Abstract
Leaf veins constitute the transport network for water and photosynthetic assimilates in vascular plants. The class III homeodomain-leucine zipper (HD-Zip III) gene family is central to the regulation of vascular development. In this research, we performed an overall analysis of the HD-Zip III genes in soybean (Glycine max L. Merr.). Our analysis included the phylogeny, conservation domains and cis-elements in the promoters of these genes. We used the quantitative reverse transcription-polymerase chain reaction to characterize the expression patterns of HD-Zip III genes in leaf vein development and analyze the effects of exogenous hormone treatments. In this study, twelve HD-Zip III genes were identified from the soybean genome and named. All soybean HD-Zip III proteins contained four highly conserved domains. GmHB15-L-1 transcripts showed steadily increasing accumulation during all stages of leaf vein development and were highly expressed in cambium cells. GmREV-L-1 and GmHB14-L-2 had nearly identical expression patterns in soybean leaf vein tissues. GmREV-L-1 and GmHB14-L-2 transcripts remained at stable high levels at all xylem developmental stages. GmREV-L-1 and GmHB14-L-2 were expressed at high levels in the vascular cambium and xylem cells. Overall, GmHB15-L-1 may be an essential regulator that is responsible for the formation or maintenance of soybean vein cambial cells. GmREV-L-1 and GmHB14-L-2 were correlated with xylem differentiation in soybean leaf veins. This study will pave the way for identifying the molecular mechanism of leaf vein development.
1. Introduction
Leaf veins provide mechanical support and are at the forefront of nutrient and water transport in photosynthesis and transpiration [1,2]. Leaf veins consist of the xylem, phloem, and cambium, which are highly organized within the vascular bundle. Cells in the cambium can differentiate to form either the phloem or the xylem. Then, the xylem and phloem are further expanded via the differentiation of cells derived from divisions in the cambium [3]. Leaves develop an adaxial side specialized for light capture, and an abaxial side specialized for gas exchange. Leaf veins are typically positioned where the adaxial and abaxial domains meet. The xylem is present in the adaxial position, and the phloem is present in the abaxial position [3,4]. The characteristics of leaf veins are directly or indirectly related to the capacity for photosynthetic carbon fixation, water absorption, and anti-interference of plants, and are thus critical factors in adapting to adverse stress [5,6]. The optimization of leaf vasculature is essential for the efficient performance of crop plants [7]. Therefore, studying the molecular mechanism of vein development is of great theoretical significance and application value. The HD-Zip III gene family regulates almost all the vascular developmental fate decisions in this process, including vascular specification, patterning, and differentiation [8,9].
HD-Zip III family members have overlapping and antagonistic roles in controlling vascular development. Five HD-Zip III genes have been found in Arabidopsis (Arabidopsis thaliana): REV, PHB/ATHB9, PHV/ATHB14, ATHB8, and ATHB15/CAN [10]. PHB and PHV perform overlapping functions with REV in controlling the abaxial–adaxial patterning of organs [11]. The REV mutant causes a reduction in interfascicular xylem fibers [12,13]. The double mutants REV PHB and REV PHV enhance the vascular defects of the REV mutant, specifically, vascular bundles with remarkably few lignified cells [10]. Loss-of-function KANADI mutants exhibit vascular patterning defects opposite to that of the rev mutant, increasing the number of xylem cells [14]. HD-Zip III and KANADI have opposing roles in ad/abaxial of the organ formation. Once vascular are formed, HD-Zip III and KANADI are required to coordinate adaxial and abaxial growth [15]. The histological analysis of the ATHB8 single mutant revealed a regular vascular system without morphological alterations [16]. A functional redundancy might explain the lack of the evident phenotypes of ATHB8 mutants within the HD-Zip III family. Slight perturbations in vascular development are seen in CNA single mutant stems; i.e., the vascular bundles are frequently poorly distributed around the stem periphery [10]. Several pieces of evidence indicate mutual antagonism among the REV, CNA, and ATHB8 genes during vascular development. The CNA and ATHB8 mutations suppress the REV phenotype. The lignification of xylem tissue and interfascicular fibers are restored in ATHB8 CNA REV triple mutants [10].
Auxin is the primary signal involved in the ontogeny of the vascular system [17]. HD-Zip III is a regulatory factor in the regulation of vascular development by auxin (Brandt et al., 2012). Cambium formation is promoted by high auxin levels activating HD-Zip III transcription factors in Arabidopsis [18]. Previous studies have proposed the auxin-flow canalization hypothesis, which states that HD-Zip III genes form an integrated feedback loop along with auxin, the auxin polar transporter PIN, and the auxin response factor (MP/ARF) [19,20]. The onset of ATHB8 expression is directly and positively regulated by MP through an auxin response element in the ATHB8 promoter and is followed by the induction of the generation and maintenance of procambial cells.
Previous studies on various mutants of vascular patterns have revealed the involvement of another new player, brassinosteroids (BRs). BR is a key regulator of the xylem, by acting to stimulate cambium cell differentiation [21]. Over expression of the BR biosynthesis gene showed increased xylem formation [22]. Brassinazole (BRZ), a specific inhibitor of BR biosynthesis, inhibits the differentiation of tracheary cells from cambium cells; such suppression is reversed by the addition of BRs [23,24]. HD-Zip III genes function in zinnia (Zinnia elegans L.) vascular differentiation in response to BR signaling. The expression of ZeHB-12 and ZeHB-10 (REV homologous transcripts) coincides with the differentiation of cambium cells into tracheary cells. The expression of ZeHB10 and ZeHB12 is repressed by BRZ but is rapidly induced by BR [25].
Although considerable evidence indicates that the HD-Zip III gene family is the essential family in controlling stem and root development, the details of individual HD-Zip III genes in the development of leaf veins have not been resolved. The structure of the leaf veins directs the mobilization of photosynthates from source to sink, and the optimization of leaf vein architecture is essential for the efficient performance of crop plants [7]. Soybean (Glycine max L. Merr.) is one of the most important crops worldwide and produces substantial amounts of proteins and oils [26]. Genome-wide investigations on the HD-Zip gene family in soybean have been reported, and gene expression in soybean roots under dehydration and salt stress has been studied [5,27]. Updating the HD-Zip III gene family in soybean has become possible with the release of a new version of the soybean genome. In this study, we aimed to identify the HD-Zip III gene family in the soybean genome and analyze their phylogenetic relationship, structural characteristics, and cis-element regions. Then, we investigated the functions of HD-Zip III genes during leaf vein development in soybean.
2. Results
2.1. Identification of GmHD-Zip III Genes and Analysis of Basic Physicochemical Properties
In this work, twelve HD-Zip III genes were found in the soybean genome and named based on their orthologous in Arabidopsis (Figure S1). According to sequence orthologous, 12 GmHD-Zip III genes were divided into three groups. The largest group was Group 1, which contained six GmHD-Zip III genes; Group 2 and 3 had two and four genes, respectively (Figure 1). We also identified interspecific orthologous genes. The orthologous gene pairs were GmHB8-L-1\GmHB8-L-2, GmHB15-L-1\GmHB15-L-2, GmHB15-L-3\GmHB15-L-4, GmHB14-L-1\GmHB14-L-2, GmHB14-L-3\GmHB14-L-4, and GmREV-L-1\GmREV-L-2 (Figure S1). As shown in Table S2, a total of 60 HD-Zip III homologous proteins were identified from five legumes. The phylogenetic tree showed that HD-Zip III proteins had a high degree of homology across all of the investigated plants (Figure 1).
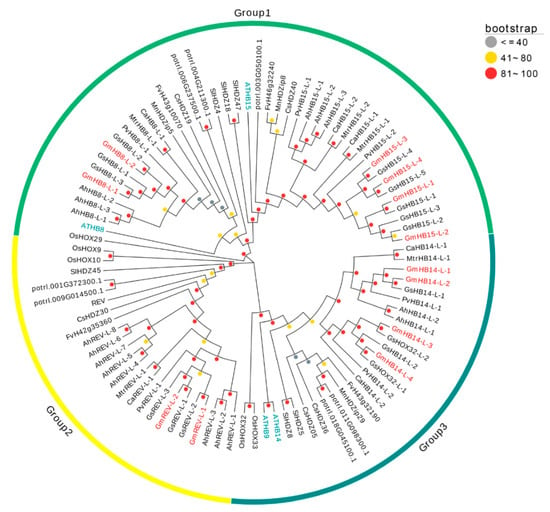
Figure 1.
Phylogenetic tree of full-length HD-Zip III proteins from soybean, chickpea, peanut, kidney bean, medicago, wild soybean, strawberry, cucumber, poplar, tomato, rice, and Arabidopsis. The 90 HD-Zip III proteins from 13 plant species can be divided into three groups. The words marked in blue are Arabidopsis HD-Zip III proteins, and those marked in red are GmHD-Zip III proteins. The colors of the circles indicate physical distances: gray represents long distances, and red means short distances.
Chromosomes 4, 5, 6, 9, 11, 12, and 15 contained only one GmHD-Zip III gene, whereas chromosome 7 contained two GmHD-Zip III genes. Chromosome 8 exhibited the highest density of GmHD-Zip III genes (Figure S2).
The results demonstrate that the HD-Zip III proteins have lengths of 838–853 amino acids and an average length of 844 amino acids. The theoretical isoelectric point value of GmREV-L-1 was the lowest, and that of GmHB14-L-3 was the highest. Protein hydrophobic and hydrophilic analysis showed that soybean HD-Zip III proteins were hydrophilic and unstable. The instability coefficients were between 45.74 and 50.47. Molecular weight analysis revealed that GmHB14-L-4 (93 958.15 Da) had the maximum protein molecular weight and that GmREV-L-1 (92 062.93 Da) had the minimum molecular weight (Table S3). Under the predicted subcellular localization of GmHD-Zip III, all of the genes in this family were located in the nucleus (Table S3).
2.2. Gene Structure, Conservative Domain, and Motif Analyses
We analyzed the conserved motifs of amino acid sequences (Figure S3). The results showed that all members of the HD-Zip III family contained almost the same motifs (1–10). Several had functional implications. For example, motif 6 specified the HD domain, motif 1 corresponded to the Zip domain, and motifs 12 and 14 represented the methionine–glutamic–lysine–histidine–leucine–alanine (MEKHLA) domain. Some motifs, such as 2, 4, and 3, specified the START domain. As revealed by the analysis of genetic structure, HD-Zip III gene structures were remarkably conserved. Each GmHD-Zip III gene had precisely 18 exons (Figure 2b). Exon length and exon–intron patterns were generally conserved.
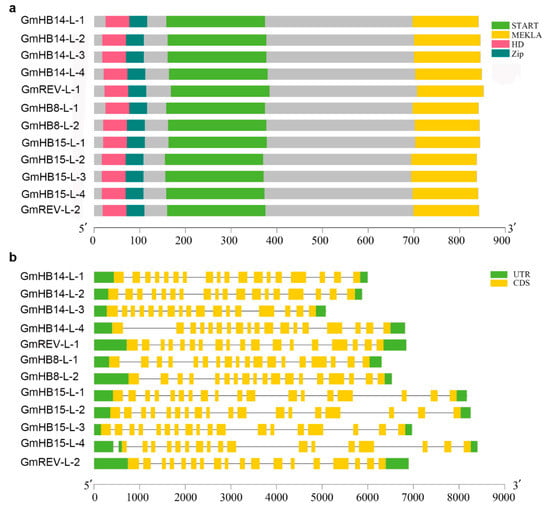
Figure 2.
Gene structure, conserved domain, and motif identification. (a) Phylogenetic relationships and protein domain prediction of HD-Zip III from soybean. The lengths of the proteins and domains can be estimated by using the scale at the bottom. (b) The gene structures of the 12 GmHD-Zip III genes. Exons and introns are shown as green and yellow boxes, respectively. Gene structures are shown in the right panel.
All of the 12 GmHD-Zip III proteins contained four highly conserved domains (Figure 2a), namely, START, MEKHLA, HD, and Zip. The closely related HD and Zip domains are the structural basis of this family of proteins, which together determine their functions as transcription factors. The START domain was predicted to be a lipid or steroid-binding receptor that can bind to small hydrophobic molecules, such as phospholipids and steroids, and is highly conserved in evolution [28]. The HD-Zip III family genes also contain an additional highly conserved MEKHLA domain. The MEKHLA domain is unique to the HD-Zip III family [29]. However, studies on the potential role of the MEKHLA domain in plants are limited. The MEKHLA domain shares high similarity with the Per-Arnt-Sim (PAS) domain [30]. The PAS domain might have originated from algae, and may be involved in the regulation of light signals received by plants and related to photosynthesis [31].
2.3. cis-Elements in GmHD-Zip III Promoters
cis-Acting elements play an important role in modulating the molecular switches of dynamic transcriptional regulation in response to developmental processes and hormonal signaling [32]. On the basis of their functions, the cis-acting elements were grouped into several classes: stress, development, and hormone-responsive elements (Table S4, Figure S4). All soybean HD-Zip III genes contained ARF elements, which are putative binding sites for auxin response factor (ARF/MP) proteins [33]. ARF proteins may bind ARF elements to activate or repress transcription of GmHD-Zip III genes (Figure 3).
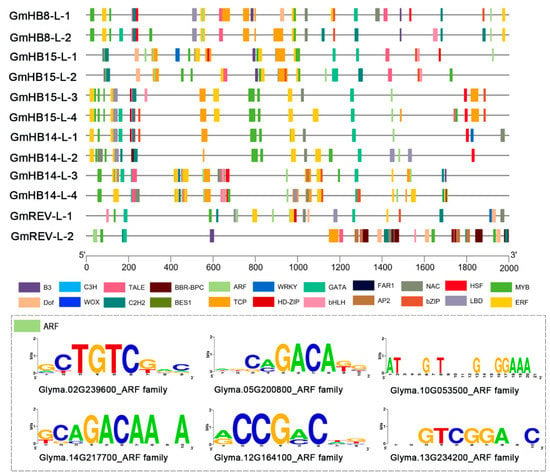
Figure 3.
Analysis of cis-acting elements in the promoter regions of GmHD-Zip III genes.
2.4. Expression Patterns of GmHD-Zip III Genes under Hormone Treatment
BRs are positive regulators of xylem differentiation, and BRZ, a specific inhibitor of BR biosynthesis, suppresses xylem differentiation [34]. We analyzed the effects of BRs and BRZ on the accumulation of GmHD-Zip III transcripts to investigate the relationships between the expression of these genes and BRs. Five GmHD-Zip III genes increased expression after BR treatment (Figure S5). GmHB8-L-1, GmHB14-L-1, GmHB14-L-2, GmREV-L-1, and GmHB15-L-2 all showed significantly upregulated expression levels. GmHB15-L-2 mRNA accumulation exhibited the greatest change. Expression analysis showed that BRZ strongly suppressed the accumulation of the transcripts of GmHB14-L-1, GmHB14-L-2, and GmREV-L-1 (Figure 4b). The other seven genes showed no significant changes. These results strongly suggest that the expression of GmHB14-L-1, GmHB14-L-2, and GmREV-L-1 can be induced or promoted by endogenous BRs. Our study also verified the promoting effect of BRs on xylem differentiation in leaf veins. The areas of xylem precursor cells, xylem cells, and lignin content in soybean leaf veins were significantly increased after 10 days of BR treatment (Figure 5a,b).
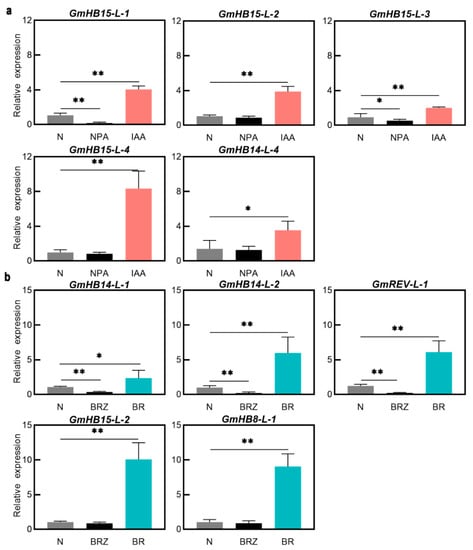
Figure 4.
(a) Expression profiles of GmHD-Zip III genes under IAA and NPA. (b) Expression profiles of GmHD-Zip III genes under BR and BRZ.Asterisks indicate significant differences between controls and treatments (* p < 0.05, ** p < 0.01).
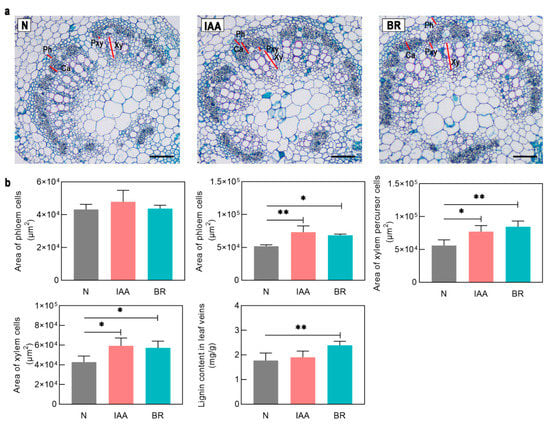
Figure 5.
(a) The anatomy of the leaf veins. Ca, cambium cell; Ph, phloem cell; TE, tracheary cell; Pxy, xylem precursor cell. All black scale bars indicate 100 μm. (b) Phenotypic analysis of soybean leaf veins under IAA and BR treatment. Asterisks indicate significant differences between controls and treatments (* p < 0.05, ** p < 0.01).
Auxin is involved in the regulation of vascular tissue development [35]. Treatment with a polar auxin transport inhibitor can mimic the vascular defect caused by HD-Zip III mutations [36]. Treatments with exogenous IAA and the auxin polar transport inhibitor NPA were applied to test whether these genes play an essential role in auxin-regulated vascular development. IAA significantly induced GmHB15-L-1, GmHB15-L-2, GmHB15-L-3, GmHB15-L-4, and GmHB14-L-4 (Figure S4), but did not induce changes in the other seven genes. GmHB15-L-2, GmHB14-L-4, and GmHB15-L-4 were not severely suppressed by NPA. NPA significantly inhibited the expression of GmHB15-L-1 and GmHB15-L-3 (Figure 4b). No absolute correlation was observed between the number of ARF elements and expression level. Although all GmHD-Zip III genes had ARF-elements, seven genes were not induced by auxin at the transcriptional level (Figure S4). These results provided support for the idea that auxin is the initiator of GmHB15-L-1 and GmHB15-L-3. The areas of leaf vein cambium cells, xylem percursor cells, and xylem cells were significantly increased after 10 days of IAA treatment (Figure 5a,b).
2.5. Expression of GmHD-Zip III Genes in Leaf Vein Development in Soybean
We analyzed the changes in the gene expression of GmHD-Zip III in samples obtained at different leaf vein developmental stages to understand the possible functions of GmHD-Zip III genes in leaf vein development in soybean. Figure 6a shows the transverse sections of leaf veins taken at five different soybean developmental stages. The results revealed that GmHB15-L-1 and GmREV-L-1 were very specifically expressed in vascular tissues at all stages, including the 0, 12, 24, 48, and 96 h stages, of vascular maturation (Figure 6a). GmHB15-L-1 was the earliest expressed gene in leaf vein development (Figure 5b). The first group genes (GmHB15-L-1, 4) exhibited similar expression patterns. As the leaf vein developed, the relative expression levels of the two genes gradually decreased. GmREV-L-1 and GmHB14-L-2 transcripts steadily accumulated during 12 to 48 h of development. As shown in Figure 6a, these time points corresponded to key xylem developmental stages. Subsequently, we examined gene expression in the cambium and the developing xylem and phloem. GmHB15-L-1 was expressed at low levels in the developing xylem and phloem but was highly expressed in the vascular cambium (Figure 7). GmREV-L-1 and GmHB14-L-2 were expressed at low levels in the developing phloem but expressed at high levels in the vascular cambium and xylem (Figure 7). The distinctive expression patterns of these genes in leaf veins imply their functional diversity in association with vascular development. These results suggest that GmHB15-L-1, GmREV-L-1, and GmHB14-L-2 play essential roles in leaf vein development.
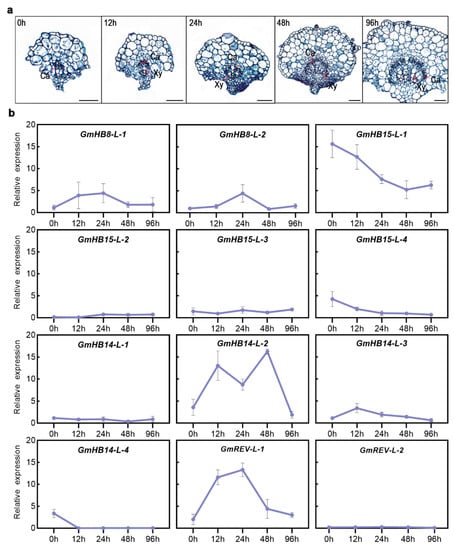
Figure 6.
(a) Transverse sections of soybean leaf veins in five different developmental stages. All black scale bars indicate 100 μm. (b) Expression profiles of 12 GmHD-Zip III genes in five stages of vein development. The normalized values of gene expression were log2-transformed and visualized in the form of heatmaps.
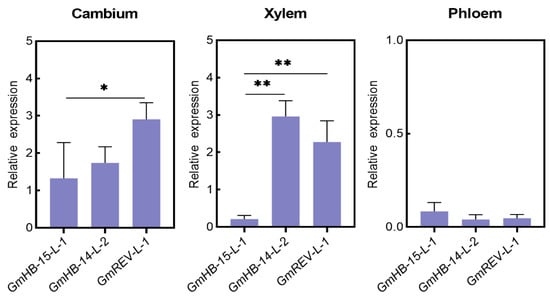
Figure 7.
Expression of soybean HD-Zip III genes in various types of vascular cells. Asterisks indicate significant differences between controls and treatments (* p < 0.05, ** p < 0.01).
3. Discussion
3.1. Phylogeny of GmHD-Zip III Genes Reflects Their Functional Conservation in Soybean
HD-Zip is a vital transcription factor family and exists only in the plant kingdom. HD-Zip transcription factors can be generally classified into the I, II, III, and IV subfamilies in accordance with their conserved sequences [37]. The majority of the reports available on HD-Zip I subfamilies respond to abiotic stresses and are crucial for maintaining plant growth under unfavorable environments [11]. The HD-Zip II gene family regulates the shade-avoiding mechanism during the photosynthetic process [38]. Genes in HD-Zip IV have well-characterized functions in trichome formation and epidermal cell differentiation [39,40]. Members of the third subfamily (HD-Zip III) control leaf polarity, embryogenesis, and vascular development [41]. The purpose of this study was to investigate the role of the HD-Zip gene family in soybean vein development. so we focused on the HD-Zip III subfamily rather than other subfamilies. In this work, 12 HD-Zip III genes were identified in the current version of the soybean genome (Figure 1). A previous work identified 11 GmHD-Zip III genes in the soybean genome (v1.01, JGI Glyma1.0) and investigated their responses to salt and dehydration stress [27]. Compared with the study of Belamkar et al., our study identified one more HD-Zip III member, namely, GLYMA_12G075800. All of the soybean HD-Zip III proteins that we identified belonged to the HD-Zip III family because they contained the four conserved domains of the HD-Zip III transcription family, START, MEKHLA, HD, and Zip. The difference between our results and the findings of Belamkar et al. may have been due to the use of the different released versions of the soybean genome. Eight HD-Zip III genes have been identified in poplar [42], four in barley [43], six in medicago [44], five in rice [45], four in strawberry [46], and five in cucumber [47]. The HD-Zip III gene family in soybean is by far more prevalent than in other plant species. The large number of GmHD-Zip III genes in soybean could have resulted from whole-genome duplication events. The soybean genome is believed to have undergone at least two independent duplications from a diploid ancestor to yield the actual polyploid plant [48].
The HD-Zip III gene family is very conserved. All of the identified HD-Zip III genes include four conserved domains [43,47,49,50]. In tomatoes and cotton, the locations and lengths of the motifs in the HD-Zip III protein sequences were also completely conserved [49,50]. The gene structures of the members of the soybean HD-Zip III family were highly similar in terms of exon and intron numbers and conserved protein motifs (Figure 2a,b). This high similarity further supported the reliability of the phylogenetic relationship analysis. The results of the phylogenetic analyses showed a high degree of the conservation of HD-Zip III protein sequences from different species. HD-Zip III genes may have similar functions in different species due to their conserved structure. Recent analyses on Arabidopsis, tomato, rice, and poplar have indicated that the HD-Zip III family is a key transcriptional regulator of vascular development and is highly expressed in the vascular tissues of roots, stems, and leaves [10,42,45,50]. The tissue-specific expression profiles obtained in this work suggested that soybean HD-Zip III genes were specifically expressed in the root, stem, and leaf (Figure S6). The first group was specifically expressed in leaves and stems. The expression level of the third group in roots was significantly higher than that in other sites. The GmHD-Zip III gene family is specifically expressed in different tissues with various subfamilies, and it may be involved in vascular development in certain sites.
3.2. Auxin Activating GmHB15-L-1 in the Regulation of Soybean Leaf Veins’ Cambium Development
The HD-Zip III family is at the center of a complex network required for initiating and maintaining plant vascular tissues [51]. However, such information is limited to roots and stems, and the general roles of HD-Zip III genes in the development of soybean leaf veins are still unknown. Despite the great variety of patterns in vascular systems, a common mechanism likely underlies the regulation of vascular tissue formation. In our study, GmHB15-L-1 was found to be a key player in the regulation of cambium formation in soybean leaf veins.
Considerable evidence indicates that auxin is the essential signal in promoting vascular cambium zone development. Decapitation of seedlings prevents the auxin supply from reaching the shoots and represses cambium activity, while application of exogenous IAA restores cambium activity [52]. Our results confirmed that auxin promoted the formation of the leaf vein cambium and showed that the area of leaf vein cambium cells increased significantly after 10 days of exogenous auxin treatment (Figure 5). In Arabidopsis, ATHB8 is a regulatory factor involved in the regulation of vascular cambium cell development by auxin [53]. The auxin element in the ATHB8 promoter is required for both ATHB8 procambial expression and auxin inducibility [20]. GmHB8-L-1 and GmHB8-L-2 had ARF elements, but these genes were not induced by auxin (Figure 3 and Figure S5). In poplar, PtrHB8 is highly expressed in the vascular cambium and developing xylem tissue [42]. This expression pattern is different from that of GmHB8-L-1,2 in soybean. The expression level of GmHB8-L-1,2 was low during vein development (Figure 5a). A likely candidate for regulating the cambium cell development is GmHB15-L-1, which is most similar to GmHB8-L-1. First, the expression of GmHB15-L-1 was significantly induced by IAA. The expression level of GmHB15-L-1 was significantly inhibited under treatment with the auxin polar transport inhibitor NPA (Figure 4a). Second, the expression pattern results revealed that GmHB15-L-1 expression levels were related to leaf vein development and showed steadily increasing transcript accumulation during all stages of leaf vein development (Figure 6b). The heat map data revealed that GmHB15-L-1 was the earliest expressed gene during vascular development (Figure 7). Third, GmHB15-L-1 was highly expressed in the vascular cambium. ZeHB-13, a homolog of the ATHB15 gene in zinnia, is localized preferentially in cambium cells. Histochemical promoter analysis using ATHB15::GUS transgenic Arabidopsis indicated that, consistent with the expression patterns of GmHB15-L-1 in soybean, ATHB15 is active specifically in the cambium [54]. Overall, these results strongly suggest that a member of the HD-Zip III family, namely, GmHB15-L-1, may be an essential transcriptional regulator responsible for cambial tissue formation or maintenance.
The positive feedback loop auxin-flow–MP–HD-Zip III–PIN1–auxin-flow is typical in cambial cell development, a crucial mechanism in determining vascular cell fates [55]. The auxin response factor MP directly bound the auxin response element in the promoter sequence of ATHB8, thereby regulating ATHB8 expression [20]. ATHB8 is required to stabilize PIN1 expression against auxin transport perturbation in cambial cells, resulting in the formation and maintenance of procambial cells [16,19]. We found that GmHB15-L-1 had ARF elements and was significantly induced by IAA, which is highly expressed in the vascular cambium. Soybean PIN1 genes show the same expression pattern in the cambial cell [56]. Whether the conserved pathway also appears in soybean remains to be clarified.
3.3. Soybean HD-Zip III Genes Perform Overlapping Functions in Promoting Xylem Differentiation in Leaf Veins
Previous studies have linked BRs to vascular development. BR treatment promotes poplar stem growth and xylem formation [21]. In cultured zinnia cells, the levels of BR increase drastically prior to tracheary cell differentiation, and this increase is indispensable for progressing to the last stage of xylem cell differentiation (Yamamoto et al., 1997). The role of BR during soybean leaf veins development is not as well explored. After BR treatment, the areas of xylem cells and lignin content in leaf veins were significantly increased (Figure 6a,b). Our results are consistent with previous reports in poplar and zinnia that point to BRs promoting the differentiation of xylem cells. HD-Zip III functions in xylem cell differentiation by responding to BR signaling [51,54]. The xylem cell-specific accumulation of ZeHB10 (the homologous gene of ATHB8) mRNA has also been observed in xylogenic cell culture [57]. The expression of ZeHB11 and ZeHB12 (REV homologous transcripts) is repressed by BRZ but is rapidly induced by BR [25]. Although HD-Zip III have been suggested to be involved in xylem cell development, the details of individual HD-Zip III functions in soybean leaf vascular differentiation have not been clarified. We characterized the expression patterns of HD-Zip III genes in leaf vein differentiation and analyzed the effects of BR and BRZ on the accumulation of GmHD-Zip III transcripts.
On the basis of the findings shown here, GmREV-L-1 and GmHB14-L-2 play pivotal roles in the differentiation of the xylem. Expression analysis showed that GmHB14-L-2 and GmREV-L-1 expression was induced by BR but was suppressed strongly by BRZ (Figure 4b). In zinnia, the rapid induction of ZeHB12 occurs upon BR treatment. Consistent with that of GmREV-L-1 in soybean, the expression of ZeHB12 is repressed by BRZ. We used qRT-PCR to check the expression patterns of GmREV-L-1 and GmHB14-L-2 in leaf veins at different developmental stages (Figure 6a). Our results showed that GmREV-L-1 and GmHB14-L-2 were expressed in all of the examined stages. Throughout these stages, the relative transcript levels of GmREV-L-1 and GmHB14-L-2 were stably maintained at high levels in leaf veins at 12, 24, and 48 h of development (Figure 6b). As shown in Figure 5a, these time-points corresponded to vital developmental stages of the xylem. In Arabidopsis, ATHB14/PHV and REV exhibit overlapping expression in the adaxial domains in vascular bundles [58]. The REV PHV double mutant enhances the vascular defects of the rev mutant, i.e., vascular bundles with remarkably few lignified cells [10]. Consistent with a previous work, this work showed that GmREV-L-1 and GmHB14-L-2 had nearly identical expression patterns in soybean vascular tissues and were primarily expressed in vascular cambium cells and developing xylem cells (Figure 7). GmHB14-L-2 may perform overlapping functions with GmREV-L-1 in vascular cell differentiation. These facts suggest that GmREV-L-1 and GmHB14-L-2 play pivotal roles in xylem differentiation.
Given the relationship between BRs and HD-Zip III genes, the START domain deserves future investigation. The START domain has lipid binding capability [59]. In plants, the START domain is predominantly found in HD-Zip gene family. Lipid/sterol ligands can directly modulate HD-Zip transcription factor activity [60,61]. Consequently, soybean HD-Zip III genes may be activated through the binding of sterols or lipids to their START domain. Given that BRs are the initiators of soybean HD-Zip III genes, they may be candidates for binding to the START domain and act as pivotal signals that activate proteins.
4. Materials and Methods
4.1. Identification and Phylogenetic Tree Construction
To identify the GmHD-Zip III genes in soybean, we used the sequences of five Arabidopsis HD-Zip III proteins [10] downloaded from the TAIR website (http://www.arabidopsis.org/ (accessed on 10 November 2021) as query sequences in the TBLASTP searches against the soybean genome (Glycine_max_v4.0), on the NCBI database (https://www.ncbi.nlm.nih.gov/genome/ (accessed on 10 November 2021) [62]. All sequences with an e-value below 10−10 were candidate soybean HD-Zip III proteins. Twelve GmHD-Zip III proteins were initially isolated in this research. Next, we performed BLASTP separately using each of the 12 HD-Zip III proteins as a query sequence in the soybean genome database. This process was repeated until no new HD-Zip III protein was found. The PLAZA database was used to obtain the orthology gene pairs for soybean (https://bioinformatics.psb.ugent.be/plaza/versions/plaza_v4_5_dicots/ (accessed on 16 June 2022)) [63]. We identified the homologous proteins of HD-Zip III in another four legumes, namely, chickpea (Cicer arietinum), peanut (Arachis hypogaea), kidney bean (Phaseolus vulgaris), and wild soybean (Glycine soja). Five Arabidopsis HD-Zip III proteins were input as query sequences into BLASTP searches with the barrel medic genome (MtrunA17r5.0-ANR), chickpea genome (A SM33114v1), peanut genome (arahy. Tifrunner. gnm1.KYV3), kidney bean genome (PhaVulg1_0), and wild soybean genome (A SM419377v2) in the NCBI database. We downloaded the protein sequences of HD-Zip III family genes that were newly identified in recent years in six plants, including strawberry (Fragaria vesca) [46], cucumber (Cucumis sativus L.) [47], poplar [42], tomato [50], medicago (Medicago truncatula) [44], and rice (Oryza sativa) [64].
The amino acid sequences of HD-Zip III were aligned using MEGA7.0 software. Phylogenetic analysis was constructed using the neighbor-joining method and 1000 bootstraps [65].
4.2. Chromosomal Mapping and Analysis of Basic Physical and Chemical Properties
All GmHD-Zip III genes were mapped on the 20 chromosomes of soybean using gene annotation (Glycine_max_v4.0_genomic.gff) acquired from NCBI by TBtools software [35]. The molecular characteristics of the soybean HD-Zip III proteins, including the theoretical predictions of isoelectric point and molecular weight, were analyzed using ExPASy Protparam Tool (https://web.expasy.org/compute_pi/ (accessed on 7 November 2021) [36]. Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/ (accessed on 1 November 2021) was used to predict the subcellular localizations of the HD-Zip III proteins.
4.3. Gene Structure, Conserved Domains, and Motif Identification
Intron/exon structures were downloaded from the NCBI genome database. We used the NCBI Conserved Domains Database (CDD) (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi/ (accessed on 10 November 2021) to predict the conserved domains of the soybean HD-Zip III proteins [66,67]. MEME-MAST programs (http://meme.nbcr.net/meme/meme.html (accessed on 15 November 2021) were used to predict the conserved protein motifs, with the motif length set to 6–200 and the e value to <1 × 10−10 [68].
4.4. Plant Transcription Factor Binding Sites in the Promoters
For this study, the 2000 bp region upstream of the annotated transcription start site for each gene was evaluated for promoter motifs [66]. TBtools was utilized to extract GmHD-Zip III genes’ CDS (Coding Sequences) in the promoter sequences and visualization mapping. We used Plantcare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (accessed on 20 December 2021) for the prediction of cis-elements and visualization mapping [49]. To identify the binding sites of transcription factors in the promoter region of each GmHD-Zip III gene, we searched via the database PlantRegMap (http://plantregmap.cbi.pku.edu.cn/binding_site_prediction.php (accessed on 16 June 2022) with the following parameter: e-value ≤ 1 × 10−6 [69]. The visualization of the motifs was performed on the LOGO website (https://weblogo.berkeley.edu/logo.cgi (accessed on 12 June 2022).
4.5. Plant Materials and Growth Conditions
The Williams 82 soybean variety was used in this study. The soybean cultivar was grown in a growth room (12 h light, 25 °C, 12 h dark, 22 °C) using a light-emitting diode. The light intensity in the growth chamber was 500 µmol m−2 s−1.
4.6. Tissue Specificity Expression Analysis
Roots, leaves, stems, shoot apical meristems, flowers, pod walls, seeds, and petioles of soybean were collected for tissue-specific expression analysis. The roots, leaves, stems, shoot apical meristems, and petioles were collected from soybean seedlings in the V2 stage (the unfolding of second trifoliate leaves). Flowers were picked three days after flowering. The pod walls and seeds were collected 14 and 30 days after flowering, and each sample was collected thrice independently. Total RNA was extracted from tissues, as described for qRT-PCR. The normalized gene expression values were log2-transformed and visualized in the form of heatmaps constructed using TBtools.
4.7. Collection of Leaf Veins Cells
To obtain the high quality RNA of the vascular tissue of soybean plants, the tissues were fixed in Carnoy–acetone fixative (/v/v = 70/30) for 12 h at 4 °C. Finally, the tissues were transparentized and mounted with t-butyl alcohol. The cells of the cambium, developing xylem, and developing phloem were collected from leaf cross-sections through laser microdissection by using a Veritas Automated Laser Capture Microdissection System (Thermo Fisher Scientific, Inc., Waltham, MA, USA). A minimum of 500 cells were collection from each sample [70,71]. The RNA extracted from laser-captured samples was extracted using the PicoPure RNA Isolation Arcturus Kit. The RNA was amplified to generate cDNA using the Target Amp 2-round aRNA amplification kit for expression analysis [72].
4.8. Leaf Anatomy
GmHD-Zip III expression was examined during leaf vein development in soybean, and leaf veins were sampled at 0, 12, 24, 48, and 96 h. Vein tissue development was observed by using cross-sections stained with safranin-O/fast green. The 0.5 cm × 1 cm sections were cut from the central leaflet of six different leaves for each treatment for fixation with FAA. Tissues were embedded in paraffin, and serially, paraffin sections were cut into 10 μm thickness. The sections were stained with safranin-O/fast green. Sections were scanned under a Nikon Eclipse 80i microscope, and images were acquired with the ACT-2U imaging software (Nikon Corporation) [73].
4.9. Exogenous Hormone Treatments
The following processes were set to elucidate the probable functions of GmHD-Zip III in leaf vein development. Seedlings that had been grown for 10 days were sprayed with 100 µmol L−1 indole-3-acetic acid (IAA) [45], 100 µmol L−1 N-1-naphthylphthalamic acid (NPA) [36], 10 mmol L−1 BR, or 5 µmol L−1 BRZ [25]. Leaf vein tissues were obtained at 4 h after these treatments. Three individual plant materials were collected, frozen in liquid nitrogen, and stored in a refrigerator at −80 °C. After 10 days of hormone treatment, soybean leaf vein tissue development was observed by using cross-sections stained with safranin-O/fast green. Image J (Image J Software, National Institutes of Health, Bethesda, MD, USA) was employed to measure the areas of xylem, phloem, and cambium of leaf veins. Leaf vein lignin content was quantitatively measured by using the lignin Kit (BC4200, Solarbio, Beijing, China) according to the manufacturer’s protocols.
4.10. Expression Analysis
Total RNA was extracted with an RNA prep pure Plant Kit (R6827-01, Omega Bio-tek, Norcross, GA, USA), and the RNA quality was determined by agarose gel electrophoresis [66]. The RNA concentration was determined by NanoDropTMOne /OneC (Thermo Fisher Scientific Inc., Waltham, MA, USA). gDNA Eraser (TaKaRa Biotechnology, Dalian, China) was used to remove the possible DNA content of the total RNA to avoid detection error when generating the level of gene expression. The first-strand cDNA (Complementary DNA) of RT was synthesized using the PrimeScriptTM RT reagent Kit with gDNA Eraser Kit from Takara.
qRT-PCR was conducted on a qRT-PCR system operated on the Quant Studiotm 7 Flex series (Thermo Fisher Scientific Inc., Waltham, MA, USA), using a qRT-PCR Kit (Takara, Dalian, China). The total reaction system had a volume of 10 µL, including 5 µL TBGreen Fastq PCR Mix (2×), 10 µmol L−1 upstream and downstream primers (0.4 µL each), and cDNA template (20 ng·μL−1), and the total reaction system was 10 µmol L−1. The reaction conditions were 95 °C for 3 min, with denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s, and extension at 72 °C for 30 s. A total of 40 PCR cycles were performed. The reaction results obtained by 2−ΔΔCT method were input to QuantStudioTM Real-Time PCR software for analysis [74]. The GmActin gene (GLYMA_19G147900) was used as the internal reference gene (Du et al., 2011), and the specific primers are shown in Table S1. The relative expression values were normalized based on the expression levels of the control. The processed data were log2 transformed, and TBtools software was used to visualize as heatmaps [75].
4.11. Statistical Analysis
Statistical analysis was carried out by using one-way ANOVA in SPSS software Version 24 (IBM SPSS Statistics, Armonk, NY, USA). The graphs were drawn by GraphPad Prism 8.0 software (GraphPad Software, Inc., La Jolla, CA, USA).
5. Conclusions
We systematically characterized the GmHD-Zip III gene family in the soybean genome and the expression profiles of all GmHD-Zip III members during leaf vein development in soybean. Twelve GmHD-Zip III genes were identified and were found to be unevenly distributed on nine chromosomes. All GmHD-Zip III gene family members had similar gene structures and motif arrangements. GmHB15-L-1 showed stable high expression during leaf vein development and was highly expressed in cambium cells. GmREV-L-1 and GmHB14-L-2 maintained stable high expression levels at all xylem developmental stages and were expressed at high levels in vascular cambium cells. Therefore, GmHB15-L-1 is an essential regulator that is responsible for soybean vein cambial cell formation. GmREV-L-1 and GmHB14-L-2 are key components in xylem differentiation. This study will pave the way for research on the different molecular mechanisms of leaf vein development. We will use transient overexpression experiment methods to study the roles of the HD-Zip III gene in soybean leaf vein development in the future.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants11131728/s1. Figure S1: Orthologous genes of soybean and Arabidopsis. List of orthologous gene pairs obtained from PLAZA. Figure S2: Chromosomal distributions of the identified soybean HD-Zip III genes. Chromosomal locations are shown from top to bottom on the corresponding chromosomes. The bar on the left side indicates chromosome sizes in megabases. The scale represents chromosome length. Figure S3: Motif of the 12 GmHD-Zip III proteins. Different colored boxes stand for different motifs. The length of each box in the figure represents the actual size of the motif in the proteins. Figure S4: Analysis of cis-acting elements in the promoter regions of GmHD-Zip III family genes. Figure S5: Expression profiles of GmHD-Zip III under IAA and BR treatment. Asterisks on top of the bars indicate statistically significant differences (* p < 0.05, ** p < 0.01). Figure S6. Expression profiles of GmHD-Zip III genes in eight soybean tissues in reads/Kb/million. The normalized values of gene expression were log2-transformed and visualized in the form of heatmaps. Table S1: Primers sequences used in qRT-PCR. Table S2: The homologous proteins of HD-ZIP III in legumes. Table S3: Basic physical and chemical information of soybean HD-ZIP III proteins. Table S4: cis-acting elements in the promoter region of GmHD-Zip III family gene.
Author Contributions
J.G. conceived and designed the experiments. Q.W., J.G., and J.C. performed the experiments. J.G. wrote the manuscript. S.L., L.F., X.T., F.Y., and W.Y. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (32071963), a grant from the International S and T Cooperation Projects of Sichuan Province (2020YFH0126), the Program on Industrial Technology System of National Soybean (CARS-04-PS19), and Chengdu Science and Technology Project (2020-YF09-00033-SN).
Data Availability Statement
The data presented in this study are available in the article and Supplementary Materials.
Acknowledgments
In this section, you can acknowledge any support given which is not covered by the author contribution or funding sections. This may include administrative and technical support, or donations in kind (e.g., materials used for experiments).
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Sack, L.; Scoffoni, C.; John, G.P.; Poorter, H.; Mason, C.M.; Mendez-Alonzo, R.; Donovan, L.A. How do leaf veins influence the worldwide leaf economic spectrum? Review and synthesis. J. Exp. Bot. 2013, 64, 4053–4080. [Google Scholar] [CrossRef] [PubMed]
- Ye, M.; Wu, M.; Zhang, H.; Zhang, Z.; Zhang, Z. High Leaf Vein Density Promotes Leaf Gas Exchange by Enhancing Leaf Hydraulic Conductance in Oryza sativa L. Front. Plant Sci. 2021, 12, 693815. [Google Scholar] [CrossRef] [PubMed]
- De Rybel, B.; Mähönen, A.P.; Helariutta, Y.; Weijers, D. Plant vascular development: From early specification to differentiation. Nat. Rev. 2016, 17, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Sack, L.; Scoffoni, C. Leaf venation: Structure, function, development, evolution, ecology and applications in the past, present and future. New Phytol. 2013, 198, 983–1000. [Google Scholar] [CrossRef] [PubMed]
- Brodribb, T.J.; Feild, T.S.; Jordan, G.J. Leaf maximum photosynthetic rate and venation are linked by hydraulics. Plant Physiol. 2007, 144, 1890–1898. [Google Scholar] [CrossRef]
- Boyce, C.K.; Brodribb, T.J.; Feild, T.S.; Zwieniecki, M.A. Angiosperm leaf vein evolution was physiologically and environmentally transformative. Proceedings 2009, 276, 1771–1776. [Google Scholar] [CrossRef]
- Mathan, J.; Bhattacharya, J.; Ranjan, A. Enhancing crop yield by optimizing plant developmental features. Development 2016, 143, 3283–3294. [Google Scholar] [CrossRef]
- Ruonala, R.; Ko, D.; Helariutta, Y. Genetic Networks in Plant Vascular Development. Annu. Rev. Genet. 2017, 51, 335–359. [Google Scholar] [CrossRef]
- Vasco, A.; Smalls, T.L.; Graham, S.W.; Cooper, E.D.; Wong, G.K.; Stevenson, D.W.; Moran, R.C.; Ambrose, B.A. Challenging the paradigms of leaf evolution: Class III HD-Zips in ferns and Lycophytes. New Phytol. 2016, 212, 745–758. [Google Scholar] [CrossRef]
- Prigge, M.J.; Otsuga, D.; Alonso, J.M.; Ecker, J.R.; Drews, G.N.; Clark, S.E. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 2005, 17, 61–76. [Google Scholar] [CrossRef]
- Sharif, R.; Raza, A.; Chen, P.; Li, Y.; El-Ballat, E.M.; Rauf, A.; Hano, C.; El-Esawi, M.A. HD-ZIP Gene Family: Potential Roles in Improving Plant Growth and Regulating Stress-Responsive Mechanisms in Plants. Genes 2021, 12, 1256. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Ye, Z.H. IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell 1999, 11, 2139–2152. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Ye, Z.H. Amphivasal vascular bundle 1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems and carpels. Plant Cell Physiol. 2004, 45, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Emery, J.F.; Floyd, S.K.; Alvarez, J.; Eshed, Y.; Hawker, N.P.; Izhaki, A.; Baum, S.F.; Bowman, J.L. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. CB 2003, 13, 1768–1774. [Google Scholar] [CrossRef]
- Huang, T.; Harrar, Y.; Lin, C.; Reinhart, B.; Newell, N.R.; Talavera-Rauh, F.; Hokin, S.A.; Barton, M.K.; Kerstetter, R.A. Arabidopsis KANADI1 acts as a transcriptional repressor by interacting with a specific cis-element and regulates auxin biosynthesis, transport, and signaling in opposition to HD-ZIPIII factors. Plant Cell 2014, 26, 246–262. [Google Scholar] [CrossRef]
- Baima, S.; Possenti, M.; Matteucci, A.; Wisman, E.; Altamura, M.M.; Ruberti, I.; Morelli, G. The Arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol. 2001, 126, 643–655. [Google Scholar] [CrossRef]
- Manuela, D.; Xu, M. Patterning a Leaf by Establishing Polarities. Front. Plant Sci. 2020, 11, 568730. [Google Scholar] [CrossRef]
- Müller, C.J.; Valdés, A.E.; Wang, G.; Ramachandran, P.; Beste, L.; Uddenberg, D.; Carlsbecker, A. PHABULOSA Mediates an Auxin Signaling Loop to Regulate Vascular Patterning in Arabidopsis. Plant Physiol. 2016, 170, 956–970. [Google Scholar] [CrossRef]
- Donner, T.J.; Sherr, I.; Scarpella, E. Regulation of preprocambial cell state acquisition by auxin signaling in Arabidopsis leaves. Development 2009, 136, 3235–3246. [Google Scholar] [CrossRef]
- Donner, T.J.; Sherr, I.; Scarpella, E. Auxin signal transduction in Arabidopsis vein formation. Plant Signal. Behav. 2010, 5, 70–72. [Google Scholar] [CrossRef][Green Version]
- Du, J.; Gerttula, S.; Li, Z.; Zhao, S.T.; Liu, Y.L.; Liu, Y.; Lu, M.Z.; Groover, A.T. Brassinosteroid regulation of wood formation in poplar. New Phytol. 2020, 225, 1516–1530. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.L.; Tang, R.J.; Wang, H.H.; Jiang, C.M.; Bao, Y.; Yang, Y.; Liang, M.X.; Sun, Z.C.; Kong, F.J.; Li, B.; et al. Overexpression of Populus trichocarpa CYP85A3 promotes growth and biomass production in transgenic trees. Plant Biotechnol. J. 2017, 15, 1309–1321. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, R.; Demura, T.; Fukuda, H. Brassinosteroids induce entry into the final stage of tracheary element differentiation in cultured Zinnia cells. Plant Cell Physiol. 1997, 38, 980–983. [Google Scholar] [CrossRef]
- Gao, J.; Yu, M.; Zhu, S.; Zhou, L.; Liu, S. Effects of exogenous 24-epibrassinolide and brassinazole on negative gravitropism and tension wood formation in hybrid poplar (Populus deltoids × Populus nigra). Planta 2019, 249, 1449–1463. [Google Scholar] [CrossRef] [PubMed]
- Ohashi-Ito, K.; Demura, T.; Fukuda, H. Promotion of transcript accumulation of novel Zinnia immature xylem-specific HD-Zip III homeobox genes by brassinosteroids. Plant Cell Physiol. 2002, 43, 1146–1153. [Google Scholar] [CrossRef] [PubMed]
- Ribichich, K.F.; Chiozza, M.; Ávalos-Britez, S.; Cabello, J.V.; Arce, A.L.; Watson, G.; Arias, C.; Portapila, M.; Trucco, F.; Otegui, M.E.; et al. Successful field performance in warm and dry environments of soybean expressing the sunflower transcription factor HB4. J. Exp. Bot. 2020, 71, 3142–3156. [Google Scholar] [CrossRef]
- Belamkar, V.; Weeks, N.T.; Bharti, A.K.; Farmer, A.D.; Graham, M.A.; Cannon, S.B. Comprehensive characterization and RNA-Seq profiling of the HD-Zip transcription factor family in soybean (Glycine max) during dehydration and salt stress. BMC Genom. 2014, 15, 950. [Google Scholar] [CrossRef]
- Ponting, C.P.; Aravind, L. START: A lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem. Sci. 1999, 24, 130–132. [Google Scholar] [CrossRef]
- McIntosh, B.E.; Hogenesch, J.B.; Bradfield, C.A. Mammalian Per-Arnt-Sim proteins in environmental adaptation. Annu. Rev. Physiol. 2010, 72, 625–645. [Google Scholar] [CrossRef]
- Blank-Landeshammer, B.; Teichert, I.; Märker, R.; Nowrousian, M.; Kück, U.; Sickmann, A. Combination of Proteogenomics with Peptide De Novo Sequencing Identifies New Genes and Hidden Posttranscriptional Modifications. mBio 2019, 10, 5. [Google Scholar] [CrossRef]
- Magnani, E.; Barton, M.K. A per-ARNT-sim-like sensor domain uniquely regulates the activity of the homeodomain leucine zipper transcription factor REVOLUTA in Arabidopsis. Plant Cell 2011, 23, 567–582. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T.J. ARF1, a transcription factor that binds to auxin response elements. Science 1997, 276, 1865–1868. [Google Scholar] [CrossRef] [PubMed]
- Nagata, N.; Asami, T.; Yoshida, S. Brassinazole, an inhibitor of brassinosteroid biosynthesis, inhibits development of secondary xylem in cress plants (Lepidium sativum). Plant Cell Physiol. 2001, 42, 1006–1011. [Google Scholar] [CrossRef]
- Scarpella, E.; Barkoulas, M.; Tsiantis, M. Control of Leaf and Vein Development by Auxin. CSH Perspect. Biol. 2010, 2, a001511. [Google Scholar] [CrossRef]
- Zhong, R.; Ye, Z.H. Alteration of auxin polar transport in the Arabidopsis ifl1 mutants. Plant Physiol. 2001, 126, 549–563. [Google Scholar] [CrossRef]
- Ariel, F.D.; Manavella, P.A.; Dezar, C.A.; Chan, R.L. The true story of the HD-Zip family. Trends Plant Sci. 2007, 12, 419–426. [Google Scholar] [CrossRef]
- Turchi, L.; Baima, S.; Morelli, G.; Ruberti, I. Interplay of HD-Zip II and III transcription factors in auxin-regulated plant development. J. Exp. Bot. 2015, 66, 5043–5053. [Google Scholar] [CrossRef]
- Javelle, M.; Vernoud, V.; Depège-Fargeix, N.; Arnould, C.; Oursel, D.; Domergue, F.; Sarda, X.; Rogowsky, P.M. Overexpression of the epidermis-specific homeodomain-leucine zipper IV transcription factor Outer Cell Layer1 in maize identifies target genes involved in lipid metabolism and cuticle biosynthesis. Plant Physiol. 2010, 154, 273–286. [Google Scholar] [CrossRef]
- Javelle, M.; Klein-Cosson, C.; Vernoud, V.; Boltz, V.; Maher, C.; Timmermans, M.; Depège-Fargeix, N.; Rogowsky, P.M. Genome-wide characterization of the HD-ZIP IV transcription factor family in maize: Preferential expression in the epidermis. Plant Physiol. 2011, 157, 790–803. [Google Scholar] [CrossRef]
- Elhiti, M.; Stasolla, C. Structure and function of homodomain-leucine zipper (HD-Zip) proteins. Plant Signal. Behav. 2009, 4, 86–88. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Song, D.; Xu, P.; Sun, J.; Li, L. A HD-ZIP III gene, PtrHB4, is required for interfascicular cambium development in Populus. Plant Biotechnol. J. 2018, 16, 808–817. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xiong, H.; Cuo, D.; Wu, X.; Duan, R. Genome-wide characterization and expression profiling of the relation of the HD-Zip gene family to abiotic stress in barley (Hordeum vulgare L.). Plant Physiol. Bio. 2019, 141, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Gao, Z.; Li, R.; Xu, Y.; Kong, Y.; Zhou, G.; Meng, C.; Hu, R. Genome-wide identification and expression profiling of HD-ZIP gene family in Medicago truncatula. Genomics 2020, 112, 3624–3635. [Google Scholar] [CrossRef]
- Itoh, J.; Hibara, K.; Sato, Y.; Nagato, Y. Developmental role and auxin responsiveness of Class III homeodomain leucine zipper gene family members in rice. Plant Physiol. 2008, 147, 1960–1975. [Google Scholar] [CrossRef]
- Wang, X.L.; Chen, X.; Yang, T.B.; Cheng, Q.; Cheng, Z.M. Genome-Wide Identification of bZIP Family Genes Involved in Drought and Heat Stresses in Strawberry (Fragaria vesca). Int. J. Genom. 2017, 2017, 3981031. [Google Scholar] [CrossRef]
- Rong, F.; Chen, F.; Huang, L.; Zhang, J.; Zhang, C.; Hou, D.; Cheng, Z.; Weng, Y.; Chen, P.; Li, Y. A mutation in class III homeodomain-leucine zipper (HD-ZIP III) transcription factor results in curly leaf (cul) in cucumber (Cucumis sativus L.). TAG. Theor. Appl. Genet. Theor. Angew. Genet. 2019, 132, 113–123. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef]
- Zhang, J.; Gao, Y.; Feng, M.; Cui, Y.; Li, S.; Liu, L.; Wang, Y.; Xu, W.; Li, F. Genome-Wide Identification of the HD-ZIP III Subfamily in Upland Cotton Reveals the Involvement of GhHB8-5D in the Biosynthesis of Secondary Wall in Fiber and Drought Resistance. Front. Plant Sci. 2021, 12, 806195. [Google Scholar] [CrossRef]
- Hong, Y.; Liu, Y.; Zhang, Y.; Jia, L.; Yang, X.; Zhang, X.; Liu, J.; Luan, Y. Genome-wide characterization of homeobox-leucine zipper gene family in tomato (Solanum lycopersicum) and functional analysis of SlHDZ34 (III sub-family member) under salinity stress. Environ. Exp. Bot. 2021, 192, 104652. [Google Scholar] [CrossRef]
- Ramachandran, P.; Carlsbecker, A.; Etchells, J.P. Class III HD-ZIPs govern vascular cell fate: An HD view on patterning and differentiation. J. Exp. Bot. 2017, 68, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Little, C.A.; MacDonald, J.E.; Olsson, O. Involvement of indole-3-acetic acid in fascicular and interfascicular cambial growth and interfascicular extraxylary fiber differentiation in Arabidopsis thaliana inflorescence stems. Int. J. Plant Sci. 2002, 163, 519–529. [Google Scholar] [CrossRef]
- Brandt, R.; Salla-Martret, M.; Bou-Torrent, J.; Musielak, T.; Stahl, M.; Lanz, C.; Ott, F.; Schmid, M.; Greb, T.; Schwarz, M.; et al. Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses. Plant J. Cell Mol. Biol. 2012, 72, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Ohashi-Ito, K.; Fukuda, H. HD-zip III homeobox genes that include a novel member, ZeHB-13 (Zinnia)/ATHB-15 (Arabidopsis), are involved in procambium and xylem cell differentiation. Plant Cell Physiol. 2003, 44, 1350–1358. [Google Scholar] [CrossRef]
- Ohashi-Ito, K.; Fukuda, H. Transcriptional regulation of vascular cell fates. Curr. Biol. CB 2010, 13, 670–676. [Google Scholar] [CrossRef]
- Wang, Y.; Chai, C.; Valliyodan, B.; Maupin, C.; Annen, B.; Nguyen, H.T. Genome-wide analysis and expression profiling of the PIN auxin transporter gene family in soybean (Glycine max). BMC Genom. 2015, 16, 951. [Google Scholar] [CrossRef]
- Ohashi-Ito, K.; Kubo, M.; Demura, T.; Fukuda, H. Class III homeodomain leucine-zipper proteins regulate xylem cell differentiation. Plant Cell Physiol. 2005, 46, 1646–1656. [Google Scholar] [CrossRef]
- McConnell, J.R.; Emery, J.; Eshed, Y.; Bao, N.; Bowman, J.; Barton, M.K. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 2001, 411, 709–713. [Google Scholar] [CrossRef]
- Schrick, K.; Nguyen, D.; Karlowski, W.M.; Mayer, K.F. START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol. 2004, 5, R41. [Google Scholar] [CrossRef]
- Schrick, K.; Bruno, M.; Khosla, A.; Cox, P.N.; Marlatt, S.A.; Roque, R.A.; Nguyen, H.C.; He, C.; Snyder, M.P.; Singh, D.; et al. Shared functions of plant and mammalian StAR-related lipid transfer (START) domains in modulating transcription factor activity. BMC Biol. 2014, 12, 70. [Google Scholar] [CrossRef]
- Alpy, F.; Tomasetto, C. Give lipids a START: The StAR-related lipid transfer (START) domain in mammals. J. Cell Sci. 2005, 118, 2791–2801. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.F.; Zhang, N.; Zhong, T.; Wang, C.; Xu, M.L.; Ye, J.R. Identification and characterization of the GH3 gene family in maize. J. Integr. Agr. 2016, 15, 249–261. [Google Scholar] [CrossRef]
- Van Bel, M.; Silvestri, F.; Weitz, E.M.; Kreft, L.; Botzki, A.; Coppens, F.; Vandepoele, K. PLAZA 5.0: Extending the scope and power of comparative and functional genomics in plants. Nucleic Acids Res. 2022, 50, D1468–D1474. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Haider, I.; Xiong, L.; Zhu, X.; Hussain, R.M.F.; Övernäs, E.; Meijer, A.H.; Zhang, G.; Wang, M.; Bouwmeester, H.J.; et al. Functional analysis of the HD-Zip transcription factor genes Oshox12 and Oshox14 in rice. PLoS ONE 2018, 13, e0199248. [Google Scholar] [CrossRef] [PubMed]
- Muleke, E.M.; Wang, Y.; Zhang, W.T.; Xu, L.; Ying, J.L.; Karanja, B.K.; Zhu, X.W.; Fan, L.X.; Ahmadzai, Z.; Liu, L.W. Genome-wide identification and expression profiling of MYB transcription factor genes in radish (Raphanus sativus L.). J. Integr. Agr. 2021, 20, 120–131. [Google Scholar] [CrossRef]
- Manoli, A.; Trevisan, S.; Quaggiotti, S.; Varotto, S. Identification and characterization of the BZR transcription factor family and its expression in response to abiotic stresses in Zea mays L. Plant Growth Regul. 2018, 89, 95–105. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, L.; Chen, Y.; Wang, S.; Fang, Y.; Zhang, X.; Wu, Y.; Xue, D. Genome-wide identification of the SOD gene family and expression analysis under drought and salt stress in barley. Plant Growth Regul. 2021, 94, 49–60. [Google Scholar] [CrossRef]
- Wang, S.X.; Shi, F.Y.; Dong, X.X.; Li, Y.X.; Zhang, Z.H.; Li, H. Genome-wide identification and expression analysis of auxin response factor (ARF) gene family in strawberry (Fragaria vesca). J. Integr. Agr. 2019, 18, 1587–1603. [Google Scholar] [CrossRef]
- Tian, F.; Yang, D.C.; Meng, Y.Q.; Jin, J.; Gao, G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2020, 48, D1104–D1113. [Google Scholar] [CrossRef]
- Song, D.; Shen, J.; Li, L. Characterization of cellulose synthase complexes in Populus xylem differentiation. New Phytol. 2010, 187, 777–790. [Google Scholar] [CrossRef]
- Zhu, Y.; Song, D.; Sun, J.; Wang, X.; Li, L. PtrHB7, a class III HD-Zip gene, plays a critical role in regulation of vascular cambium differentiation in Populus. Mol. Plant 2013, 6, 1331–1343. [Google Scholar] [CrossRef] [PubMed]
- Hannapel, D.J.; Sharma, P.; Lin, T. Phloem-mobile messenger RNAs and root development. Front. Plant Sci. 2013, 4, 257. [Google Scholar] [CrossRef] [PubMed]
- Behr, M.; Legay, S.; Žižková, E.; Motyka, V.; Dobrev, P.I.; Hausman, J.F.; Lutts, S.; Guerriero, G. Studying Secondary Growth and Bast Fiber Development: The Hemp Hypocotyl Peeks behind the Wall. Front. Plant Sci. 2016, 7, 1733. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nature 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Guang, Y.; Luo, S.; Ahammed, G.J.; Xiao, X.; Li, J.; Zhou, Y.; Yang, Y. The OPR gene family in watermelon: Genome-wide identification and expression profiling under hormone treatments and root-knot nematode infection. Plant Biol. 2021, 23 (Suppl. S1), 80–88. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).