Expression and Functional Analyses of the WIP Gene Family in Arabidopsis
Abstract
:1. Introduction
2. Results
2.1. Expression Pattern of the WIP Gene Family
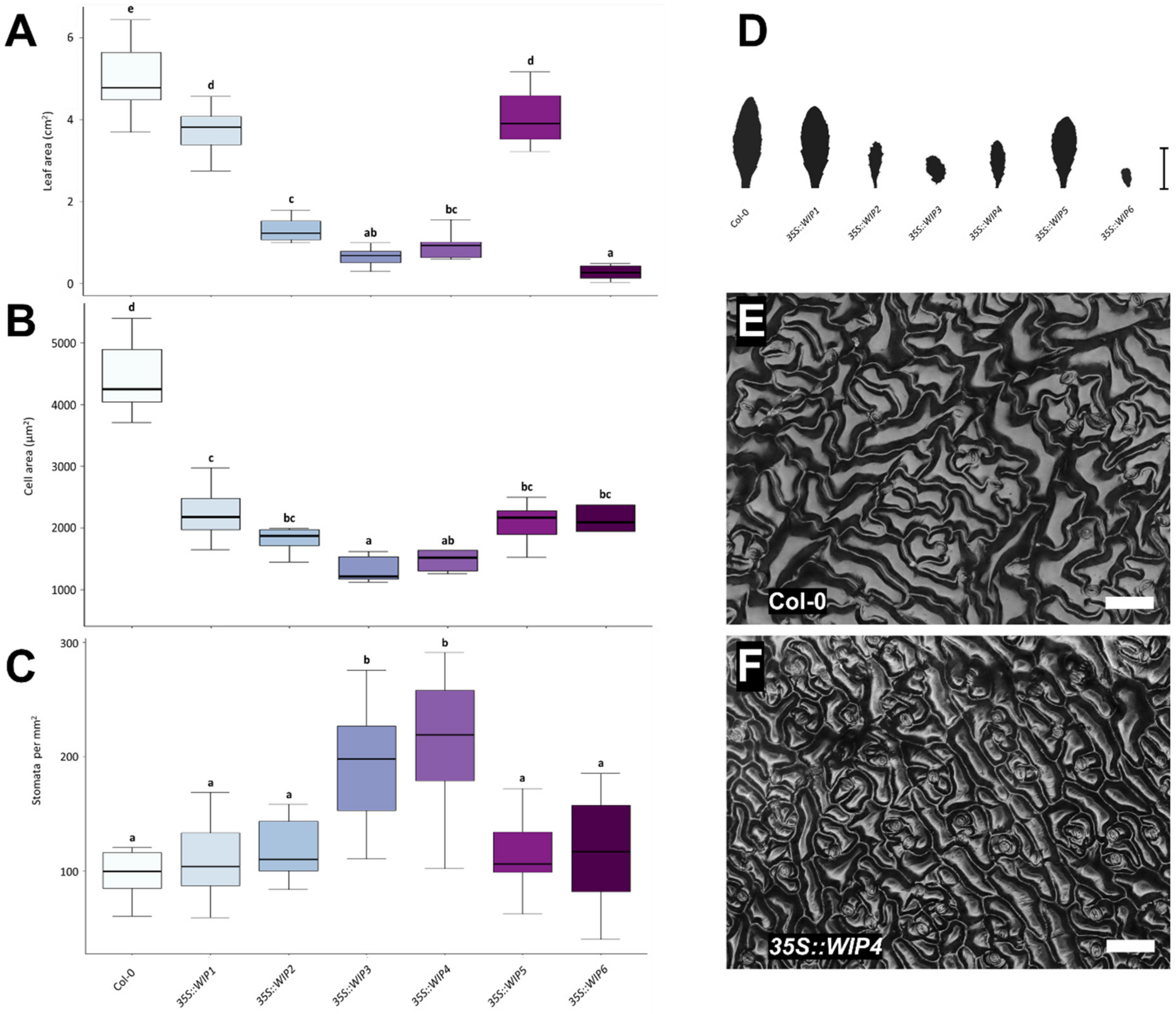
2.2. Overexpression of the WIP Gene Family Causes Leaf and Cell Size Reduction
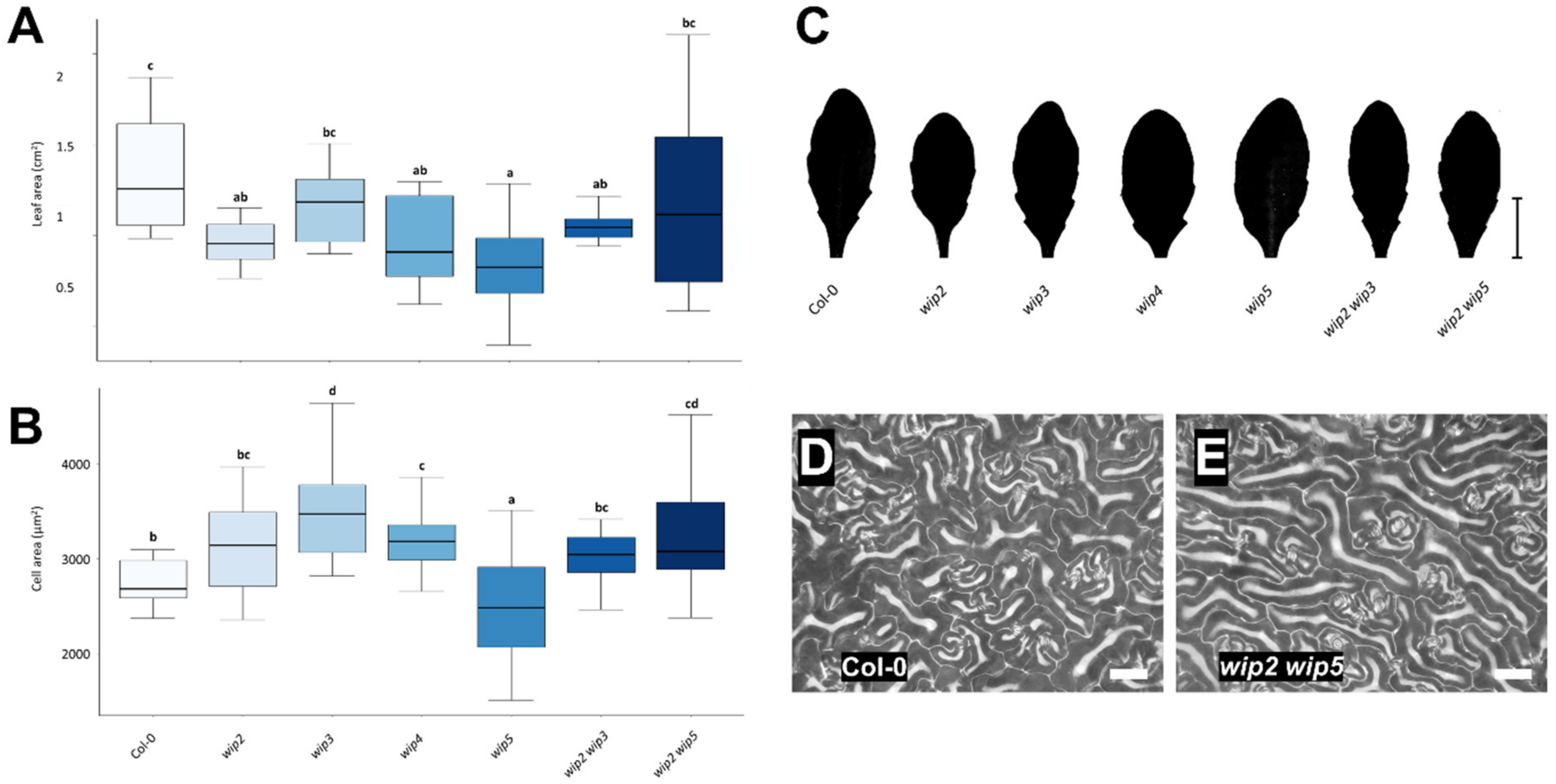
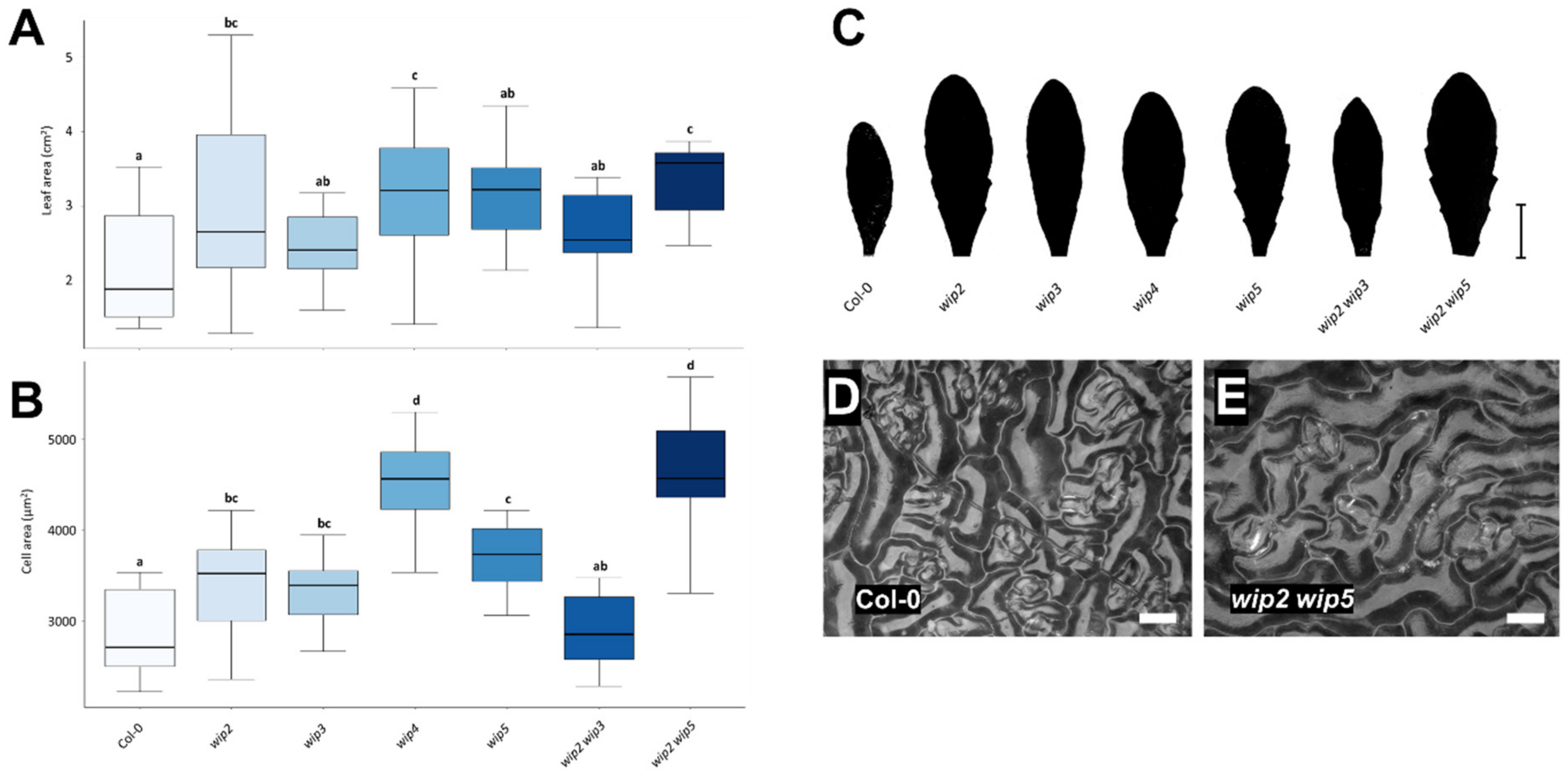
2.3. Loss of WIP Function Affects Leaf and Cell Size
2.4. Wip Phenotype in Stems
3. Discussion
3.1. Expression Patterns of WIP Family Promoters
3.2. WIPs Alter Leaf Shape and Size, and Leaf Cell Size
3.3. Loss of WIP Function Affects Leaf and Cell Size
4. Materials and Methods
4.1. Plant Material and Constructs
4.2. Growth Conditions
4.3. Leaf and Cell Analyses
4.4. Expression Analyses
4.5. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Appelhagen, I.; Huep, G.; Lu, G.H.; Strompen, G.; Weisshaar, B.; Sagasser, M. Weird fingers: Functional analysis of WIP domain proteins. FEBS Lett. 2010, 584, 3116–3122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Englbrecht, C.C.; Schoof, H.; Böhm, S. Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genom. 2004, 5, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sagasser, M.; Lu, G.H.; Hahlbrock, K.; Weisshaar, B. A. thaliana Transparent testa 1 is involved in seed coat development and defines the WIP subfamily of plant zinc finger proteins. Genes Dev. 2002, 16, 138–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, A.; Troadec, C.; Boualem, A.; Rajab, M.; Fernandez, R.; Morin, H.; Pitrat, M.; Dogimont, C.; Bendahmane, A. A transposon-induced epigenetic change leads to sex determination in melon. Nature 2009, 461, 1135–1138. [Google Scholar] [CrossRef]
- Ren, G.; Li, L.; Huang, Y.; Wang, Y.; Zhang, W.; Zheng, R.; Zhong, C.; Wang, X. GhWIP2, a WIP zinc finger protein, suppresses cell expansion in Gerbera hybrida by mediating crosstalk between gibberellin, abscisic acid, and auxin. New Phytol. 2018, 219, 728–742. [Google Scholar] [CrossRef] [Green Version]
- Jones, V.A.S.; Dolan, L. MpWIP regulates air pore complex development in the liverwort Marchantia polymorpha. Development 2017, 144, 1472–1476. [Google Scholar] [CrossRef] [Green Version]
- Petricka, J.J.; Clay, N.K.; Nelson, T.M. Vein patterning screens and the defectively organized tributaries mutants in Arabidopsis thaliana. Plant J. 2008, 56, 251–263. [Google Scholar] [CrossRef]
- Crawford, B.C.W.; Ditta, G.; Yanofsky, M.F. The NTT Gene Is Required for Transmitting-Tract Development in Carpels of Arabidopsis thaliana. Curr. Biol. 2007, 17, 1101–1108. [Google Scholar] [CrossRef] [Green Version]
- Marsch-Martínez, N.; Zúñiga-Mayo, M.V.; Herrera-Ubaldo, H.; Ouwerkerk, B.P.; Pablo-Villa, J.; Lozano-Sotomayor, P.; Greco, R.; Ballester, P.; Balanzá, V.; Kuijt, J.S.; et al. The NTT transcription factor promotes replum development in Arabidopsis fruits. Plant J. 2014, 80, 69–81. [Google Scholar] [CrossRef]
- Chung, K.S.; Lee, J.H.; Lee, J.S.; Ahn, J.H. Fruit indehiscence caused by enhanced expression of NO TRANSMITTING TRACT in Arabidopsis thaliana. Mol. Cells 2013, 35, 519–525. [Google Scholar] [CrossRef] [Green Version]
- Herrera-Ubaldo, H.; Lozano-Sotomayor, P.; Ezquer, I.; Di Marzo, M.; Montes, R.A.C.; Gómez-Felipe, A.; Pablo-Villa, J.; Diaz-Ramirez, D.; Ballester, P.; Ferrándiz, C.; et al. New roles of NO TRANSMITTING TRACT and SEEDSTICK during medial domain development in Arabidopsis fruits. Development 2019, 146, dev172395. [Google Scholar] [CrossRef] [Green Version]
- Crawford, B.C.W.; Sewell, J.; Golembeski, G.; Roshan, C.; Long, J.A.; Yanofsky, M.F. Genetic control of distal stem cell fate within root and embryonic meristems. Science 2015, 347, 655–659. [Google Scholar] [CrossRef] [Green Version]
- Roldan, M.V.G.; Izhaq, F.; Verdenaud, M.; Eleblu, J.; Haraghi, A.; Sommard, V.; Chambrier, P.; Latrasse, D.; Jégu, T.; Benhamed, M.; et al. Integrative genome-wide analysis reveals the role of WIP proteins in inhibition of growth and development. Commun. Biol. 2020, 3, 239. [Google Scholar] [CrossRef]
- Horiguchi, G.; Tsukaya, H. Organ size regulation in plants: Insights from compensation. Front. Plant Sci. 2011, 2, 24. [Google Scholar] [CrossRef] [Green Version]
- Du, Y.; Roldan, M.V.G.; Haraghi, A.; Haili, N.; Izhaq, F.; Verdenaud, M.; Boualem, A.; Bendahmane, A. Spatially expressed WIP genes control Arabidopsis embryonic root development. Nat. Plants 2022, 8, 635–645. [Google Scholar] [CrossRef]
- Becker, D.; Kemper, E.; Schell, J.; Masterson, R. New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 1992, 20, 1195–1197. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Lazcano-Ramírez, H.G.; Gómez-Felipe, A.; Díaz-Ramírez, D.; Durán-Medina, Y.; Sánchez-Segura, L.; De Folter, S.; Marsch-Martínez, N. Non-destructive Plant Morphometric and Color Analyses Using an Optoelectronic 3D Color Microscope. Front. Plant Sci. 2018, 9, 1409. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, F.; Yu, Y.; Feng, L.I.; Jia, J.; Liu, B.O.; Li, B.; Guo, H.; Zhai, J. A Comprehensive Online Database for Exploring ∼20,000 Public Arabidopsis RNA-Seq Libraries. Mol. Plant 2020, 13, 1231–1233. [Google Scholar] [CrossRef]
- Rstudio Team. RStudio: Integrated Development for R; Rstudio PBC: Boston, MA, USA, 2022. [Google Scholar]
- Tissier, A.F.; Marillonnet, S.; Klimyuk, V.; Patel, K.; Torres, M.A.; Murphy, G.; Jones, J.D. Multiple independent defective suppressor-mutator transposon insertions in Arabidopsis: A tool for functional genomics. Plant Cell 1999, 11, 1841–1852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R.; et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleinboelting, N.; Huep, G.; Kloetgen, A.; Viehoever, P.; Weisshaar, B. GABI-Kat SimpleSearch: New features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res. 2012, 40, D1211–D1215. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diaz-Ramirez, D.; Diaz-Garcia, U.S.; Magdaleno-Garcia, G.; Huep, G.; Appelhagen, I.; Sagasser, M.; Marsch-Martinez, N. Expression and Functional Analyses of the WIP Gene Family in Arabidopsis. Plants 2022, 11, 2010. https://doi.org/10.3390/plants11152010
Diaz-Ramirez D, Diaz-Garcia US, Magdaleno-Garcia G, Huep G, Appelhagen I, Sagasser M, Marsch-Martinez N. Expression and Functional Analyses of the WIP Gene Family in Arabidopsis. Plants. 2022; 11(15):2010. https://doi.org/10.3390/plants11152010
Chicago/Turabian StyleDiaz-Ramirez, David, Ury Sarai Diaz-Garcia, Guadalupe Magdaleno-Garcia, Gunnar Huep, Ingo Appelhagen, Martin Sagasser, and Nayelli Marsch-Martinez. 2022. "Expression and Functional Analyses of the WIP Gene Family in Arabidopsis" Plants 11, no. 15: 2010. https://doi.org/10.3390/plants11152010
APA StyleDiaz-Ramirez, D., Diaz-Garcia, U. S., Magdaleno-Garcia, G., Huep, G., Appelhagen, I., Sagasser, M., & Marsch-Martinez, N. (2022). Expression and Functional Analyses of the WIP Gene Family in Arabidopsis. Plants, 11(15), 2010. https://doi.org/10.3390/plants11152010





