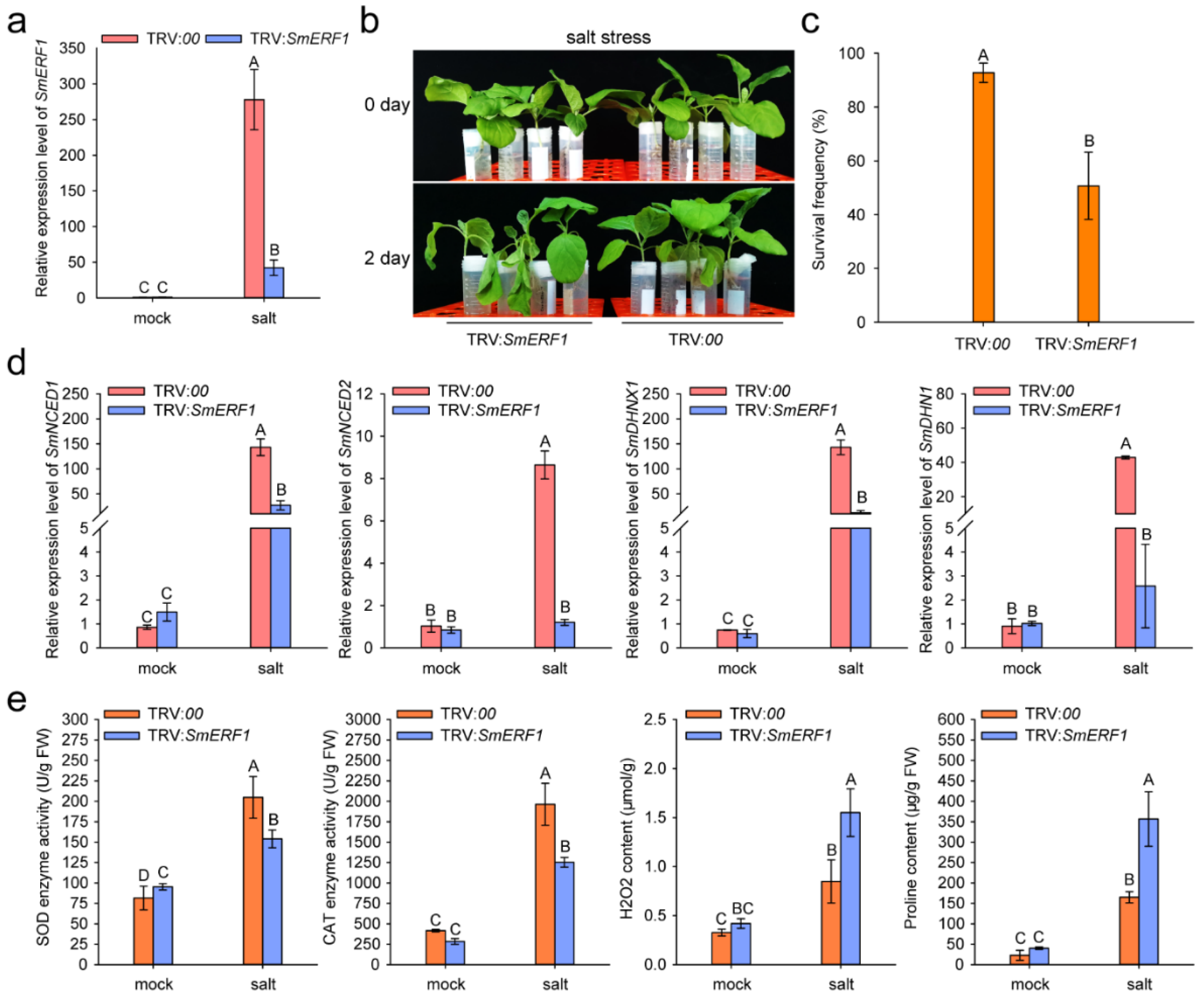
2.7. SmERF1 Silencing Enhanced the Susceptibility of Eggplant to Salt Stress
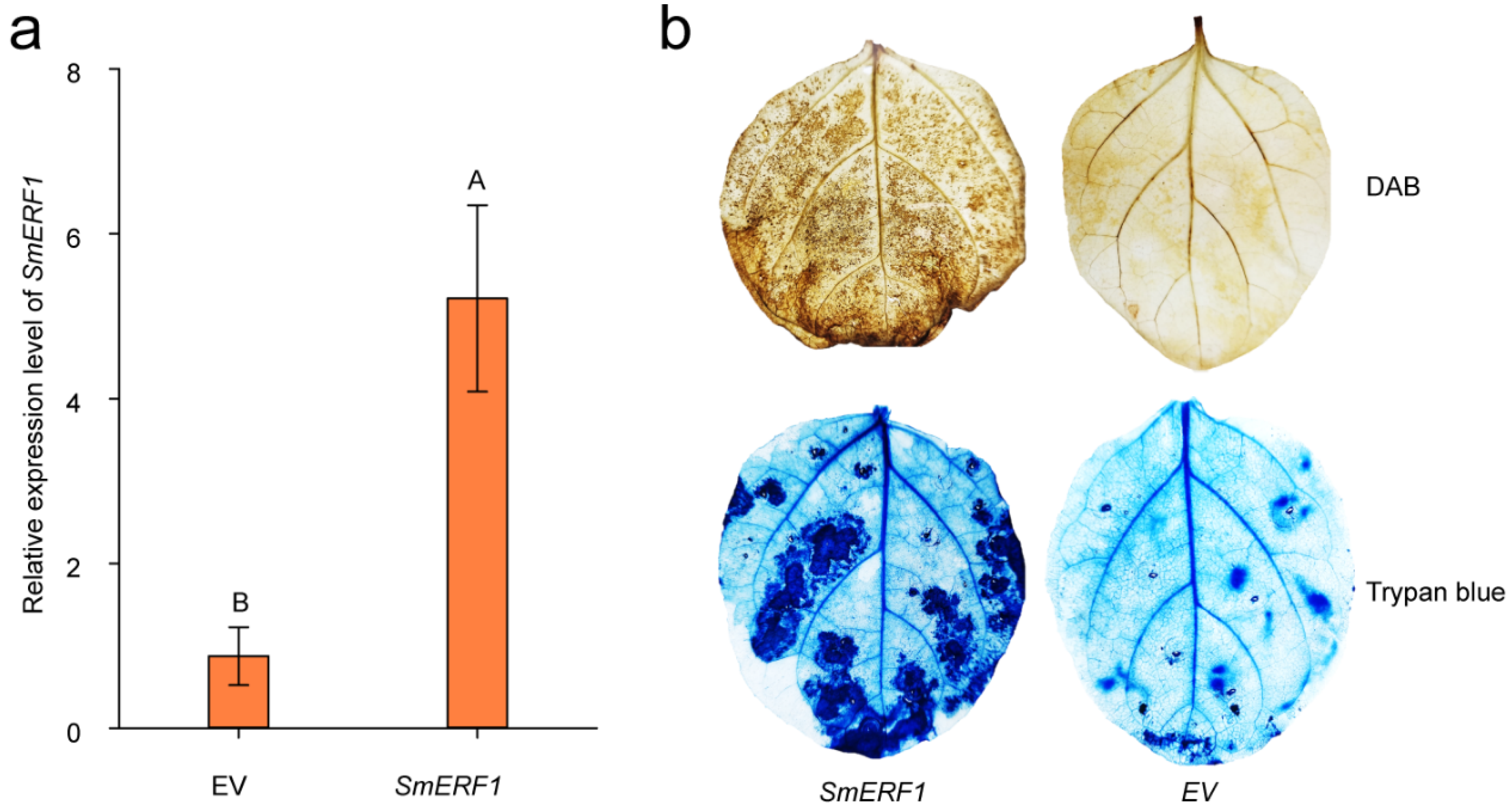
2.8. The Transient Overexpression of SmERF1 Triggers Cell Death in Eggplant Leaves
3. Discussion
4. Materials and Methods
4.1. Plant Material and Salt Treatment
4.2. Total RNA Extraction, cDNA Library Construction, and Illumina Sequencing
4.3. mRNA-Seq Read Processing and Assembly
4.4. Sequencing Data Processing and Analysis
4.5. Vector Construction
4.6. Agrobacterium tumefaciens Cultivation and Infiltration and Subcellular Localization Analysis of SmERF1
4.7. VIGS Assay
4.8. qPCR Assay
4.9. Physiological Index Measurement
4.10. Histochemical Staining
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Munns, R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002, 25, 239–250. [Google Scholar] [CrossRef]
- Akrami, M.; Arzani, A. Inheritance of fruit yield and quality in melon (Cucumis melo L.) grown under field salinity stress. Sci. Rep. 2019, 9, 7249. [Google Scholar] [CrossRef]
- Munns, R.; Gilliham, M. Salinity tolerance of crops—What is the cost? New Phytol. 2015, 208, 668–673. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Dai, X.; Wang, H.; Wang, F.; Tang, D.; Jiang, C.; Zhang, X.; Guo, W.; Lei, Y.; Ma, C.; et al. Transcriptomic Profiling Provides Molecular Insights into Hydrogen Peroxide-Enhanced Arabidopsis Growth and Its Salt Tolerance. Front. Plant Sci. 2022, 13, 866063. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Lu, B.; Liu, L.; Duan, W.; Meng, Y.; Li, J.; Zhang, K.; Sun, H.; Zhang, Y.; Dong, H.; et al. Exogenous melatonin improves the salt tolerance of cotton by removing active oxygen and protecting photosynthetic organs. BMC Plant Biol. 2021, 21, 331. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Chang, J.; Chen, H.; Wang, Z.; Gu, X.; Wei, C.; Zhang, Y.; Ma, J.; Yang, J.; Zhang, X. Exogenous Melatonin Confers Salt Stress Tolerance to Watermelon by Improving Photosynthesis and Redox Homeostasis. Front. Plant Sci. 2017, 8, 295. [Google Scholar] [CrossRef]
- Lei, P.; Liu, Z.; Hu, Y.; Kim, H.; Liu, S.; Liu, J.; Xu, L.; Li, J.; Zhao, Y.; Yu, Z.; et al. Transcriptome analysis of salt stress responsiveness in the seedlings of wild and cultivated Ricinus communis L. J. Biotechnol. 2021, 327, 106–116. [Google Scholar] [CrossRef]
- Negi, P.; Pandey, M.; Dorn, K.M.; Nikam, A.A.; Devarumath, R.M.; Srivastava, A.K.; Suprasanna, P. Transcriptional reprogramming and enhanced photosynthesis drive inducible salt tolerance in sugarcane mutant line M4209. J. Exp. Bot. 2020, 71, 6159–6173. [Google Scholar] [CrossRef]
- Bundo, M.; Martin-Cardoso, H.; Pesenti, M.; Gomez-Ariza, J.; Castillo, L.; Frouin, J.; Serrat, X.; Nogues, S.; Courtois, B.; Grenier, C.; et al. Integrative Approach for Precise Genotyping and Transcriptomics of Salt Tolerant Introgression Rice Lines. Front. Plant Sci. 2021, 12, 797141. [Google Scholar] [CrossRef]
- Chen, H.; Hu, L.; Wang, L.; Wang, S.; Cheng, X. Genome-wide identification and expression profiles of AP2/ERF transcription factor family in mung bean (Vigna radiata L.). J. Appl. Genet. 2022, 63, 223–236. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef]
- Licausi, F.; Giorgi, F.M.; Zenoni, S.; Osti, F.; Pezzotti, M.; Perata, P. Genomic and transcriptomic analysis of the AP2/ERF superfamily in Vitis vinifera. BMC Genom. 2010, 11, 719. [Google Scholar] [CrossRef]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef]
- Masaki, T.; Mitsui, N.; Tsukagoshi, H.; Nishii, T.; Morikami, A.; Nakamura, K. ACTIVATOR of Spomin::LUC1/WRINKLED1 of Arabidopsis thaliana transactivates sugar-inducible promoters. Plant Cell Physiol. 2005, 46, 547–556. [Google Scholar] [CrossRef]
- Welsch, R.; Maass, D.; Voegel, T.; Dellapenna, D.; Beyer, P. Transcription factor RAP2.2 and its interacting partner SINAT2: Stable elements in the carotenogenesis of Arabidopsis leaves. Plant Physiol. 2007, 145, 1073–1085. [Google Scholar] [CrossRef]
- Sharma, V.; Goel, P.; Kumar, S.; Singh, A.K. An apple transcription factor, MdDREB76, confers salt and drought tolerance in transgenic tobacco by activating the expression of stress-responsive genes. Plant Cell Rep. 2019, 38, 221–241. [Google Scholar] [CrossRef]
- Sun, M.; Shen, Y.; Chen, Y.; Wang, Y.; Cai, X.; Yang, J.; Jia, B.; Dong, W.; Chen, X.; Sun, X. Osa-miR1320 targets the ERF transcription factor OsERF096 to regulate cold tolerance via JA-mediated signaling. Plant Physiol. 2022, 189, 2500–2516. [Google Scholar] [CrossRef]
- Park, S.J.; Park, S.; Kim, Y.; Hyeon, D.Y.; Park, H.; Jeong, J.; Jeong, U.; Yoon, Y.S.; You, D.; Kwak, J.; et al. Ethylene responsive factor34 mediates stress-induced leaf senescence by regulating salt stress-responsive genes. Plant Cell Environ. 2022, 45, 1719–1733. [Google Scholar] [CrossRef]
- Wang, D.; Sun, Z.; Hu, X.; Xiong, J.; Hu, L.; Xu, Y.; Tang, Y.; Wu, Y. The key regulator LcERF056 enhances salt tolerance by modulating reactive oxygen species-related genes in Lotus corniculatus. BMC Plant Biol. 2021, 21, 605. [Google Scholar] [CrossRef]
- Huang, S.; Ma, Z.; Hu, L.; Huang, K.; Zhang, M.; Zhang, S.; Jiang, W.; Wu, T.; Du, X. Involvement of rice transcription factor OsERF19 in response to ABA and salt stress responses. Plant Physiol. Biochem. 2021, 167, 22–30. [Google Scholar] [CrossRef]
- Zhang, Y.; Ming, R.; Khan, M.; Wang, Y.; Dahro, B.; Xiao, W.; Li, C.; Liu, J.H. ERF9 of Poncirus trifoliata (L.) Raf. undergoes feedback regulation by ethylene and modulates cold tolerance via regulating a glutathione S-transferase U17 gene. Plant Biotechnol. J. 2022, 20, 183–200. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, L.; Wong, D.C.J.; Wang, Y.; Zhu, Z.; Xu, G.; Wang, Q.; Li, S.; Liang, Z.; Xin, H. The ethylene response factor VaERF092 from Amur grape regulates the transcription factor VaWRKY33, improving cold tolerance. Plant J. 2019, 99, 988–1002. [Google Scholar]
- Bolt, S.; Zuther, E.; Zintl, S.; Hincha, D.K.; Schmulling, T. ERF105 is a transcription factor gene of Arabidopsis thaliana required for freezing tolerance and cold acclimation. Plant Cell Environ. 2017, 40, 108–120. [Google Scholar] [CrossRef]
- Li, T.; Wu, Z.; Xiang, J.; Zhang, D.; Teng, N. Overexpression of a novel heat-inducible ethylene-responsive factor gene LlERF110 from Lilium longiflorum decreases thermotolerance. Plant Sci. 2022, 319, 111246. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhao, X.; Burger, M.; Wang, Y.; Chory, J. Two interacting ethylene response factors regulate heat stress response. Plant Cell 2021, 33, 338–357. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhao, X.; Ren, Z.; Abou-Elwafa, S.F.; Pu, X.; Zhu, Y.; Dou, D.; Su, H.; Cheng, H.; Liu, Z.; et al. ZmERF21 directly regulates hormone signaling and stress-responsive gene expression to influence drought tolerance in maize seedlings. Plant Cell Environ. 2022, 45, 312–328. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ni, D.; Shen, J.; Deng, S.; Xuan, H.; Wang, C.; Xu, J.; Zhou, L.; Guo, N.; Zhao, J.; et al. Genome-Wide Identification of the AP2/ERF Gene Family and Functional Analysis of GmAP2/ERF144 for Drought Tolerance in Soybean. Front. Plant Sci. 2022, 13, 848766. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Zhang, X.W.; Bi, S.Q.; You, C.X.; Wang, X.F.; Hao, Y.J. The ERF transcription factor MdERF38 promotes drought stress-induced anthocyanin biosynthesis in apple. Plant J. 2020, 101, 573–589. [Google Scholar] [CrossRef]
- Jin, Y.; Pan, W.; Zheng, X.; Cheng, X.; Liu, M.; Ma, H.; Ge, X. OsERF101, an ERF family transcription factor, regulates drought stress response in reproductive tissues. Plant Mol. Biol. 2018, 98, 51–65. [Google Scholar] [CrossRef]
- Qi, Y.; Yang, Z.; Sun, X.; He, H.; Guo, L.; Zhou, J.; Xu, M.; Luo, M.; Chen, H.; Tian, Z. Heterologous overexpression of StERF3 triggers cell death in Nicotiana benthamiana. Plant Sci. 2022, 315, 111149. [Google Scholar] [CrossRef]
- Li, Y.; Liu, K.; Tong, G.; Xi, C.; Liu, J.; Zhao, H.; Wang, Y.; Ren, D.; Han, S. MPK3/MPK6-mediated phosphorylation of ERF72 positively regulates resistance to Botrytis cinerea through directly and indirectly activating the transcription of camalexin biosynthesis enzymes. J. Exp. Bot. 2022, 73, 413–428. [Google Scholar] [CrossRef]
- Yang, H.; Shen, F.; Wang, H.; Zhao, T.; Zhang, H.; Jiang, J.; Xu, X.; Li, J. Functional analysis of the SlERF01 gene in disease resistance to S. lycopersici. BMC Plant Biol. 2020, 20, 376. [Google Scholar] [CrossRef]
- Feng, K.; Hou, X.L.; Xing, G.M.; Liu, J.X.; Duan, A.Q.; Xu, Z.S.; Li, M.Y.; Zhuang, J.; Xiong, A.S. Advances in AP2/ERF super-family transcription factors in plant. Crit Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF Transcription Factor Regulatory Networks in Hormone and Abiotic Stress Responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, J.; Li, Z.; Qiao, J.; Quan, R.; Wang, J.; Huang, R.; Qin, H. SALT AND ABA RESPONSE ERF1 improves seed germination and salt tolerance by repressing ABA signaling in rice. Plant Physiol. 2022, 189, 1110–1127. [Google Scholar] [CrossRef]
- Chen, G.; Zheng, D.; Feng, N.; Zhou, H.; Mu, D.; Zhao, L.; Shen, X.; Rao, G.; Meng, F.; Huang, A. Physiological mechanisms of ABA-induced salinity tolerance in leaves and roots of rice. Sci. Rep. 2022, 12, 8228. [Google Scholar] [CrossRef]
- Liu, J.; Shu, D.; Tan, Z.; Ma, M.; Guo, N.; Gao, S.; Duan, G.; Kuai, B.; Hu, Y.; Li, S.; et al. The Arabidopsis IDD14 transcription factor interacts with bZIP-type ABFs/AREBs and cooperatively regulates ABA-mediated drought tolerance. New Phytol. 2022. [Google Scholar] [CrossRef]
- Verslues, P.E.; Agarwal, M.; Katiyar-Agarwal, S.; Zhu, J.; Zhu, J.K. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 2006, 45, 523–539. [Google Scholar] [CrossRef]
- Lefebvre, V.; North, H.; Frey, A.; Sotta, B.; Seo, M.; Okamoto, M.; Nambara, E.; Marion-Poll, A. Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 2006, 45, 309–319. [Google Scholar] [CrossRef]
- Barrero, J.M.; Rodriguez, P.L.; Quesada, V.; Piqueras, P.; Ponce, M.R.; Micol, J.L. Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant Cell Environ. 2006, 29, 2000–2008. [Google Scholar] [CrossRef]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef] [Green Version]
- Sato, H.; Takasaki, H.; Takahashi, F.; Suzuki, T.; Iuchi, S.; Mitsuda, N.; Ohme-Takagi, M.; Ikeda, M.; Seo, M.; Yamaguchi-Shinozaki, K.; et al. Arabidopsis thaliana NGATHA1 transcription factor induces ABA biosynthesis by activating NCED3 gene during dehydration stress. Proc. Natl. Acad. Sci. USA 2018, 115, E11178–E11187. [Google Scholar] [CrossRef]
- Hannachi, S.; Steppe, K.; Eloudi, M.; Mechi, L.; Bahrini, I.; Van Labeke, M.C. Salt Stress Induced Changes in Photosynthesis and Metabolic Profiles of One Tolerant (‘Bonica’) and One Sensitive (‘Black Beauty’) Eggplant Cultivars (Solanum melongena L.). Plants 2022, 11, 590. [Google Scholar] [CrossRef]
- Wei, Q.; Wang, J.; Wang, W.; Hu, T.; Hu, H.; Bao, C. A high-quality chromosome-level genome assembly reveals genetics for important traits in eggplant. Hortic. Res. 2020, 7, 153. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Di, F.; Wang, T.; Ding, Y.; Chen, X.; Wang, H.; Li, J.; Liu, L. Genetic Mapping Combined with a Transcriptome Analysis to Screen for Candidate Genes Responsive to Abscisic Acid Treatment in Brassica napus Embryos During Seed Germination. DNA Cell Biol. 2020, 39, 533–547. [Google Scholar] [CrossRef]
- Zhao, Y.; Ai, X.; Wang, M.; Xiao, L.; Xia, G. A putative pyruvate transporter TaBASS2 positively regulates salinity tolerance in wheat via modulation of ABI4 expression. BMC Plant Biol. 2016, 16, 109. [Google Scholar] [CrossRef]
- Mota, A.P.Z.; Oliveira, T.N.; Vinson, C.C.; Williams, T.C.R.; Costa, M.; Araujo, A.C.G.; Danchin, E.G.J.; Grossi-de-Sa, M.F.; Guimaraes, P.M.; Brasileiro, A.C.M. Contrasting Effects of Wild Arachis Dehydrin Under Abiotic and Biotic Stresses. Front. Plant Sci. 2019, 10, 497. [Google Scholar] [CrossRef]
- Chung, S.; Parish, R.W. Combinatorial interactions of multiple cis-elements regulating the induction of the Arabidopsis XERO2 dehydrin gene by abscisic acid and cold. Plant J. 2008, 54, 15–29. [Google Scholar] [CrossRef]
- Li, S.; Wang, N.; Ji, D.; Zhang, W.; Wang, Y.; Yu, Y.; Zhao, S.; Lyu, M.; You, J.; Zhang, Y.; et al. A GmSIN1/GmNCED3s/GmRbohBs Feed-Forward Loop Acts as a Signal Amplifier That Regulates Root Growth in Soybean Exposed to Salt Stress. Plant Cell 2019, 31, 2107–2130. [Google Scholar] [CrossRef]
- Zheng, H.; Jing, L.; Jiang, X.; Pu, C.; Zhao, S.; Yang, J.; Guo, J.; Cui, G.; Tang, J.; Ma, Y.; et al. The ERF-VII transcription factor SmERF73 coordinately regulates tanshinone biosynthesis in response to stress elicitors in Salvia miltiorrhiza. New Phytol. 2021, 231, 1940–1955. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Wang, H.; Gao, Y.; Bi, Y.; Xiong, X.; Yan, Y.; Wang, J.; Li, D.; Song, F. ERF Transcription Factor OsBIERF3 Positively Contributes to Immunity against Fungal and Bacterial Diseases but Negatively Regulates Cold Tolerance in Rice. Int. J. Mol. Sci. 2022, 23, 606. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Jiang, Y.; Zou, Y.; Long, X.; Wu, X.; Ren, Y.; Li, Y.; Li, H.L. Genome-wide investigation of the AP2/ERF gene family in ginger: Evolution and expression profiling during development and abiotic stresses. BMC Plant Biol. 2021, 21, 561. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.E.; Bang, S.W.; Kim, S.H.; Seo, J.S.; Yoon, H.B.; Kim, Y.S.; Kim, J.K. Overexpression of OsERF83, a Vascular Tissue-Specific Transcription Factor Gene, Confers Drought Tolerance in Rice. Int. J. Mol. Sci. 2021, 22, 7656. [Google Scholar] [CrossRef]
- Zhao, M.J.; Yin, L.J.; Ma, J.; Zheng, J.C.; Wang, Y.X.; Lan, J.H.; Fu, J.D.; Chen, M.; Xu, Z.S. The Roles of GmERF135 in Improving Salt Tolerance and Decreasing ABA Sensitivity in Soybean. Front. Plant Sci. 2019, 10, 940. [Google Scholar] [CrossRef]
- Zhang, G.; Chen, M.; Li, L.; Xu, Z.; Chen, X.; Guo, J.; Ma, Y. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J. Exp. Bot. 2009, 60, 3781–3796. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, H.; Zhang, Q.; Liu, Q.; Zhai, H.; Zhao, N.; He, S. An AP2/ERF gene, IbRAP2-12, from sweetpotato is involved in salt and drought tolerance in transgenic Arabidopsis. Plant Sci. 2019, 281, 19–30. [Google Scholar] [CrossRef]
- Seok, H.Y.; Tran, H.T.; Lee, S.Y.; Moon, Y.H. AtERF71/HRE2, an Arabidopsis AP2/ERF Transcription Factor Gene, Contains Both Positive and Negative Cis-Regulatory Elements in Its Promoter Region Involved in Hypoxia and Salt Stress Responses. Int. J. Mol. Sci. 2022, 23, 5310. [Google Scholar] [CrossRef]
- Qu, Y.; Nong, Q.; Jian, S.; Lu, H.; Zhang, M.; Xia, K. An AP2/ERF Gene, HuERF1, from Pitaya (Hylocereus undatus) Positively Regulates Salt Tolerance. Int. J. Mol. Sci. 2020, 21, 4586. [Google Scholar] [CrossRef]
- Jin, J.H.; Zhang, H.X.; Ali, M.; Wei, A.M.; Luo, D.X.; Gong, Z.H. The CaAP2/ERF064 Regulates Dual Functions in Pepper: Plant Cell Death and Resistance to Phytophthora capsici. Genes 2019, 10, 541. [Google Scholar] [CrossRef]
- Zheng, Y.; He, S.; Cai, W.; Shen, L.; Huang, X.; Yang, S.; Huang, Y.; Lu, Q.; Wang, H.; Guan, D.; et al. CaAIL1 Acts Positively in Pepper Immunity against Ralstonia solanacearum by Repressing Negative Regulators. Plant Cell Physiol. 2021, 62, 1702–1717. [Google Scholar] [CrossRef]
- Yang, L.; Jin, J.; Fan, D.; Hao, Q.; Niu, J. Transcriptome Analysis of Jujube (Ziziphus jujuba Mill.) Response to Heat Stress. Int. J. Genomics. 2021, 2021, 3442277. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Shen, L.; Yang, S.; Yang, F.; Guan, D.; He, S. CaCBL1 Acts as a Positive Regulator in Pepper Response to Ralstonia solanacearum. Mol. Plant Microbe. Interact. 2020, 33, 945–957. [Google Scholar] [CrossRef]
- Shen, L.; Liu, Z.; Yang, S.; Yang, T.; Liang, J.; Wen, J.; Liu, Y.; Li, J.; Shi, L.; Tang, Q.; et al. Pepper CabZIP63 acts as a positive regulator during Ralstonia solanacearum or high temperature-high humidity challenge in a positive feedback loop with CaWRKY40. J. Exp. Bot. 2016, 67, 2439–2451. [Google Scholar] [CrossRef]
- Li, Y.; Feng, Z.; Wei, H.; Cheng, S.; Hao, P.; Yu, S.; Wang, H. Silencing of GhKEA4 and GhKEA12 Revealed Their Potential Functions Under Salt and Potassium Stresses in Upland Cotton. Front. Plant Sci. 2021, 12, 789775. [Google Scholar] [CrossRef]
- Choi, D.S.; Hwang, I.S.; Hwang, B.K. Requirement of the cytosolic interaction between PATHOGENESIS-RELATED PROTEIN10 and LEUCINE-RICH REPEAT PROTEIN1 for cell death and defense signaling in pepper. Plant Cell 2012, 24, 1675–1690. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, L.; Zhao, E.; Liu, R.; Yang, X. Transcriptome Analysis of Eggplant under Salt Stress: AP2/ERF Transcription Factor SmERF1 Acts as a Positive Regulator of Salt Stress. Plants 2022, 11, 2205. https://doi.org/10.3390/plants11172205
Shen L, Zhao E, Liu R, Yang X. Transcriptome Analysis of Eggplant under Salt Stress: AP2/ERF Transcription Factor SmERF1 Acts as a Positive Regulator of Salt Stress. Plants. 2022; 11(17):2205. https://doi.org/10.3390/plants11172205
Chicago/Turabian StyleShen, Lei, Enpeng Zhao, Ruie Liu, and Xu Yang. 2022. "Transcriptome Analysis of Eggplant under Salt Stress: AP2/ERF Transcription Factor SmERF1 Acts as a Positive Regulator of Salt Stress" Plants 11, no. 17: 2205. https://doi.org/10.3390/plants11172205
APA StyleShen, L., Zhao, E., Liu, R., & Yang, X. (2022). Transcriptome Analysis of Eggplant under Salt Stress: AP2/ERF Transcription Factor SmERF1 Acts as a Positive Regulator of Salt Stress. Plants, 11(17), 2205. https://doi.org/10.3390/plants11172205




