High-Throughput Sequencing Discloses the Cucumber Mosaic Virus (CMV) Diversity in Slovakia and Reveals New Hosts of CMV from the Papaveraceae Family
Abstract
1. Introduction
2. Results
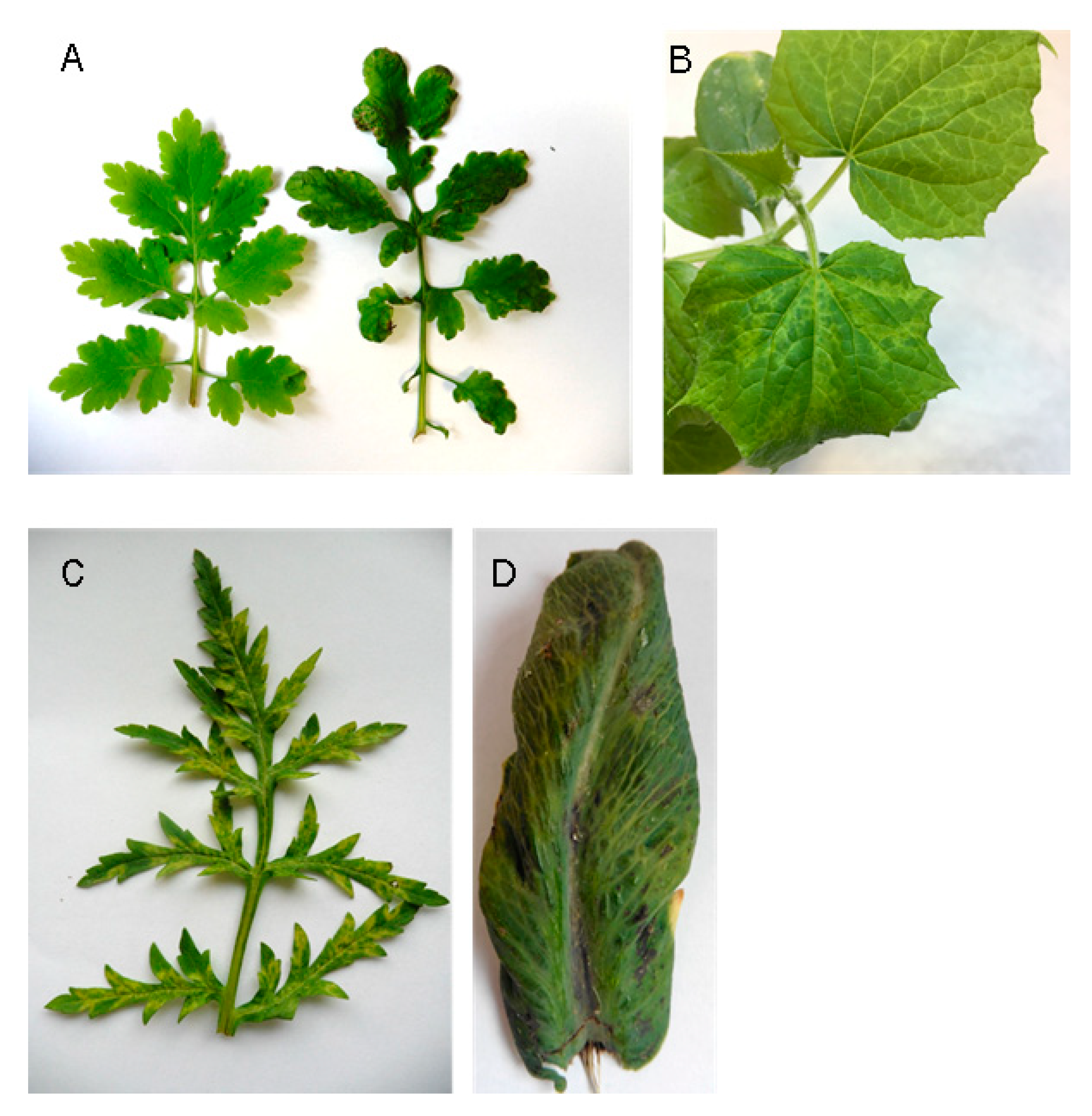
2.1. HTS Enabled to Identify CMV within Complex Viromes Reveals New CMV Hosts from the Papaveracea Family
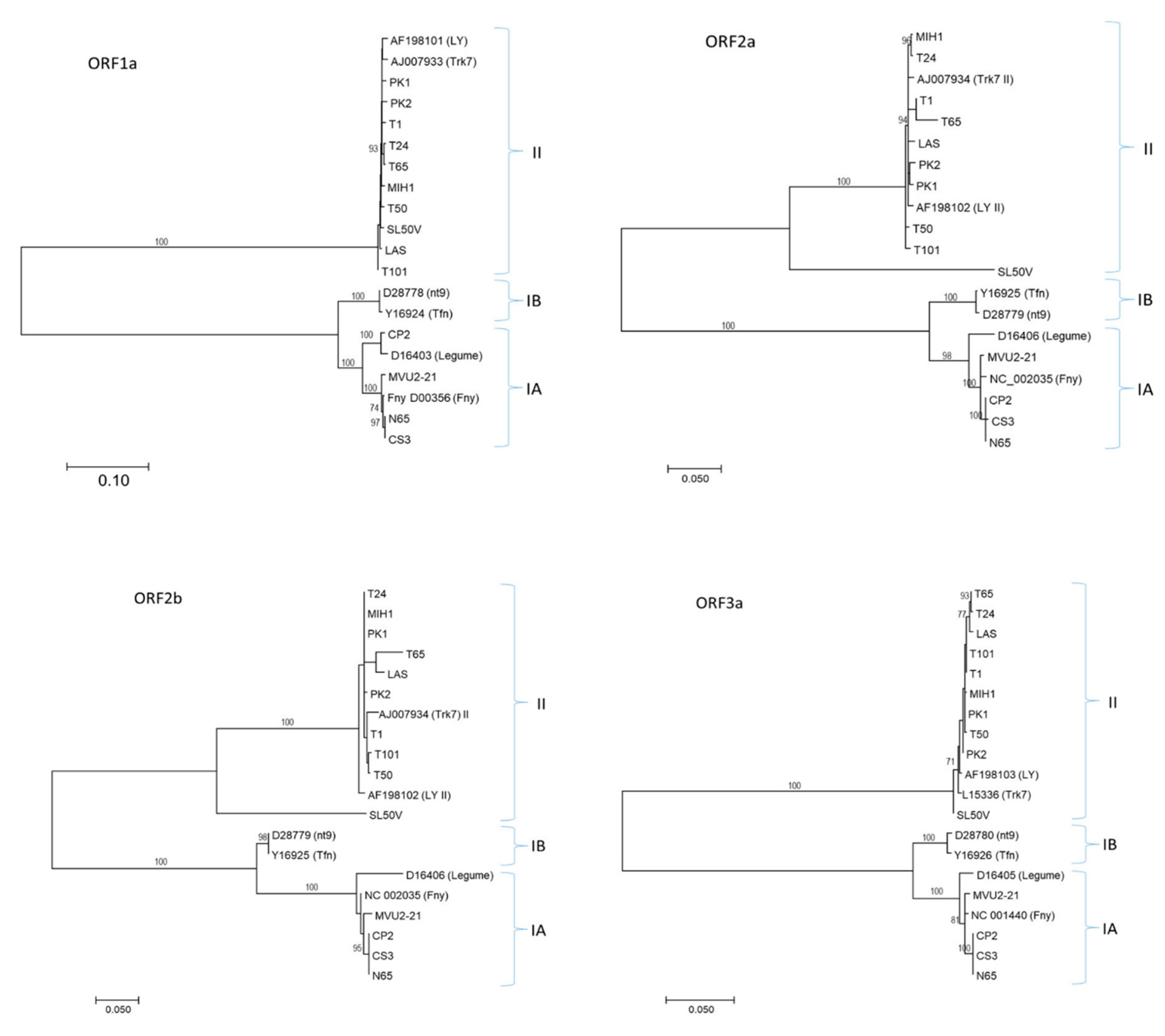
2.2. Slovak CMV Isolates Belong to Two Different Genetic Groups
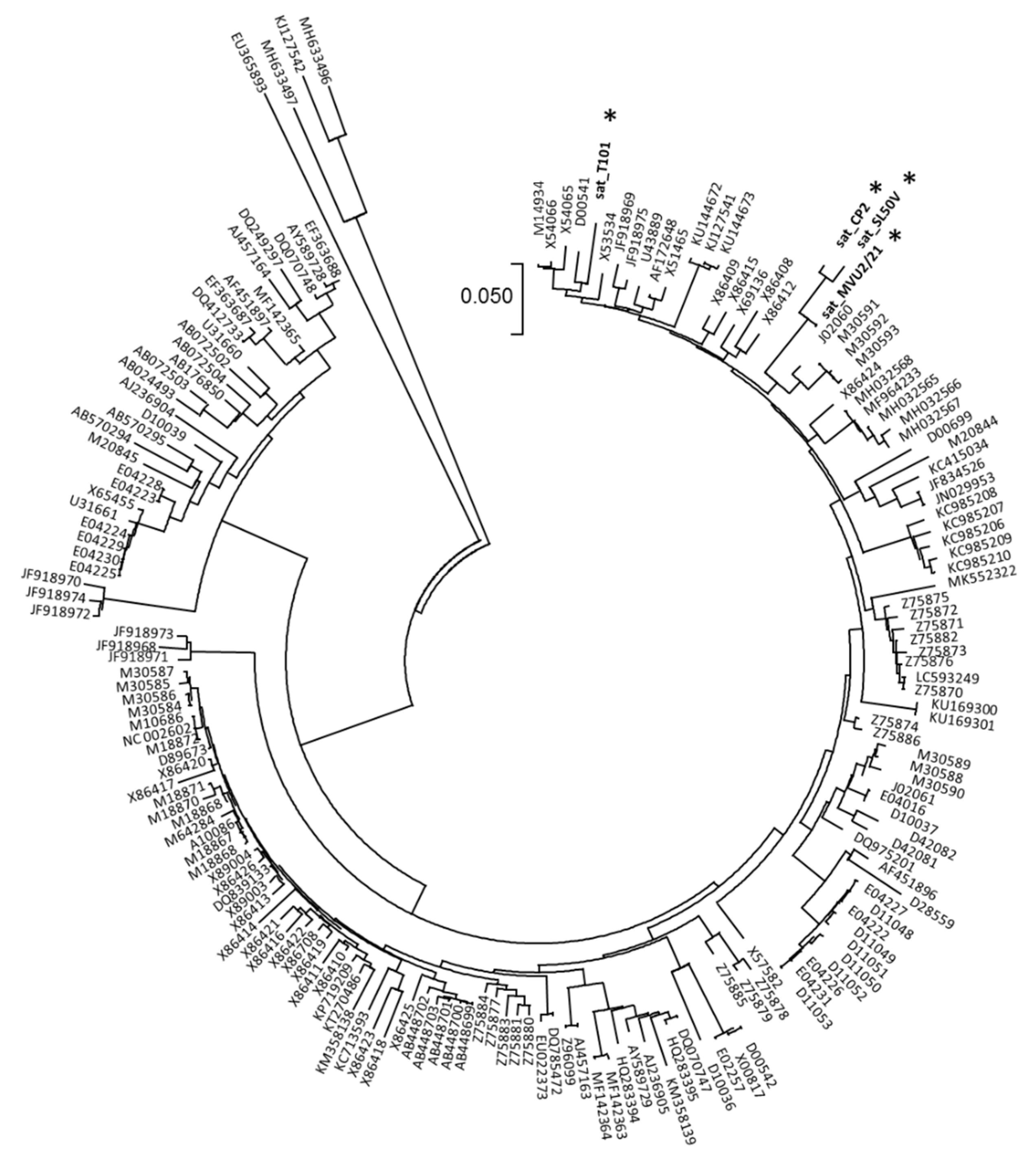
2.3. Satellite RNAs Were Found in Association with Some CMV Variants
3. Discussion
4. Materials and Methods
4.1. Samples and HTS Analysis
4.2. Mechanical Transmission and Detection of Satellite CMV
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jones, R.A.; Naidu, R.A. Global Dimensions of Plant Virus Diseases: Current Status and Future Perspectives. Annu. Rev. Virol. 2019, 6, 387–409. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.A. Plant virus emergence and evolution: Origins, new encounter scenarios, factors driving emergence, effects of changing world conditions, and prospects for control. Virus Res. 2009, 141, 113–130. [Google Scholar] [CrossRef] [PubMed]
- Doolittle, S.P. A new infectious mosaic disease of cucumber. Phytopathology 1916, 6, 145–147. [Google Scholar]
- Jagger, I.C. Experiments with the cucumber mosaic disease. Phytopathology 1916, 6, 148–151. [Google Scholar]
- Mochizuki, T.; Ohki, S.T. Cucumber mosaic virus: Viral genes as virulence determinants. Mol. Plant Pathol. 2012, 13, 217–225. [Google Scholar] [CrossRef]
- Roossinck, M.J. Evolutionary History of Cucumber Mosaic Virus Deduced by Phylogenetic Analyses. J. Virol. 2002, 76, 3382–3387. [Google Scholar] [CrossRef]
- Bujarski, J.; Figlerowicz, M.; Gallitelli, D.; Roossinck, J.; Scott, W. Bromoviridae. In Virus Taxonomy, Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier: Amsterdam, The Netherlands, 2012; pp. 965–976. [Google Scholar] [CrossRef]
- García-Arenal, F.; Palukaitis, P. Cucumber Mosaic Virus. In Encyclopedia of Virology, 3rd ed.; Mahy, B.W.J., van Regenmortel, M.H.V., Eds.; Academic Press: Oxford, UK, 2008; pp. 614–619. [Google Scholar]
- Palukaitis, P.; Roossinck, M.J.; Dietzgen, R.G.; Francki, R.I. Cucumber MOSAIC Virus. Adv. Vir. Res. 1992, 41, 281–348. [Google Scholar] [CrossRef]
- Jacquemond, M. Cucumber Mosaic Virus. Adv. Vir. Res. 2012, 84, 439–504. [Google Scholar] [CrossRef]
- Roossinck, M.J.; Zhang, L.; Hellwald, K.-H. Rearrangements in the 5′ Nontranslated Region and Phylogenetic Analyses of Cucumber Mosaic Virus RNA 3 Indicate Radial Evolution of Three Subgroups. J. Virol. 1999, 73, 6752–6758. [Google Scholar] [CrossRef]
- Palukaitis, P.; García-Arenal, F. Cucumoviruses. Adv. Vir. Res. 2003, 62, 241–323. [Google Scholar] [CrossRef]
- Li, N.; Yu, C.; Yin, Y.; Gao, S.; Wang, F.; Jiao, C.; Yao, M. Pepper Crop Improvement Against Cucumber Mosaic Virus (CMV): A Review. Front. Plant Sci. 2020, 11, 598798. [Google Scholar] [CrossRef] [PubMed]
- Ouedraogo, R.S.; Pita, J.S.; Somda, I.P.; Traore, O.; Roossinck, M.J. Impact of Cultivated Hosts on the Recombination of Cucumber Mosaic Virus. J. Virol. 2019, 93, e01770-18. [Google Scholar] [CrossRef] [PubMed]
- Apalowo, O.A.; Adediji, A.O.; Balogun, O.S.; Fakolujo, T.I.; Archibong, J.M.; Izuogu, N.B.; Abdelgawad, M.A.; Ghoneim, M.M.; Mustapha, S.; Qashqari, F.S.I.; et al. Genetic Structure of Cucumber Mosaic Virus From Natural Hosts in Nigeria Reveals High Diversity and Occurrence of Putative Novel Recombinant Strains. Front. Microbiol. 2022, 13, 753054. [Google Scholar] [CrossRef] [PubMed]
- Nouri, S.; Falk, B.W.; Groves, R.L. A new satellite RNA is associated with natural infections of cucumber mosaic virus in succulent snap bean. Arch. Virol. 2012, 157, 375–377. [Google Scholar] [CrossRef] [PubMed]
- Liao, Q.; Zhu, L.; DU, Z.; Zeng, R.; Feng, J.; Chen, J. Satellite RNA-mediated Reduction of Cucumber Mosaic Virus Genomic RNAs Accumulation in Nicotiana tabacum. Acta Biochim. et Biophys. Sin. 2007, 39, 217–223. [Google Scholar] [CrossRef]
- Giakountis, A.; Tsarmpopoulos, I.; Chatzivassiliou, E.K. Cucumber mosaic virus Isolates from Greek Legumes are Associated with Satellite RNAs that are Necrogenic for Tomato. Plant Dis. 2018, 102, 2268–2276. [Google Scholar] [CrossRef]
- Singhal, P.; Nabi, S.U.; Yadav, M.K.; Dubey, A. Mixed infection of plant viruses: Diagnostics, interactions and impact on host. J. Plant Dis. Prot. 2021, 128, 353–368. [Google Scholar] [CrossRef]
- González, L.E.; Peiró, R.; Rubio, L.; Galipienso, L. Persistent Southern Tomato Virus (STV) Interacts with Cucumber Mosaic and/or Pepino Mosaic Virus in Mixed- Infections Modifying Plant Symptoms, Viral Titer and Small RNA Accumulation. Microorganisms 2021, 9, 689. [Google Scholar] [CrossRef]
- Heo, K.-J.; Kwon, S.-J.; Kim, M.-K.; Kwak, H.-R.; Han, S.-J.; Kwon, M.-J.; Rao, A.L.N.; Seo, J.-K. Newly emerged resistance-breaking variants of cucumber mosaic virus represent ongoing host-interactive evolution of an RNA virus. Virus Evol. 2020, 6, veaa070. [Google Scholar] [CrossRef]
- Ali, A.; Li, H.; Schneider, W.L.; Sherman, D.J.; Gray, S.; Smith, D.; Roossinck, M.J. Analysis of Genetic Bottlenecks during Horizontal Transmission of Cucumber Mosaic Virus. J. Virol. 2006, 80, 8345–8350. [Google Scholar] [CrossRef]
- Nouri, S.; Arevalo, R.; Falk, B.W.; Groves, R.L. Genetic Structure and Molecular Variability of Cucumber mosaic virus Isolates in the United States. PLoS ONE 2014, 9, e96582. [Google Scholar] [CrossRef] [PubMed]
- Hasiów-Jaroszewska, B.; Chrzanowski, M.; Budzyńska, D.; Rymelska, N.; Borodynko-Filas, N. Genetic diversity, distant phylogenetic relationships and the occurrence of recombination events among cucumber mosaic virus isolates from zucchini in Poland. Arch. Virol. 2017, 162, 1751–1756. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, M.; Ashfaq, M.; Mukhtar, T.; Abbasi, N.A. Current status andgenetic variability of cucumber mosaic cucumovirus (CMV) isolatesinfecting major cucurbits and solanaceous vegetables in Pothwarregion of Pakistan. Pak. J. Agric. Sci. 2020, 57, 1353–1361. [Google Scholar] [CrossRef]
- Valachas, C.A.; Giantsis, I.A.; Sareli, K.; Winter, S.; Zelezniakof, E.; Pentheroudaki, Z.; Chatzivassiliou, E.K. Molecular analysis of Greek isolates of cucumber mosaic virus from vegetables shows a low prevalence of satellite RNAs and suggests the presence of host-associated virus strains. Arch. Virol. 2021, 166, 2199–2208. [Google Scholar] [CrossRef] [PubMed]
- Valadez-Ramírez, P.; Paz-Román, J.; Guzmán, M.; Buenrostro-Nava, M.; Ochoa-Martínez, D. New records of Cucumber mosaic virus isolates and associated-satellite RNA in Colima, Mexico. Rev. Mex. Fitopatol. 2019, 37, 357–364. [Google Scholar] [CrossRef]
- Stanković, I.; Vučurović, A.; Zečević, K.; Petrović, B.; Nikolić, D.; Delibašić, G. Characterization of cucumber mosaic virus and its satellite RNAs associated with tomato lethal necrosis in Serbia. Eur. J. Plant Pathol. 2021, 160, 301–313. [Google Scholar] [CrossRef]
- Sinha, S.; Samad, A. Characterization and evolutionary analysis of Cucumber mosaic virus isolate infecting Salvia sclarea in India. 3 Biotech 2021, 11, 468. [Google Scholar] [CrossRef]
- Santosa, A.I.; Ertunc, F. Characterization of two Cucumber mosaic virus isolates infecting Allium cepa in Turkey. Phytopathol. Mediterr. 2021, 60, 13–21. [Google Scholar] [CrossRef]
- Pecman, A.; Kutnjak, D.; Gutiérrez-Aguirre, I.; Adams, I.; Fox, A.; Boonham, N.; Ravnikar, M. Next Generation Sequencing for Detection and Discovery of Plant Viruses and Viroids: Comparison of Two Approaches. Front. Microbiol. 2017, 8, 1998. [Google Scholar] [CrossRef]
- Tian, A.; Miyashita, S.; Ando, S.; Takahashi, H. Single Amino Acid Substitutions in the Cucumber Mosaic Virus 1a Protein Induce Necrotic Cell Death in Virus-Inoculated Leaves without Affecting Virus Multiplication. Viruses 2020, 12, 91. [Google Scholar] [CrossRef]
- Akhtar, K.P.; Anwer, M.; Saleem, M.Y.; Yousaf, S.; Ullah, N.; Cheema, H.M.N.; Sarwar, N. Identification of natural weed hosts of Cucumber mosaic virus subgroup-I and the absence of seed transmission in weed hosts in Pakistan. J. Hortic. Sci. Biotechnol. 2019, 94, 468–474. [Google Scholar] [CrossRef]
- Andika, I.B.; Wei, S.; Cao, C.; Salaipeth, L.; Kondo, H.; Sun, L. Phytopathogenic fungus hosts a plant virus: A naturally occurring cross-kingdom viral infection. Proc. Natl. Acad. Sci. USA 2017, 114, 12267–12272. [Google Scholar] [CrossRef] [PubMed]
- Gallitelli, D. The ecology of Cucumber mosaic virus and sustainable agriculture. Virus Res. 2000, 71, 9–21. [Google Scholar] [CrossRef]
- Komuro, Y. Studies on cucumber mosaic virus. Jpn. J. Phytopathol. 1958, 23, 235–239. [Google Scholar] [CrossRef]
- Kubelková, D.; Špak, J. Virus diseases of poppy (Papaver somniferum) and some further species from the family Papaveraceae. Plant Prot. Sci. 1999, 35, 33–36. [Google Scholar] [CrossRef]
- Kobyłko, T.; Dańda, P.; Hasiów, B.; Borodynko-Filas, N.; Pospieszny, H. First Report of Cucumber mosaic virus on Lavandula angustifolia in Poland. Plant Dis. 2008, 92, 978. [Google Scholar] [CrossRef]
- Alexander, H.M.; Mauck, K.E.; Whitfield, A.E.; Garrett, K.A.; Malmstrom, C.M. Plant-virus interactions and the agro-ecological interface. Eur. J. Plant Pathol. 2014, 138, 529–547. [Google Scholar] [CrossRef]
- Pinke, G.; Pál, R.W.; Tóth, K.; Karácsony, P.; Czúcz, B.; Botta-Dukát, Z. Weed vegetation of poppy (Papaver somniferum) fields in Hungary: Effects of management and environmental factors on species composition. Weed Res. 2011, 51, 621–630. [Google Scholar] [CrossRef]
- Sacristán, S.; Fraile, A.; García-Arenal, F. Population Dynamics of Cucumber mosaic virus in Melon Crops and in Weeds in Central Spain. Phytopathology® 2004, 94, 992–998. [Google Scholar] [CrossRef]
- Lin, H.-X.; Rubio, L.; Smythe, A.B.; Falk, B.W. Molecular Population Genetics of Cucumber Mosaic Virus in California: Evidence for Founder Effects and Reassortment. J. Virol. 2004, 78, 6666–6675. [Google Scholar] [CrossRef]
- Rubio, L.; Galipienso, L.; Ferriol, I. Detection of Plant Viruses and Disease Management: Relevance of Genetic Diversity and Evolution. Front. Plant Sci. 2020, 11, 1092. [Google Scholar] [CrossRef] [PubMed]
- Moreno, A.B.; Lopez-Moya, J.J. When viruses play team sports: Mixed infections in plants. Phytopathology 2020, 110, 29–48. [Google Scholar] [CrossRef] [PubMed]
- Glasa, M.; Hančinský, R.; Šoltys, K.; Predajňa, L.; Tomašechová, J.; Hauptvogel, P.; Mrkvová, M.; Mihálik, D.; Candresse, T. Molecular Characterization of Potato Virus Y (PVY) Using High-Throughput Sequencing: Constraints on Full Genome Reconstructions Imposed by Mixed Infection Involving Recombinant PVY Strains. Plants 2021, 10, 753. [Google Scholar] [CrossRef] [PubMed]
- Glasa, M.; Šoltys, K.; Vozárová, Z.; Predajňa, L.; Sihelská, N.; Šubr, Z.; Candresse, T. High intra-host cherry virus a population heterogeneity in cherry trees in slovakia. J. Plant Pathol. 2017, 99, 745–752. [Google Scholar] [CrossRef]
- Glasa, M.; Predajna, L.; Soltys, K.; Sihelska, N.; Nagyova, A.; Wetzel, T.; Sabanadzovic, S. Analysis of Grapevine rupestris stem pitting-associated virus (GRSPaV) in Slovakia Reveals Differences in Intra-Host Population Diversity and Naturally Occurring Recombination Events. Plant Pathol. J. 2017, 33, 34–42. [Google Scholar] [CrossRef][Green Version]
- Betancourt, M.; Fraile, A.; García-Arenal, F. Cucumber mosaic virus satellite RNAs that induce similar symptoms in melon plants show large differences in fitness. J. Gen. Virol. 2011, 92, 1930–1938. [Google Scholar] [CrossRef]
- He, L.; Wang, Q.; Gu, Z.; Liao, Q.; Palukaitis, P.; Du, Z. A conserved RNA structure is essential for a satellite RNA-mediated inhibition of helper virus accumulation. Nucleic Acids Res. 2019, 47, 8255–8271. [Google Scholar] [CrossRef]
- Montasser, M.S.; Tousignant, M.E.; Kaper, J.M. Viral Satellite RNAs for the Prevention of Cucumber Mosaic Virus (CMV) Disease in Field-Grown Pepper and Melon Plants. Plant Dis. 1998, 82, 1298–1303. [Google Scholar] [CrossRef][Green Version]
- Wang, Y.; Gaba, V.; Yang, J.; Palukaitis, P.; Gal-On, A. Characterization of Synergy Between Cucumber mosaic virus and Potyviruses in Cucurbit Hosts. Phytopathology® 2002, 92, 51–58. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
| Isolate | Original Host | Locality, Year of Sampling | GenBank Accession Number | Presence of CMV Satellite (GenBank Accession Number) | Other Viruses Identified by HTS |
|---|---|---|---|---|---|
| CP2 | Cucurbita pepo | Pezinok, 2019 | MZ298665 a, MZ298675 b, MZ298685 c | Yes (ON493796) | PRSV, ZYMV, WMV |
| CS3 | Cucumis sativum | Cífer, 2018 | MZ298666, MZ298676, MZ298686 | No | ZYMV, WMV |
| LAS | Chelidonium majus | Bratislava, 2017 | MZ298667, MZ298677, MZ298687 | No | None |
| MIH1 | Capsicum annum | Čachtice, 2017 | MZ298668, MZ298678, MZ298688 | No | PCV-2 |
| MVU2-21 | Cucumis melo | Velke Ulany, 2021 | ON409882, ON409886, ON493791 | Yes (ON493795) | WMV, ZYMV, CmEV |
| N65 | Capsicum annum | Čachtice, 2017 | ON409883, ON409887, ON493792 | No | WMV, PCV-2, BPEV |
| PK1 | Papaver somniferum | Pezinok, 2017 | MN792886, MN792887, MN792888 | No | TuMV |
| PK2 | Papaver rhoeas | Pezinok, 2017 | ON409881, ON409885, ON493790 | No | TuMV, TuYV |
| SL50V | Solanum lycopersicum | Pezinok, 2018 | MZ298669, MZ298679, MZ298689 | Yes (ON493794) | PVY |
| T1 | Solanum lycopersicum | Paňa, 2017 | MZ298671, MZ298681, MZ298691 | No | PVY |
| T24 | Solanum lycopersicum | Nitra, 2017 | MZ298672, MZ298682, MZ298692 | No | PVY |
| T50 | Solanum lycopersicum | Plavecký Mikuláš, 2017 | MZ298673, MZ298683, MZ298693 | No | PVM, PVY |
| T65 | Solanum lycopersicum | Sološnica, 2017 | MZ298674, MZ298684, MZ298694 | No | PVM, PVY |
| T101 | Solanum lycopersicum | Pezinok, 2019 | MZ298670, MZ298680, MZ298690 | Yes (ON493793) | PVY |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mrkvová, M.; Hančinský, R.; Predajňa, L.; Alaxin, P.; Achs, A.; Tomašechová, J.; Šoltys, K.; Mihálik, D.; Olmos, A.; Ruiz-García, A.B.; et al. High-Throughput Sequencing Discloses the Cucumber Mosaic Virus (CMV) Diversity in Slovakia and Reveals New Hosts of CMV from the Papaveraceae Family. Plants 2022, 11, 1665. https://doi.org/10.3390/plants11131665
Mrkvová M, Hančinský R, Predajňa L, Alaxin P, Achs A, Tomašechová J, Šoltys K, Mihálik D, Olmos A, Ruiz-García AB, et al. High-Throughput Sequencing Discloses the Cucumber Mosaic Virus (CMV) Diversity in Slovakia and Reveals New Hosts of CMV from the Papaveraceae Family. Plants. 2022; 11(13):1665. https://doi.org/10.3390/plants11131665
Chicago/Turabian StyleMrkvová, Michaela, Richard Hančinský, Lukáš Predajňa, Peter Alaxin, Adam Achs, Jana Tomašechová, Katarína Šoltys, Daniel Mihálik, Antonio Olmos, Ana Belén Ruiz-García, and et al. 2022. "High-Throughput Sequencing Discloses the Cucumber Mosaic Virus (CMV) Diversity in Slovakia and Reveals New Hosts of CMV from the Papaveraceae Family" Plants 11, no. 13: 1665. https://doi.org/10.3390/plants11131665
APA StyleMrkvová, M., Hančinský, R., Predajňa, L., Alaxin, P., Achs, A., Tomašechová, J., Šoltys, K., Mihálik, D., Olmos, A., Ruiz-García, A. B., & Glasa, M. (2022). High-Throughput Sequencing Discloses the Cucumber Mosaic Virus (CMV) Diversity in Slovakia and Reveals New Hosts of CMV from the Papaveraceae Family. Plants, 11(13), 1665. https://doi.org/10.3390/plants11131665







