Endophytic Bacillus subtilis SR22 Triggers Defense Responses in Tomato against Rhizoctonia Root Rot
Abstract
:1. Introduction
2. Results
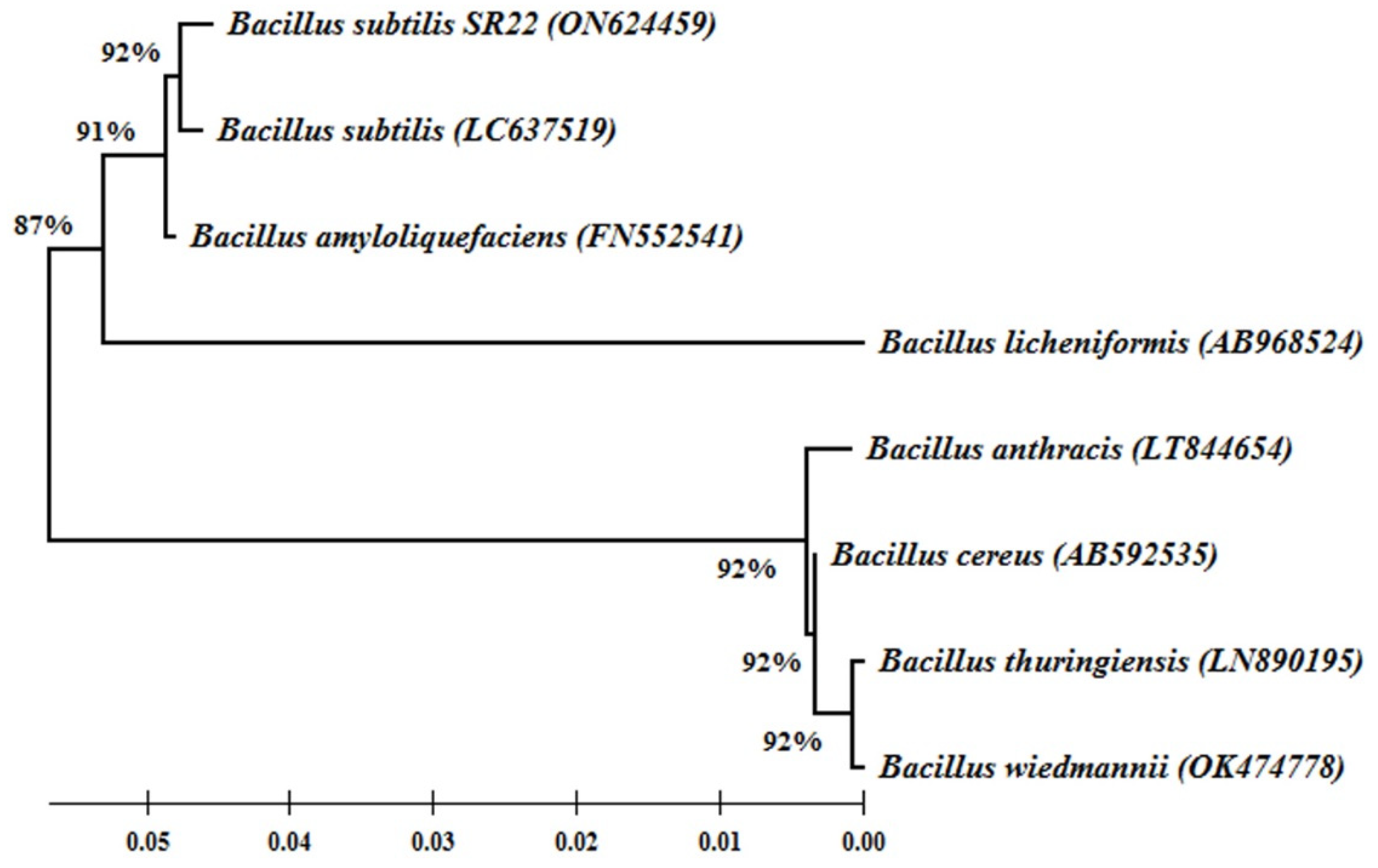
2.1. Molecular Identification and Phylogeny of the Endophytic Bacteria SR22
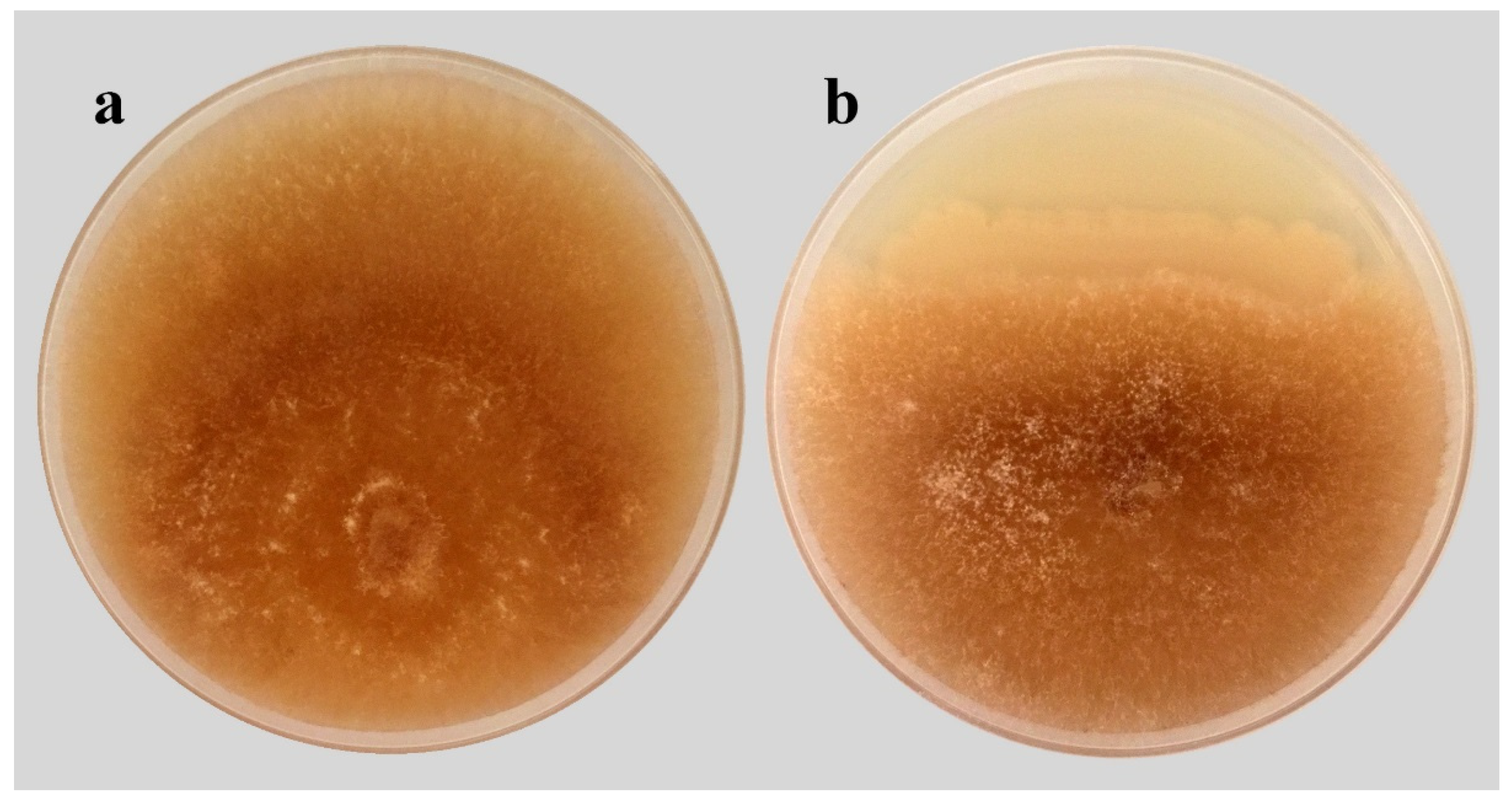
2.2. Antagonism Assay of B. subtilis SR22 against R. solani In Vitro
2.3. Gas Chromatography-Mass Spectrometry (GC-MS)
2.4. Effect on Transcriptional Expression of the Defense-Related Genes
2.5. Effect on Disease Incidence and Severity
2.6. Effect on Growth Parameters
2.7. Effect on Phenolic Content and Activity of Antioxidant Enzymes
2.8. Pearson Correlation Coefficient between the Studied Variables
2.9. Principal Component Analysis of Studied Variables
3. Discussion
4. Materials and Methods
4.1. Tomato Cultivar and Used Microorganisms
4.2. Antagonism Assay of B. subtilis SR22 against R. solani In Vitro
4.3. GC-MS
4.4. Greenhouse Experiment
4.4.1. qRT-PCR
4.4.2. Disease Assessment
4.4.3. Plant Growth Evaluation
4.4.4. Estimation of Total Phenolic Content and Enzymes Activities
4.5. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Collins, E.J.; Bowyer, C.; Tsouza, A.; Chopra, M. Tomatoes: An Extensive Review of the Associated Health Impacts of Tomatoes and Factors That Can Affect Their Cultivation. Biology 2022, 11, 239. [Google Scholar] [CrossRef] [PubMed]
- FAOSTAT Food and Agriculture Organization of the United Nations. 2022. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 5 June 2022).
- Al-Askar, A.A.; Ghoneem, K.M.; Rashad, Y.M.; Abdulkhair, W.M.; Hafez, E.E.; Shabana, Y.M.; Baka, Z.A. Occurrence and distribution of tomato seed-borne mycoflora in Saudi Arabia and its correlation with the climatic variables. Microb. Biotechnol. 2014, 7, 556–569. [Google Scholar] [CrossRef] [PubMed]
- Nefzi, A.; Jabnoun-Khiareddine, H.; Aydi Ben Abdallah, R.; Ammar, N.; Medimagh-Saïdana, S.; Haouala, R.; Daami-Remadi, M. Suppressing Fusarium Crown and Root Rot infections and enhancing the growth of tomato plants by Lycium arabicum Schweinf. Ex Boiss. extracts. S. Afr. J. Bot. 2017, 113, 288–299. [Google Scholar] [CrossRef]
- Goudjal, Y.; Toumatia, O.; Yekkour, A.; Sabaou, N.; Mathieu, F.; Zitouni, A. Biocontrol of Rhizoctonia solani damping-off and promotion of tomato plant growth by endophytic actinomycetes isolated from native plants of Algerian Sahara. Microbiol. Res. 2014, 169, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Tsror, L. Biology, epidemiology and management of Rhizoctonia solani on potato. J. Phytopathol. 2010, 158, 649–658. [Google Scholar] [CrossRef]
- Naseri, B. Epidemics of Rhizoctonia Root Rot in Association with Biological and Physicochemical Properties of Field Soil in Bean Crops. J. Phytopathol. 2013, 161, 397–404. [Google Scholar] [CrossRef]
- Gondal, A.S.; Rauf, A.; Naz, F. Anastomosis Groups of Rhizoctonia solani associated with tomato foot rot in Pothohar Region of Pakistan. Sci. Rep. 2019, 9, 3910. [Google Scholar] [CrossRef]
- Le Cointe, R.; Simon, T.E.; Delarue, P.; Hervé, M.; Leclerc, M.; Poggi, S. Reducing the use of pesticides with site-specific application: The chemical control of Rhizoctonia solani as a case of study for the management of soil-borne diseases. PLoS ONE 2016, 11, e0163221. [Google Scholar] [CrossRef]
- Rashad, Y.M.; El-Sharkawy, H.H.A.; Elazab, N.T. Ascophyllum nodosum Extract and Mycorrhizal Colonization Synergistically Trigger Immune Responses in Pea Plants against Rhizoctonia Root Rot, and Enhance Plant Growth and Productivity. J. Fungi 2022, 8, 268. [Google Scholar] [CrossRef]
- Rashad, Y.M.; Aseel, D.G.; Hafez, E.E. Antifungal potential and defense gene induction in maize against Rhizoctonia root rot by seed extract of Ammi visnaga (L.) Lam. Phytopathol. Mediterr. 2018, 57, 73–88. [Google Scholar] [CrossRef]
- AYDIN, M.H. Rhizoctonia Solani and Its Biological Control. Türkiye Tarımsal. Araştırmalar. Derg. 2022, 9, 118–135. [Google Scholar] [CrossRef]
- Abdel-Fattah, G.M.; El-Haddad, S.A.; Hafez, E.E.; Rashad, Y.M. Induction of defense responses in common bean plants by arbuscular mycorrhizal fungi. Microbiol. Res. 2011, 166, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Toral, L.; Rodríguez, M.; Béjar, V.; Sampedro, I. Crop protection against botrytis cinerea by rhizhosphere biological control agent bacillus velezensis XT1. Microorganisms 2020, 8, 992. [Google Scholar] [CrossRef]
- Rashad, Y.M.; Abbas, M.A.; Soliman, H.M.; Abdel-Fattah, G.G.; Abdel-Fattah, G.M. Synergy between endophytic Bacillus amyloliquefaciens GGA and arbuscular mycorrhizal fungi induces plant defense responses against white rot of garlic and improves host plant growth. Phytopathol. Mediterr. 2020, 59, 169–186. [Google Scholar] [CrossRef]
- Sharma, A.; Kaushik, N.; Sharma, A.; Bajaj, A.; Rasane, M.; Shouche, Y.S.; Marzouk, T.; Djébali, N. Screening of Tomato Seed Bacterial Endophytes for Antifungal Activity Reveals Lipopeptide Producing Bacillus siamensis Strain NKIT9 as a Potential Bio-Control Agent. Front. Microbiol. 2021, 12, 1228. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M. Bacillus spp.: A promising biocontrol agent of root, foliar, and postharvest diseases of plants. In Bacilli and Agrobiotechnology; Islam, M.T., Rahman, M., Pandey, P., Jha, C.K., Aeron, A., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 113–141. ISBN 9783319444093. [Google Scholar]
- Liu, H.; Carvalhais, L.C.; Crawford, M.; Singh, E.; Dennis, P.G.; Pieterse, C.M.J.; Schenk, P.M. Inner plant values: Diversity, colonization and benefits from endophytic bacteria. Front. Microbiol. 2017, 8, 2552. [Google Scholar] [CrossRef]
- Kandel, S.L.; Joubert, P.M.; Doty, S.L. Bacterial endophyte colonization and distribution within plants. Microorganisms 2017, 5, 77. [Google Scholar] [CrossRef] [Green Version]
- Rat, A.; Naranjo, H.D.; Krigas, N.; Grigoriadou, K.; Maloupa, E.; Alonso, A.V.; Schneider, C.; Papageorgiou, V.P.; Assimopoulou, A.N.; Tsafantakis, N.; et al. Endophytic Bacteria From the Roots of the Medicinal Plant Alkanna tinctoria Tausch (Boraginaceae): Exploration of Plant Growth Promoting Properties and Potential Role in the Production of Plant Secondary Metabolites. Front. Microbiol. 2021, 12, 633488. [Google Scholar] [CrossRef]
- Rajendran, L.; Samiyappan, R. Endophytic Bacillus Species Confer Increased Resistance in Cotton against Damping off Disease Caused by Rhizoctonia solani. Plant. Pathol. J. 2008, 7, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Fadiji, A.E.; Babalola, O.O. Elucidating Mechanisms of Endophytes Used in Plant Protection and Other Bioactivities with Multifunctional Prospects. Front. Bioeng. Biotechnol. 2020, 8, 467. [Google Scholar] [CrossRef]
- Yu, Y.; Gui, Y.; Li, Z.; Jiang, C.; Guo, J.; Niu, D. Induced Systemic Resistance for Improving Plant Immunity by Beneficial Microbes. Plants 2022, 11, 386. [Google Scholar] [CrossRef] [PubMed]
- Rashad, Y.M.; Fekry, W.M.E.; Sleem, M.M.; Elazab, N.T. Effects of Mycorrhizal Colonization on Transcriptional Expression of the Responsive Factor JERF3 and Stress-Responsive Genes in Banana Plantlets in Response to Combined Biotic and Abiotic Stresses. Front. Plant. Sci. 2021, 12, 742628. [Google Scholar] [CrossRef]
- Sansinenea, E.; Ortiz, A. Secondary metabolites of soil Bacillus spp. Biotechnol. Lett. 2011, 33, 1523–1538. [Google Scholar] [CrossRef] [PubMed]
- Martínez, G.; Regente, M.; Jacobi, S.; Del Rio, M.; Pinedo, M.; de la Canal, L. Chlorogenic acid is a fungicide active against phytopathogenic fungi. Pestic. Biochem. Physiol. 2017, 140, 30–35. [Google Scholar] [CrossRef]
- Abd El-Hameed, R.H.; Sayed, A.I.; Mahmoud Ali, S.; Mosa, M.A.; Khoder, Z.M.; Fatahala, S.S. Synthesis of novel pyrroles and fused pyrroles as antifungal and antibacterial agents. J. Enzyme Inhib. Med. Chem. 2021, 36, 2183–2198. [Google Scholar] [CrossRef]
- Wu, Z.; Huang, Y.; Li, Y.; Dong, J.; Liu, X.; Li, C. Biocontrol of Rhizoctonia solani via Induction of the Defense Mechanism and Antimicrobial Compounds Produced by Bacillus subtilis SL-44 on Pepper (Capsicum annuum L.). Front. Microbiol. 2019, 10, 2676. [Google Scholar] [CrossRef] [Green Version]
- Kidwai, M.; Ahmad, I.Z.; Chakrabarty, D. Class III peroxidase: An indispensable enzyme for biotic/abiotic stress tolerance and a potent candidate for crop improvement. Plant. Cell Rep. 2020, 39, 1381–1393. [Google Scholar] [CrossRef]
- Breen, S.; Williams, S.J.; Outram, M.; Kobe, B.; Solomon, P.S. Emerging Insights into the Functions of Pathogenesis-Related Protein 1. Trends Plant. Sci. 2017, 22, 871–879. [Google Scholar] [CrossRef]
- Rashad, Y.; Aseel, D.; Hammad, S.; Elkelish, A. Rhizophagus irregularis and Rhizoctonia solani differentially elicit systemic transcriptional expression of polyphenol biosynthetic pathways genes in sunflower. Biomolecules 2020, 10, 379. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Zhou, J.; Li, C.; Ma, Y. Antifungal and plant growth promotion activity of volatile organic compounds produced by Bacillus amyloliquefaciens. Microbiologyopen 2019, 8, e00813. [Google Scholar] [CrossRef] [Green Version]
- Gohil, R.B.; Raval, V.H.; Panchal, R.R.; Rajput, K.N. Plant Growth-Promoting Activity of Bacillus sp. PG-8 Isolated from Fermented Panchagavya and Its Effect on the Growth of Arachis hypogea. Front. Agron. 2022, 4, 1–13. [Google Scholar] [CrossRef]
- Radhakrishnan, R.; Hashem, A.; Abd_Allah, E.F. Bacillus: A Biological Tool for Crop Improvement through Bio-Molecular Changes in Adverse Environments. Front. Physiol. 2017, 8, 667. [Google Scholar] [CrossRef] [PubMed]
- Samaniego-Gámez, B.Y.; Garruña, R.; Tun-Suárez, J.M.; Kantun-Can, J.; Reyes-Ramírez, A.; Cervantes-Díaz, L. Bacillus spp. Inoculation improves photosystem II efficiency and enhances photosynthesis in pepper plants. Chil. J. Agric. Res. 2016, 76, 409–416. [Google Scholar] [CrossRef] [Green Version]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal Rna Genes for Phylogenetics. In PCR Protocols; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Carling, D.E.; Pope, E.J.; Brainard, K.A.; Carter, D.A. Characterization of mycorrhizal isolates of Rhizoctonia solani from an orchid, including AG-12, a new anastomosis group. Phytopathology 1999, 89, 942–946. [Google Scholar] [CrossRef] [Green Version]
- Malik, C.P.; Singh, M.B. Estimation of total phenols. In Plant Enzymology and Histo-Enzymology; Kalyani Publishers: New Delhi, India, 1980. [Google Scholar]
- Maxwell, D.P.; Bateman, D.F. Changes in the activities of some oxidases in extracts of Rhizoctonia-infected bean hypocotyls in relation to lesion maturation. Phytopathology 1967, 57, 132–136. [Google Scholar]
- Galeazzi, M.A.M.; Sgarbieri, V.C.; Constantinides, S.M. Isolation, purification and physicochemical characterization of polyphenol oxidase (PPO) from a dwarf variety of banana (Musa cavendishii, L). J. Food Sci. 1981, 46, 150–155. [Google Scholar] [CrossRef]
- CoStat Software. Available online: https://www.cohortsoftware.com/index.html (accessed on 13 May 2022).
- Haynes, W. Tukey’s Test. In Encyclopedia of Systems Biology; Dubitzky, W., Wolkenhauer, O., Cho, K.-H., Yokota, H., Eds.; Springer: New York, NY, USA, 2013; pp. 2303–2304. ISBN 978-1-4419-9863-7. [Google Scholar]
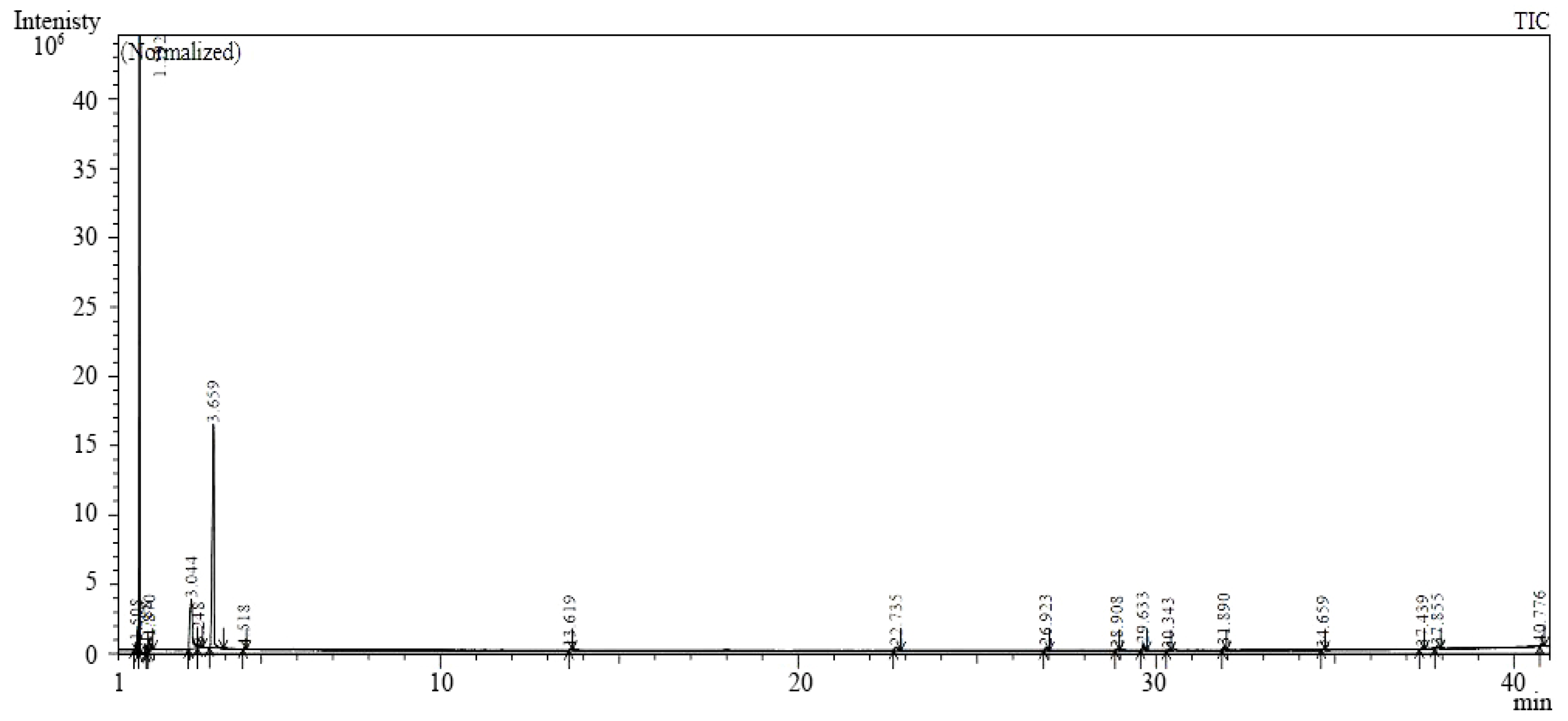


| Peak # | Retention Time (min) | Peak Area (%) | Compound Name |
|---|---|---|---|
| 1 | 1.508 | 1.50 | Propyl thioglycolic acid |
| 2 | 1.592 | 33.87 | Chlorogenic acid |
| 3 | 1.788 | 1.09 | 2,3-Butanediol |
| 4 | 1.870 | 1.90 | 1-Hexadecanol |
| 5 | 3.044 | 13.70 | Pyrrolo [1,2-a]pyrazine-1,4-dione, hexahydro |
| 6 | 3.248 | 0.57 | Acetamide |
| 7 | 3.659 | 41.47 | Propane, 2-ethoxy- |
| 8 | 4.518 | 0.22 | Propanamide |
| 9 | 13.619 | 0.27 | 2-Piperidinone |
| 10 | 22.735 | 0.89 | 3-Isopropoxy-1,1,1,7,7,7-hexamethyl-3,5,5-tri |
| 11 | 26.923 | 0.61 | Benzeneethanamine, N-[(pentafluorophenyl) |
| 12 | 28.908 | 0.20 | (S)-(+)-1,2-Propanediol |
| 13 | 29.633 | 1.03 | Phthalic acid, 3,5-dimethylphenyl 4-isopropyl |
| 14 | 30.343 | 0.08 | Phthalic acid, di(3,5-dimethylphenyl) ester |
| 15 | 31.890 | 0.59 | 1,7-Di(3-ethylphenyl)-2,2,4,4,6,6-hexamethyl |
| 16 | 34.659 | 0.13 | 1,2-Diphenyltetramethyldisilane |
| 17 | 37.439 | 0.22 | 2,5-Piperazinedione, 3,6-bis(2-methylpropyl) |
| 18 | 37.855 | 0.54 | 3-(2-N-Acetyl-N-methylaminoethyl)indol |
| 19 | 40.776 | 0.35 | Bis[di(trimethylsiloxy)phenylsiloxy]trimethyl |
| 20 | 41.311 | 0.80 | Heptasiloxane, hexadecamethyl- |
| Treatment | Disease Incidence (%) | Disease Severity (%) | Severity Reduction (%) |
|---|---|---|---|
| C | 0.0 c | 0.0 c | 0.0 b |
| P | 100.0 a | 65.0 ± 3.2 a | 0.0 b |
| B | 0.0 c | 0.0 c | 0.0 b |
| P+B | 60.3 ± 2.5 b | 30.0 ± 2.4 b | 53.8 ± 2.0 a |
| Treatment | Shoot Length (cm) | Root Length (cm) | Shoot Dry Weight (g) | Root Dry Weight (g) | No. of Leaves |
|---|---|---|---|---|---|
| C | 21.5 ± 2.1 b | 13.5 ± 0.9 b | 1.43 ± 0.07 b | 0.33 ± 0.04 b | 7.7 ± 0.6 a |
| P | 15.7 ± 3.0 c | 10.1 ± 0.7 c | 0.88 ± 0.09 d | 0.24 ± 0.03 c | 7.3 ± 0.5 a |
| B | 29.1 ± 1.5 a | 16.8 ± 1.0 a | 1.97 ± 0.10 a | 0.39 ± 0.03 a | 8.0 ± 0.7 a |
| P+B | 20.8 ± 1.8 b | 12.7 ± 0.9 b | 1.18 ± 0.06 c | 0.30 ± 0.05 b | 7.6 ± 0.3 a |
| Treatment | Phenolic Content (mg.g−1 Fresh wt) | POD (∆A470 min−1 g−1 Fresh wt) | PPO (∆A420 min−1 g−1 Fresh wt) |
|---|---|---|---|
| C | 125.3 ± 2.2 d | 1.419 ± 0.07 d | 1.374 ± 0.06 d |
| P | 290.7 ± 2.7 b | 2.993 ± 0.06 b | 1.984 ± 0.04 b |
| B | 221.5 ± 3.1 c | 2.215 ± 0.04 c | 1.775 ± 0.09 c |
| P+B | 387.0 ± 5.4 a | 3.157 ± 0.08 a | 2.204 ± 0.08 a |
| Shoot Length | Root Length | Shoot Dry Weight | Root Dry Weight | Number of Leaves | Phenolic Content | Peroxidase | Polyphenol Oxidase | |
|---|---|---|---|---|---|---|---|---|
| Shoot length | 1 | |||||||
| Root length | 0.91 *** | 1 | ||||||
| Shoot dry weight | 0.90 *** | 0.82 ** | 1 | |||||
| Root dry weight | 0.72 ** | 0.62 * | 0.73 ** | 1 | ||||
| Number of leaves | 0.35 ns | 0.44 ns | 0.52 ns | 0.38 ns | 1 | |||
| Phenolic content | −0.28 ns | −0.31 ns | −0.25 ns | −0.38 ns | −0.16 ns | 1 | ||
| Peroxidase | −0.38 ns | −0.41 ns | −0.37 ns | −0.50 ns | −0.24 ns | 0.96 *** | 1 | |
| Polyphenol oxidase | −0.22 ns | −0.27 ns | −0.18 ns | −0.36 ns | −0.17 ns | 0.98 *** | 0.97 *** | 1 |
| Gene Description | Abbrev. | Accession No. | Sequence (5′-3′) |
|---|---|---|---|
| Jasmonate and ethylene-responsive factor 3 | JERF3-F JERF3-R | AY383630 | GCCATTTGCCTTCTCTGCTTC GCAGCAGCATCCTTGTCTGA |
| Peroxidase | POD-F POD-R | X94943 | CCTTGTTGGTGGGCACACAA GGCCACCAGTGGAGTTGAAA |
| Pathogenesis-related protein 1 | PR1-F PR1-R | M69247 | ACTTGGCATCCCGAGCACAA CTCGGACACCCACAATTGCA |
| β-actin | β-actin-F β-actin -R | GTGGGCCGCTCTAGGCACCAA | |
| CTCTTTGATGTCACGCACGATTTC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rashad, Y.M.; Abdalla, S.A.; Sleem, M.M. Endophytic Bacillus subtilis SR22 Triggers Defense Responses in Tomato against Rhizoctonia Root Rot. Plants 2022, 11, 2051. https://doi.org/10.3390/plants11152051
Rashad YM, Abdalla SA, Sleem MM. Endophytic Bacillus subtilis SR22 Triggers Defense Responses in Tomato against Rhizoctonia Root Rot. Plants. 2022; 11(15):2051. https://doi.org/10.3390/plants11152051
Chicago/Turabian StyleRashad, Younes M., Sara A. Abdalla, and Mohamed M. Sleem. 2022. "Endophytic Bacillus subtilis SR22 Triggers Defense Responses in Tomato against Rhizoctonia Root Rot" Plants 11, no. 15: 2051. https://doi.org/10.3390/plants11152051
APA StyleRashad, Y. M., Abdalla, S. A., & Sleem, M. M. (2022). Endophytic Bacillus subtilis SR22 Triggers Defense Responses in Tomato against Rhizoctonia Root Rot. Plants, 11(15), 2051. https://doi.org/10.3390/plants11152051







