Gb_ANR-47 Enhances the Resistance of Gossypium barbadense to Fusarium oxysporum f. sp. vasinfectum (FOV) by Regulating the Content of Proanthocyanidins
Abstract
:1. Introduction
2. Results
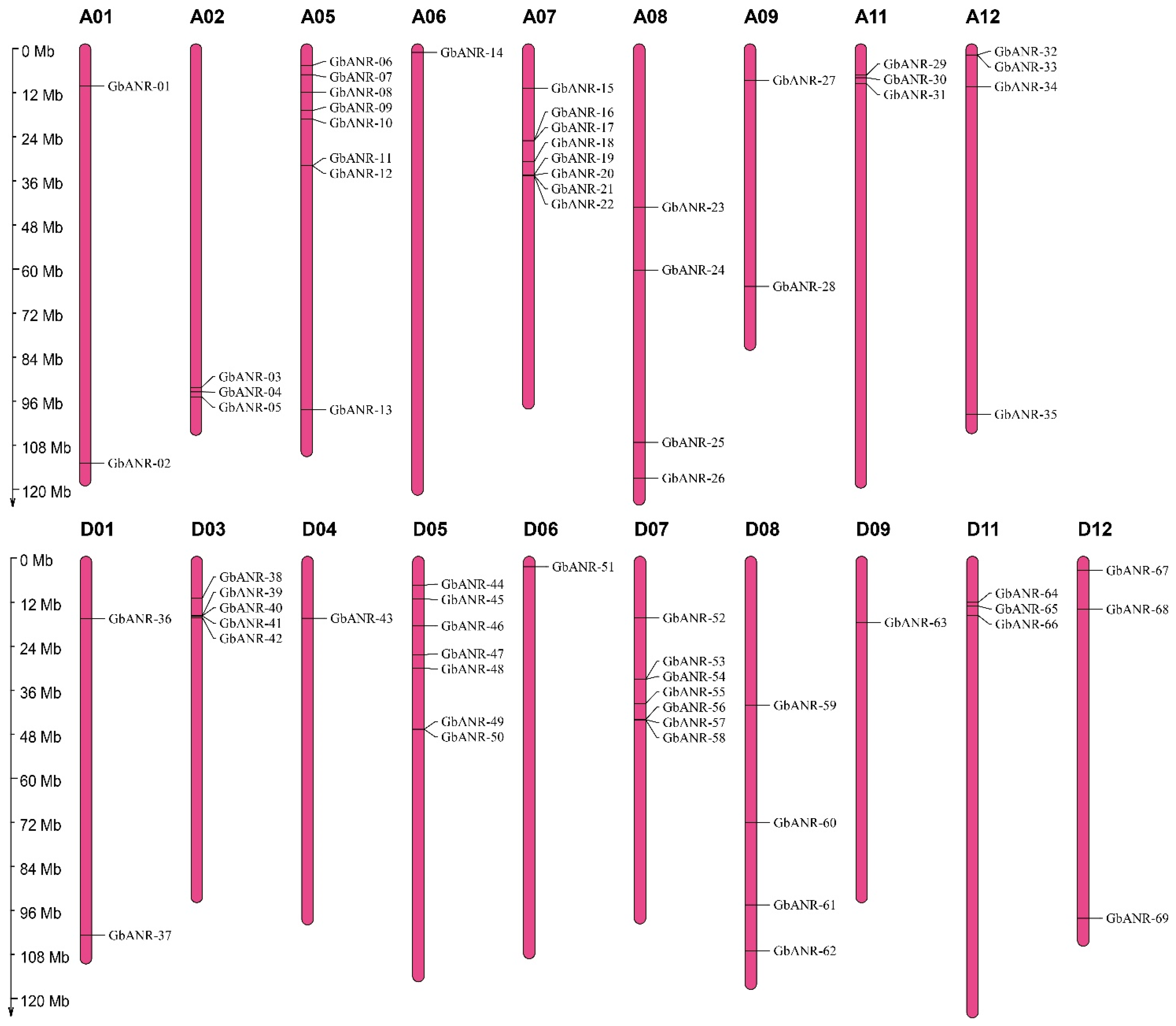
2.1. Identification of the ANR Gene Family Members in Cotton
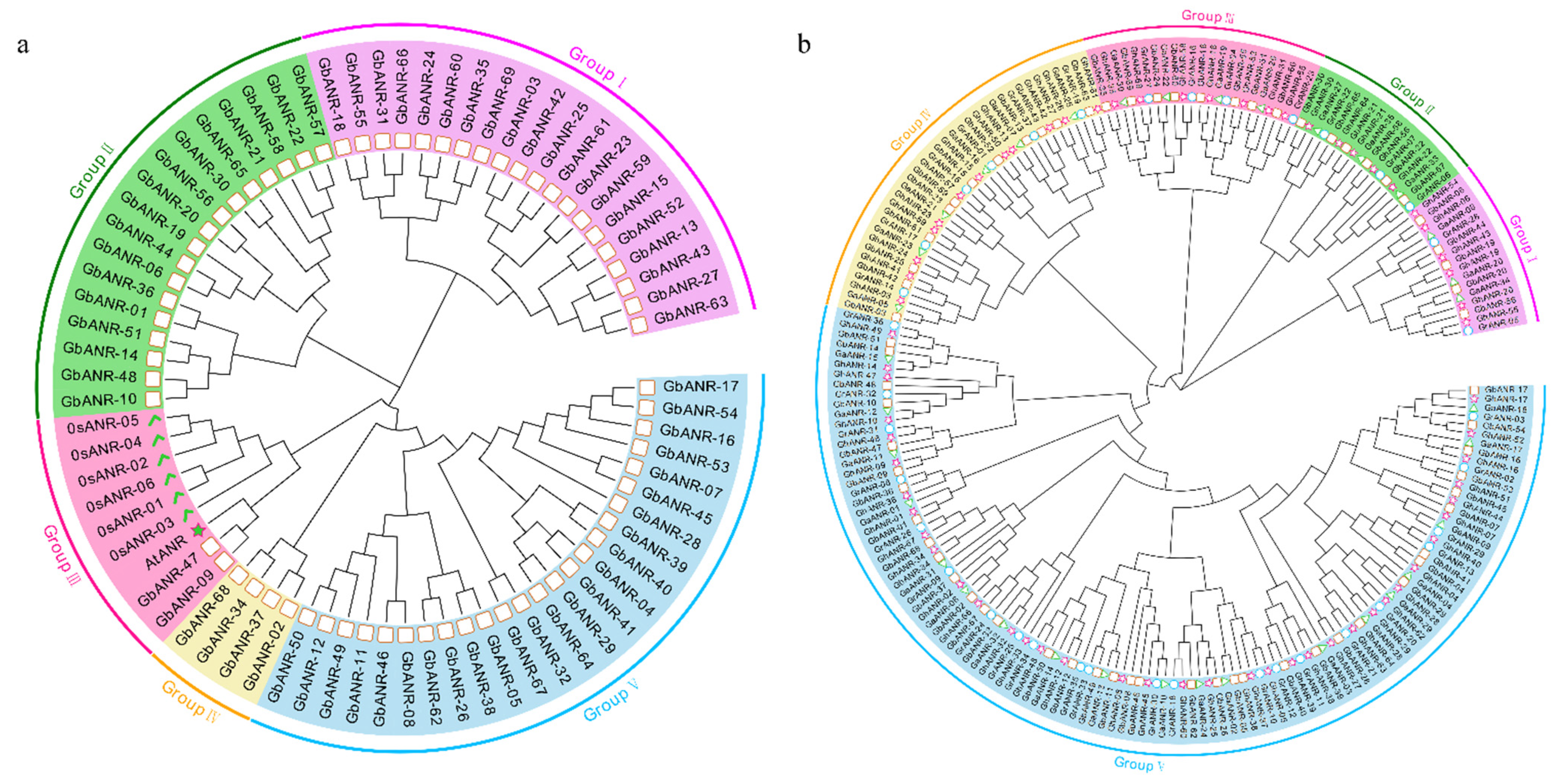
2.2. Evolutionary Analysis of the ANR Genes in Cotton
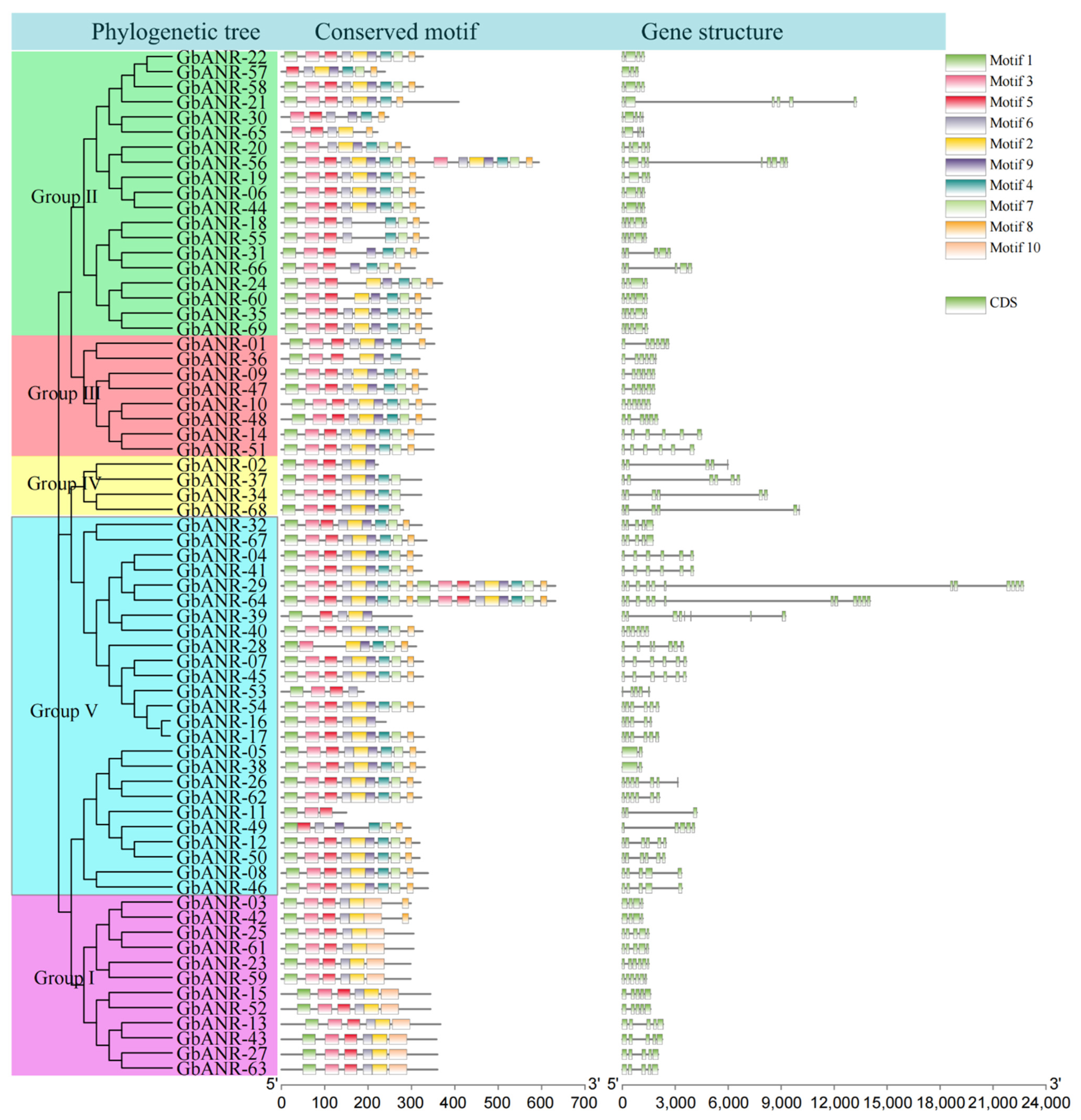
2.3. Evolutionary Tree, Gene Structure, and Motif Analysis of Cotton ANR Genes
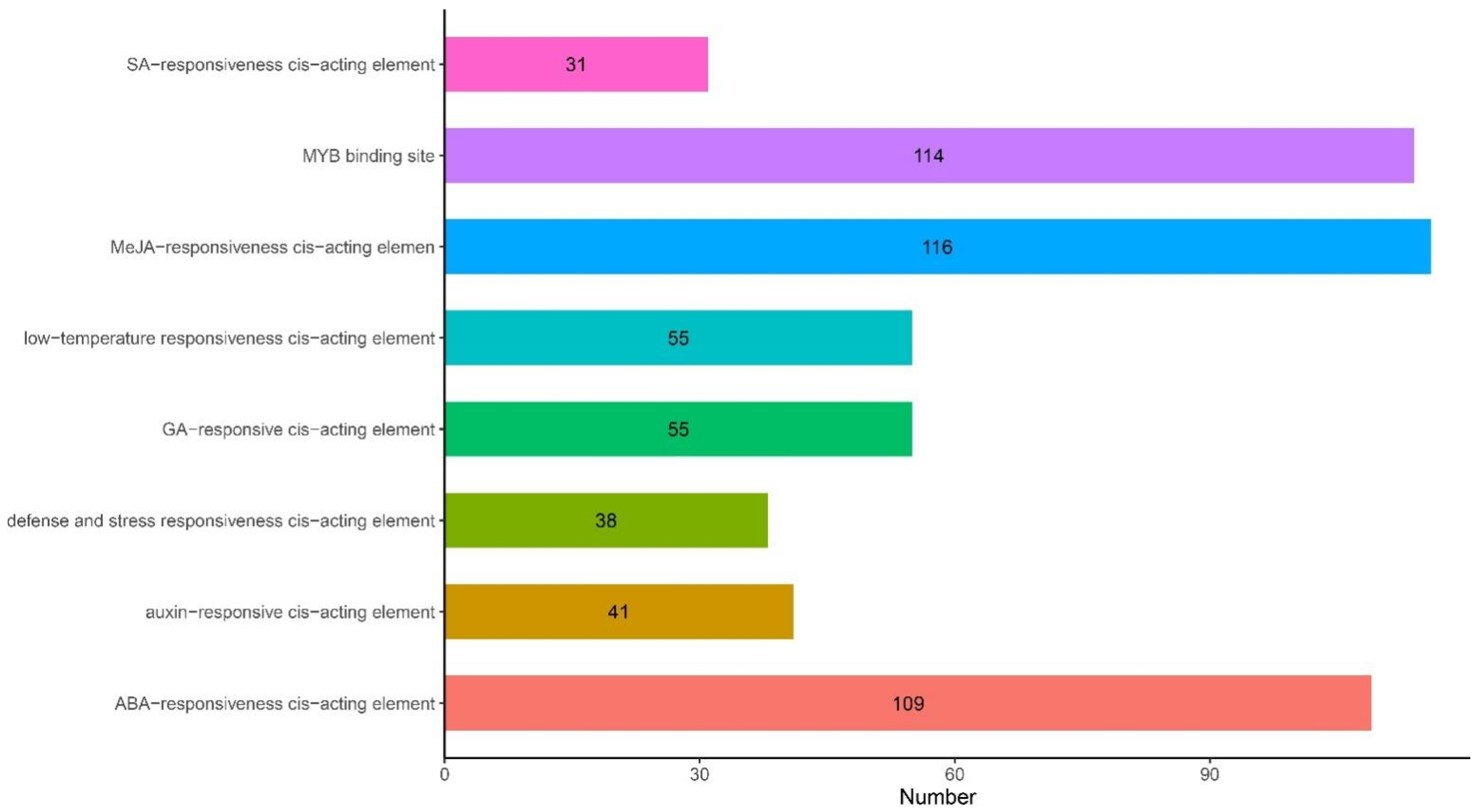
2.4. Analysis of Cis-Acting Elements of ANR Genes
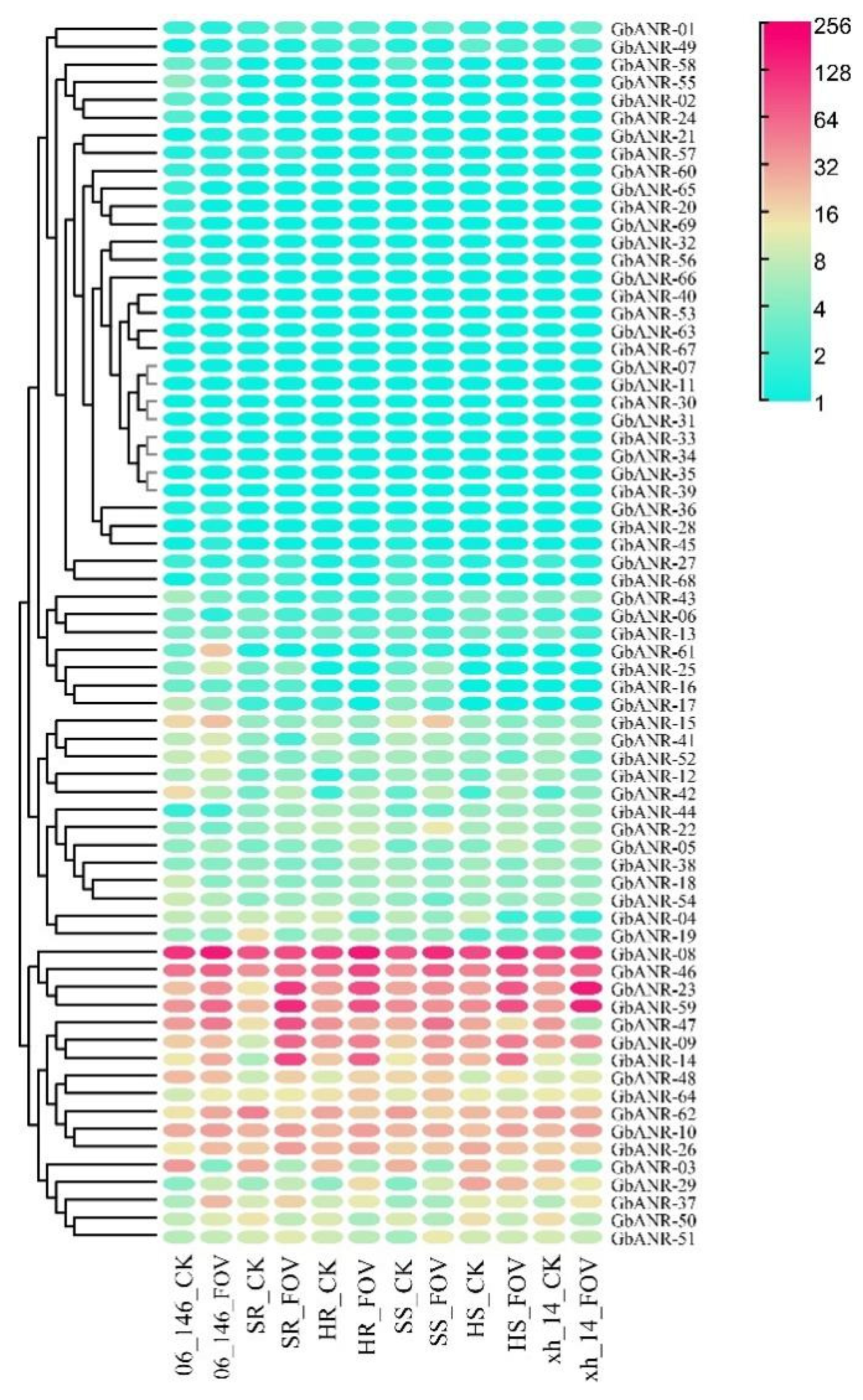
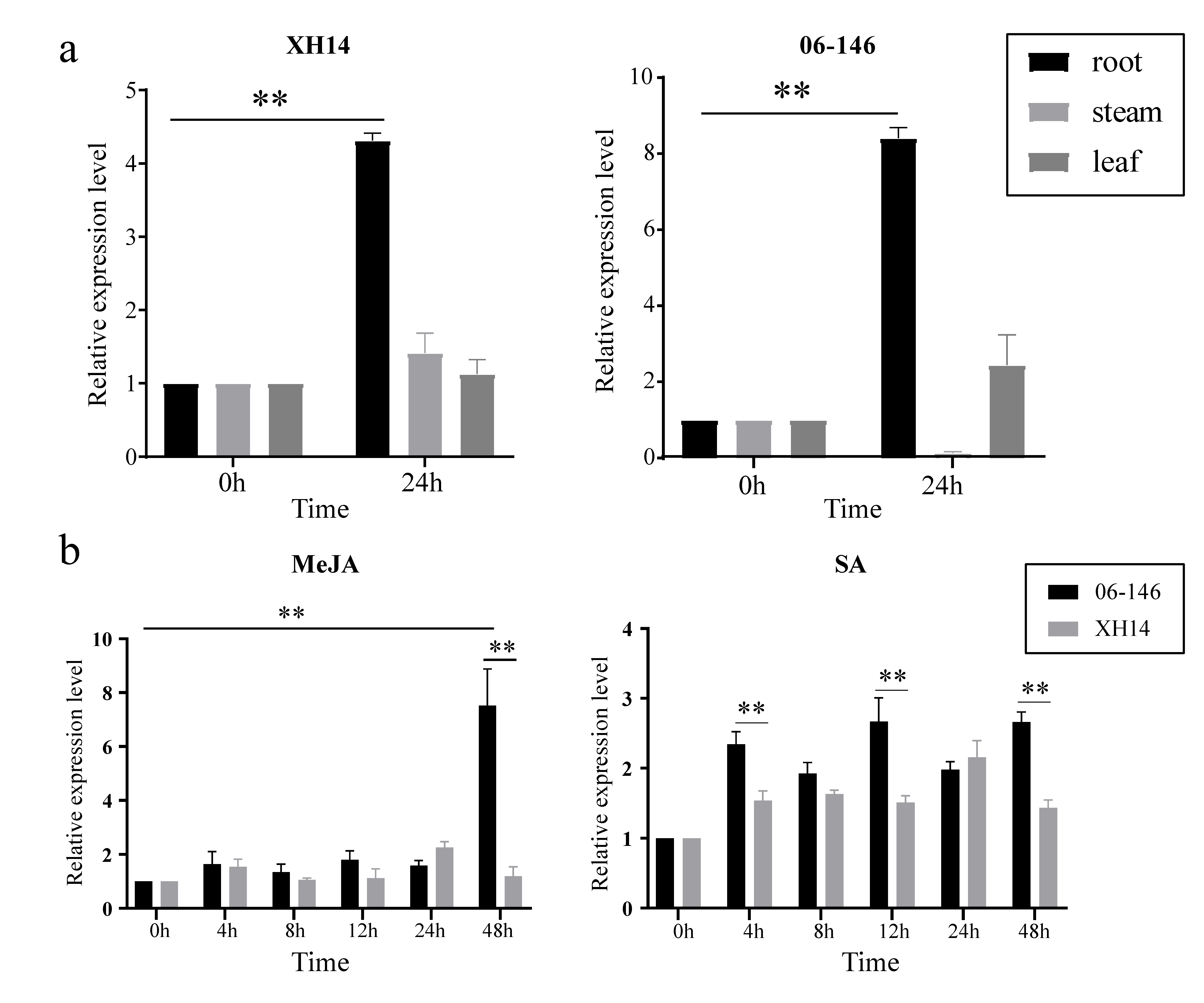
2.5. ANR Gene Expression Analysis of G. barbadense
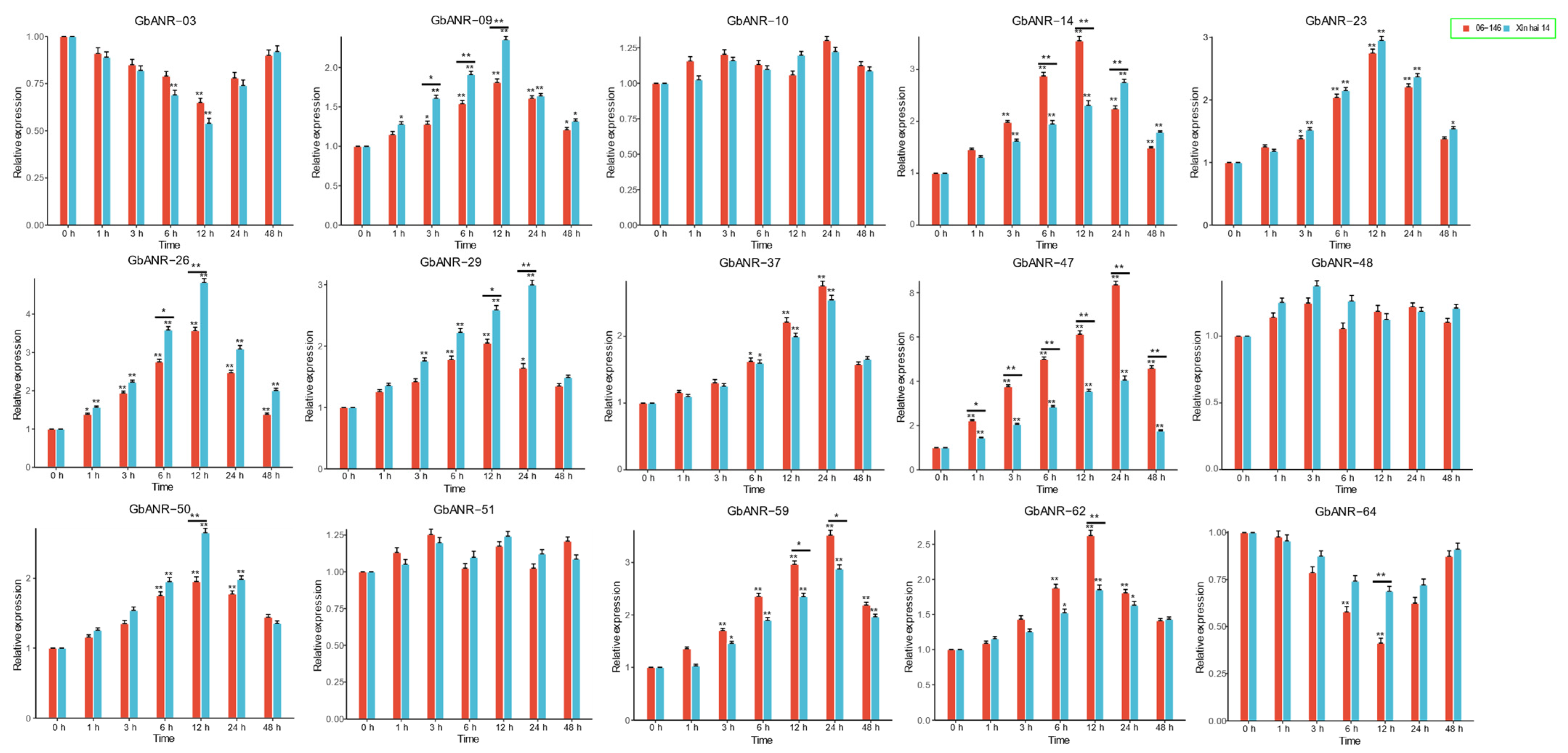
2.6. qRT–PCR
2.7. VIGS
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Identification and Bioinformatics Analysis of the ANR Genes in Cotton
4.3. Phylogenetic and Collinearity Analysis of ANR Gene Family Members in Cotton
4.4. Chromosomal Location, Gene Structure, and Motif Analysis of the ANR Gene Family
Members in Cotton
4.5. Analysis of Cis-Acting Elements Upstream of the G. barbadense ANR Gene
4.6. RNA Sequencing (RNA-seq) Analysis
4.7. qRT–PCR
4.8. VIGS
4.9. Determination of Anthocyanin Content
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yu, J.; Hui, Y.; Chen, J.; Yu, H.; Gao, X.; Zhang, Z.; Li, Q.; Zhu, S.; Zhao, T. Whole-genome resequencing of 240 Gossypium barbadense accessions reveals genetic variation and genes associated with fiber strength and lint percentage. Theor. Appl. Genet. 2021, 134, 3249–3261. [Google Scholar] [CrossRef]
- Zhu, Y.; Abdelraheem, A.; Lujan, P.; Idowu, J.; Sullivan, P.; Nichols, R.; Wedegaertner, T.; Zhang, J. Detection and Characterization of Fusarium Wilt (Fusarium oxysporum f. sp. vasinfectum) Race 4 Causing Fusarium Wilt of Cotton Seedlings in New Mexico. Plant Dis. 2021, 105, 3353–3367. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Chen, Q.; Chen, D.; Zhan, L.; Zeng, K.; Gu, A.; Zhou, J.; Zhang, Y.; Zhu, Y.; Gao, W.; et al. The susceptibility of sea-island cotton recombinant inbred lines to Fusarium oxysporum f. sp. vasinfectum infection is characterized by altered expression of long noncoding RNAs. Sci. Rep. 2019, 9, 2894. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.; Zhang, H. A preliminary study on cotton yield loss caused by Fusarium wilt disease. Acta Phytopathol. Sin. 1986, 16, 245–251. [Google Scholar]
- Zhang, J.; Sanogo, S.; Ma, Z.; Qu, Y. Breeding, Genetics, and Quantitative Trait Locus Mapping for Fusarium Wilt Resistance in Cotton. Crop. Sci. 2015, 55, 2435–2452. [Google Scholar] [CrossRef]
- Hijaz, F.; Al-Rimawi, F.; Manthey, J.A.; Killiny, N. Phenolics, flavonoids and antioxidant capacities in Citrus species with different degree of tolerance to Huanglongbing. Plant Signal. Behav. 2020, 15, 1752447. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; Du, Y.; Yu, X.; Shi, J.; Yuan, X.; Liu, X.; Zhang, H.; Zhang, Z.; Yan, N. Dynamics of antioxidant activities, metabolites, phenolic acids, flavonoids, and phenolic biosynthetic genes in germinating Chinese wild rice (Zizania latifolia). Food Chem. 2020, 318, 126483. [Google Scholar] [CrossRef]
- Zhou, Y.; Jiao, Y.; Sun, Y.; Gao, S. In vitro production and distribution of flavonoids in Glycyrrhiza uralensis Fisch. J. Food Sci. Technol. 2020, 57, 1553–1564. [Google Scholar] [CrossRef]
- Mohammed, S.A.A.; Suresh, M. Anti-Fungal Efficacy and Mechanisms of Flavonoids. Antibiotics 2020, 9, 45. [Google Scholar]
- Achika, J.I.; Ayo, R.G.; Oyewale, A.O.; Habila, J.D. Flavonoids with antibacterial and antioxidant potentials from the stem bark of Uapaca heudelotti. Heliyon 2020, 6, e03381. [Google Scholar] [CrossRef]
- Yahia, Y.; Benabderrahim, M.A.; Tlili, N.; Hannachi, H.; Ayadi, L.; Elfalleh, W. Comparison of Three Extraction Protocols for the Characterization of Caper (Capparis spinosa L.) Leaf Extracts: Evaluation of Phenolic Acids and Flavonoids by Liquid Chromatography—Electrospray Ionization—Tandem Mass Spectrometry (LC–ESI–MS) and the Antioxidant Activity. Anal. Lett. 2020, 53, 1366–1377. [Google Scholar]
- Xu, F.; Cao, S.; Wang, C.; Wang, K.; Wei, Y.; Shao, X.; Wang, H. Antimicrobial activity of flavonoids from Sedum aizoon L. against Aeromonas in culture medium and in frozen pork. Food Sci. Nutr. 2019, 7, 3224–3232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.; Vimolmangkang, S.; Soria-Guerra, R.E.; Korban, S.S. Introduction of apple ANR genes into tobacco inhibits expression of both CHI and DFR genes in flowers, leading to loss of anthocyanin. J. Exp. Bot. 2012, 63, 2437–2447. [Google Scholar] [CrossRef] [Green Version]
- Luo, P. Cloning and Functional Analysis of Genes Related to Rose Flavonoid Synthesis. Ph.D. Thesis, Huazhong Agricultural University, Wuhan, China, 2016. [Google Scholar]
- Ikuo, K.; Yasutaka, K.; Akira, S.; Takashi, T. Changes in L-Phenylalanine Ammonia-lyase Activity and Anthocyanin Synthesis during Berry Ripening of Three Grape Cultivars. J. Jpn. Soc. Hortic. Sci. 1983, 52, 273–279. [Google Scholar]
- Hu, Y.; Chen, J.; Fang, L.; Zhang, Z.; Ma, W.; Niu, Y.; Ju, L.; Deng, J.; Zhao, T.; Lian, J.; et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 2019, 4, 51. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Wang, Z.; Li, F.; Ye, W.; Wang, J.; Song, G.; Yue, Z.; Cong, L.; Shang, H.; Zhu, S.; et al. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 2012, 44, 7052. [Google Scholar] [CrossRef]
- Du, X.; Huang, G.; He, S.; Yang, Z.; Sun, G.; Ma, X.; Li, N.; Zhang, X.; Sun, J.; Liu, M.; et al. Resequencing of 243 diploid cotton accessions based on an updated A genome identifies the genetic basis of key agronomic traits. Nat. Genet. 2018, 6, 50. [Google Scholar] [CrossRef]
- Wang, M.; Tu, L.; Yuan, D.; Zhu, D.; Shen, C.; Li, J.; Liu, F.; Pei, L.; Wang, P.; Zhao, G.; et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 2019, 2, 51. [Google Scholar] [CrossRef] [Green Version]
- Tu, X.; Tan, J.; Guo, K.; Li, Z.; Zhang, X. Flavonoid Pathway in Cotton Fiber Development. Sci. Sin. Vitae 2014, 44, 758–765. [Google Scholar]
- Trainin, T.; Harel, B.R.; Bar, Y.I.; Ben, S.Z.; Yahalomi, R.; Borochov, N.H.; Ophir, R.; Sherman, A.; Doron, F.A.; Holland, D. Fine Mapping of the “black” Peel Color in Pomegranate (Punica granatum L.) Strongly Suggests That a Mutation in the Anthocyanidin Reductase (ANR) Gene Is Responsible for the Trait. Front. Plant Sci. 2021, 12, 642019. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, H.; Peng, Q.; Tang, Y.; Xia, G.; Wu, J.; Xie, D. Functional characterization of an anthocyanidin reductase gene from the fibers of upland cotton (Gossypium hirsutum). Planta 2015, 241, 1075–1089. [Google Scholar] [CrossRef] [PubMed]
- Pessi, G.; Haas, D. Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the quorum-sensing regulators LasR and RhlR in Pseudomonas aeruginosa. J. Bacteriol. 2000, 182, 6940–6949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Wang, P.; Gao, W.; Long, Y.; Wang, Y.; Geng, S.; Su, X.; Jiao, Y.; Chen, Q.; Qu, Y. Genome-wide identification of the DUF668 gene family in cotton and expression profiling analysis of GhDUF668 in Gossypium hirsutum under adverse stress. BMC Genom. 2021, 22, 395. [Google Scholar] [CrossRef]
- Suganuma, T.; Workman, J.L. Features of the PHF8/KIAA1718 histone demethylase. Cell Res. 2010, 20, 861–862. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Wang, Y.; Zhang, N.; Wu, Z.; Zeng, Q.; Wu, J.; Wu, X.; Wang, L.; Zhang, J.; Qi, Y. Genome-wide systematic characterization of the HAK/KUP/KT gene family and its expression profile during plant growth and in response to low-K+ stress in Saccharum. BMC Plant Biol. 2020, 20, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, L.; Zuo, D.; Wang, X.; Cheng, H.; Zhang, Y.; Wang, Q.; Song, G.; Ma, Z. Genome-wide identification of the expansin gene family reveals that expansin genes are involved in fibre cell growth in cotton. BMC Plant Biol. 2020, 20, 223. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Li, M.; Wang, S.; Yin, H. Revisiting the Role of Plant Transcription Factors in the Battle against Abiotic Stress. Int. J. Mol. Sci. 2018, 19, 1634. [Google Scholar] [CrossRef] [Green Version]
- Seo, E.; Choi, D. Functional studies of transcription factors involved in plant defenses in the genomics era. Brief. Funct. Genom. 2015, 14, 260–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Po, H.; Wu, Z.; Zhi, C.; Meng, F.; Lei, W.; Dao, X. JAV1 Controls Jasmonate-Regulated Plant Defense. Mol. Cell 2013, 50, 504–515. [Google Scholar]
- Han, X.; Zhang, M.; Yang, M.; Hu, Y. Arabidopsis JAZ Proteins Interact with and Suppress RHD6 Transcription Factor to Regulate Jasmonate-Stimulated Root Hair Development. Plant Cell 2020, 32, 1049–1062. [Google Scholar] [CrossRef]
- Zhi, Y.; Yu, H.; Jia, Y.; Sheng, Y.; Kun, Z.; Dong, W.; Qing, Q.; Zhan, B.; Yan, L.; Ying, L.; et al. Jasmonate Signaling Enhances RNA Silencing and Antiviral Defense in Rice. Cell Host Microbe 2020, 28, 89–103. [Google Scholar]
- Zhao, N.; Wang, W.; Grover, C.E.; Jiang, K.; Pan, Z.; Guo, B.; Zhu, J.; Su, Y.; Wang, M.; Nie, H.; et al. Genomic and GWAS analyses demonstrate phylogenomic relationships of Gossypium barbadense in China and selection for fibre length, lint percentage and Fusarium wilt resistance. Plant Biotechnol. J. 2021, 20, 691. [Google Scholar] [CrossRef]
- Ni, J.; Premathilake, A.T.; Gao, Y.; Yu, W.; Tao, R.; Teng, Y.; Bai, S. Ethylene-activated PpERF105 induces the expression of the repressor-type R2R3-MYB gene PpMYB140 to inhibit anthocyanin biosynthesis in red pear fruit. Plant J. Cell Mol. Biol. 2020, 105, 167–181. [Google Scholar] [CrossRef]
- Shen, Y.; Sun, T.; Pan, Q.; Anupol, N.; Chen, H.; Shi, J.; Liu, F.; Deqiang, D.; Wang, C.; Zhao, J.; et al. RrMYB5- and RrMYB10-regulated flavonoid biosynthesis plays a pivotal role in feedback loop responding to wounding and oxidation in Rosa rugosa. Plant Biotechnol. J. 2019, 17, 2078–2095. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Liu, L.; Ma, K.; Wang, W.; Lv, H.; Gao, M.; Wang, X.; Zhang, X.; Ren, S.; Zhang, N.; et al. The Jasmonate-Induced bHLH Gene SlJIG Functions in Terpene Biosynthesis and Resistance to Insects and Fungus. J. Integr. Plant Biol. 2022, 64, 1102–1115. [Google Scholar] [CrossRef]
- Lister, C.; Lancaster, J.; Walker, J. Phenylalanine Ammonia-Lyase Activity and Anthocyanin Synthesis in Ripening Strawberry Fruit. J. Am. Soc. Hortic. Sci. 1996, 121, 25–30. [Google Scholar]
- Yu, J.; Jung, S.; Cheng, C.; Ficklin, S.P.; Lee, T.; Zheng, P.; Jones, D.; Percy, R.G.; Main, D. CottonGen: A genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 2014, 42, D1229–D1236. [Google Scholar] [CrossRef] [Green Version]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [Green Version]
- Mariethoz, J.; Alocci, D.; Gastaldello, A.; Horlacher, O.; Gasteiger, E.; Rojas-Macias, M.; Karlsson, N.G.; Packer, N.H.; Lisacek, F. Glycomics@ExPASy: Bridging the Gap. Mol. Cell. Proteom. 2018, 11, 17. [Google Scholar] [CrossRef] [Green Version]
- Chang, T.; Wu, L.; Lee, T.; Chen, S.; Huang, H.; Horng, J. EuLoc: A web-server for accurately predict protein subcellular localization in eukaryotes by incorporating various features of sequence segments into the general form of Chou’s PseAAC. J. Comput.-Aided Mol. Des. 2013, 1, 91–103. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 7, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 1, 93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 8, 13. [Google Scholar] [CrossRef]
- Tanino, Y.; Kodama, M.; Daicho, H.; Miyauchi, Y.; Yasumoto, T.; Yamada, Y.; Kyotani, N.; Kurahashi, S.; Ushiyama, M.; Kimura, T.; et al. Selection of Laboratory Procedures to Detect Toxigenic by the 2-Step Method. Rinsho Biseibutshu Jinsoku Shindan Kenkyukai Shi 2017, 1, 9–14. [Google Scholar]
- Akio Amorim, L.L.; da Fonseca Dos Santos, R.; Pacifico Bezerra Neto, J.; Guida-Santos, M.; Crovella, S.; Maria Benko-Iseppon, A. Transcription Factors Involved in Plant Resistance to Pathogens. Curr. Protein Pept. Sci. 2017, 18, 335–351. [Google Scholar] [CrossRef]

| Gene Name | Gene ID | Open Reading Frame/bp | Protein Length/aa | Relative Molecular Weight(r)/kDa | Theoretical Isoelectric Point (pI) | Subcellular Localization |
|---|---|---|---|---|---|---|
| GbANR-01 | Gb_A01G0696 | 1062 | 353 | 38.66 | 5.65 | cytoplasm |
| GbANR-02 | Gb_A01G2252 | 672 | 223 | 24.64 | 7.54 | nucleus |
| GbANR-03 | Gb_A02G1539 | 900 | 299 | 33.45 | 5.31 | chloroplast |
| GbANR-04 | Gb_A02G1557 | 975 | 324 | 35.44 | 8.71 | cytoplasm |
| GbANR-05 | Gb_A02G1593 | 996 | 331 | 36.10 | 6.36 | cytoplasm |
| GbANR-06 | Gb_A05G0439 | 987 | 328 | 35.84 | 5.83 | cytoplasm |
| GbANR-07 | Gb_A05G0715 | 984 | 327 | 35.62 | 5.77 | cytoplasm |
| GbANR-08 | Gb_A05G1219 | 1017 | 338 | 37.23 | 6.33 | cytoplasm |
| GbANR-09 | Gb_A05G1702 | 1011 | 336 | 36.27 | 5.54 | cytoplasm |
| GbANR-10 | Gb_A05G1949 | 1068 | 355 | 39.65 | 5.67 | cytoplasm |
| GbANR-11 | Gb_A05G2812 | 453 | 150 | 16.27 | 8.84 | extracellular space |
| GbANR-12 | Gb_A05G2814 | 960 | 319 | 35.12 | 6.66 | cytoplasm |
| GbANR-13 | Gb_A05G3871 | 1104 | 367 | 41.08 | 6.06 | nucleus |
| GbANR-14 | Gb_A06G0087 | 1056 | 351 | 39.04 | 5.45 | cytoplasm |
| GbANR-15 | Gb_A07G0794 | 1035 | 344 | 38.50 | 6.48 | nucleus |
| GbANR-16 | Gb_A07G1334 | 726 | 241 | 25.89 | 8.10 | organelle membrane |
| GbANR-17 | Gb_A07G1335 | 990 | 329 | 35.89 | 6.46 | cytoplasm |
| GbANR-18 | Gb_A07G1511 | 1020 | 339 | 36.68 | 7.04 | endomembrane system |
| GbANR-19 | Gb_A07G1592 | 990 | 329 | 36.12 | 5.55 | cytoplasm |
| GbANR-20 | Gb_A07G1593 | 891 | 296 | 32.25 | 5.02 | cytoplasm |
| GbANR-21 | Gb_A07G1594 | 1230 | 409 | 46.21 | 9.22 | endomembrane system |
| GbANR-22 | Gb_A07G1596 | 984 | 327 | 35.88 | 5.94 | cytoplasm |
| GbANR-23 | Gb_A08G1080 | 897 | 298 | 33.21 | 5.02 | cytoplasm |
| GbANR-24 | Gb_A08G1297 | 1116 | 371 | 41.31 | 5.68 | nucleus |
| GbANR-25 | Gb_A08G1918 | 918 | 305 | 34.32 | 5.34 | nucleus |
| GbANR-26 | Gb_A08G2417 | 966 | 321 | 35.37 | 5.62 | endomembrane system |
| GbANR-27 | Gb_A09G0335 | 1083 | 360 | 39.91 | 6.04 | cytoplasm |
| GbANR-28 | Gb_A09G1409 | 936 | 311 | 34.18 | 8.38 | cytoplasm |
| GbANR-29 | Gb_A11G0776 | 1899 | 632 | 70.31 | 5.88 | cytoplasm |
| GbANR-30 | Gb_A11G0849 | 744 | 247 | 27.51 | 8.57 | cytoplasm |
| GbANR-31 | Gb_A11G1008 | 1017 | 338 | 36.70 | 5.81 | cytoplasm |
| GbANR-32 | Gb_A12G0107 | 975 | 324 | 36.05 | 5.89 | cytoplasm |
| GbANR-33 | Gb_A12G0108 | 306 | 101 | 10.96 | 8.78 | extracellular space |
| GbANR-34 | Gb_A12G0520 | 972 | 323 | 35.73 | 6.66 | cytoplasm |
| GbANR-35 | Gb_A12G2641 | 1041 | 346 | 37.76 | 6.51 | plasma membrane |
| GbANR-36 | Gb_D01G0734 | 960 | 319 | 34.87 | 5.85 | nucleus |
| GbANR-37 | Gb_D01G2338 | 972 | 323 | 35.86 | 5.79 | endomembrane system |
| GbANR-38 | Gb_D03G0433 | 996 | 331 | 36.05 | 7.12 | cytoplasm |
| GbANR-39 | Gb_D03G0545 | 906 | 301 | 33.94 | 6.02 | nucleus |
| GbANR-40 | Gb_D03G0546 | 981 | 326 | 35.66 | 5.73 | organelle membrane |
| GbANR-41 | Gb_D03G0549 | 975 | 324 | 35.45 | 8.94 | cytoplasm |
| GbANR-42 | Gb_D03G0557 | 900 | 299 | 33.42 | 5.40 | chloroplast |
| GbANR-43 | Gb_D04G0591 | 1077 | 358 | 39.86 | 7.56 | chloroplast |
| GbANR-44 | Gb_D05G0439 | 990 | 329 | 35.96 | 5.39 | cytoplasm |
| GbANR-45 | Gb_D05G0704 | 984 | 327 | 35.60 | 5.77 | cytoplasm |
| GbANR-46 | Gb_D05G1207 | 1017 | 338 | 37.19 | 6.33 | cytoplasm |
| GbANR-47 | Gb_D05G1725 | 1011 | 336 | 36.34 | 5.55 | cytoplasm |
| GbANR-48 | Gb_D05G1976 | 1068 | 355 | 39.75 | 5.41 | cytoplasm |
| GbANR-49 | Gb_D05G2804 | 897 | 298 | 33.41 | 8.89 | mitochondrion |
| GbANR-50 | Gb_D05G2805 | 960 | 319 | 35.06 | 6.61 | cytoplasm |
| GbANR-51 | Gb_D06G0087 | 1056 | 351 | 38.94 | 5.34 | cytoplasm |
| GbANR-52 | Gb_D07G0801 | 1035 | 344 | 38.36 | 5.98 | nucleus |
| GbANR-53 | Gb_D07G1332 | 573 | 190 | 21.55 | 7.68 | cytoplasm |
| GbANR-54 | Gb_D07G1333 | 990 | 329 | 35.79 | 7.04 | cytoplasm |
| GbANR-55 | Gb_D07G1524 | 1020 | 339 | 36.64 | 6.78 | endomembrane system |
| GbANR-56 | Gb_D07G1612 | 1785 | 594 | 64.74 | 4.92 | cytoplasm |
| GbANR-57 | Gb_D07G1616 | 720 | 239 | 26.62 | 6.08 | cytoplasm |
| GbANR-58 | Gb_D07G1617 | 984 | 327 | 35.72 | 6.46 | cytoplasm |
| GbANR-59 | Gb_D08G1045 | 882 | 293 | 33.14 | 5.02 | cytoplasm |
| GbANR-60 | Gb_D08G1399 | 1035 | 344 | 38.30 | 5.54 | endomembrane system |
| GbANR-61 | Gb_D08G1912 | 918 | 305 | 34.41 | 5.48 | cytoplasm |
| GbANR-62 | Gb_D08G2407 | 972 | 323 | 35.58 | 5.94 | cytoplasm |
| GbANR-63 | Gb_D09G0306 | 1083 | 360 | 40.00 | 5.97 | cytoplasm |
| GbANR-64 | Gb_D11G0798 | 1899 | 632 | 70.34 | 6.56 | cytoplasm |
| GbANR-65 | Gb_D11G0879 | 669 | 222 | 24.52 | 6.45 | cytoplasm |
| GbANR-66 | Gb_D11G1037 | 927 | 308 | 33.72 | 5.58 | cytoplasm |
| GbANR-67 | Gb_D12G0116 | 1008 | 335 | 37.19 | 6.16 | cytoplasm |
| GbANR-68 | Gb_D12G0517 | 846 | 281 | 30.91 | 6.93 | cytoplasm |
| GbANR-69 | Gb_D12G2647 | 1044 | 347 | 37.92 | 6.75 | endomembrane system |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, X.; Zhao, J.; Gao, W.; Zu, Q.; Chen, Q.; Li, C.; Qu, Y. Gb_ANR-47 Enhances the Resistance of Gossypium barbadense to Fusarium oxysporum f. sp. vasinfectum (FOV) by Regulating the Content of Proanthocyanidins. Plants 2022, 11, 1902. https://doi.org/10.3390/plants11151902
Su X, Zhao J, Gao W, Zu Q, Chen Q, Li C, Qu Y. Gb_ANR-47 Enhances the Resistance of Gossypium barbadense to Fusarium oxysporum f. sp. vasinfectum (FOV) by Regulating the Content of Proanthocyanidins. Plants. 2022; 11(15):1902. https://doi.org/10.3390/plants11151902
Chicago/Turabian StyleSu, Xuening, Jieyin Zhao, Wenju Gao, Qianli Zu, Quanjia Chen, Chunping Li, and Yanying Qu. 2022. "Gb_ANR-47 Enhances the Resistance of Gossypium barbadense to Fusarium oxysporum f. sp. vasinfectum (FOV) by Regulating the Content of Proanthocyanidins" Plants 11, no. 15: 1902. https://doi.org/10.3390/plants11151902
APA StyleSu, X., Zhao, J., Gao, W., Zu, Q., Chen, Q., Li, C., & Qu, Y. (2022). Gb_ANR-47 Enhances the Resistance of Gossypium barbadense to Fusarium oxysporum f. sp. vasinfectum (FOV) by Regulating the Content of Proanthocyanidins. Plants, 11(15), 1902. https://doi.org/10.3390/plants11151902





