Evaluating Plant Gene Models Using Machine Learning
Abstract
:1. Introduction
2. Results
2.1. Feature Table Construction
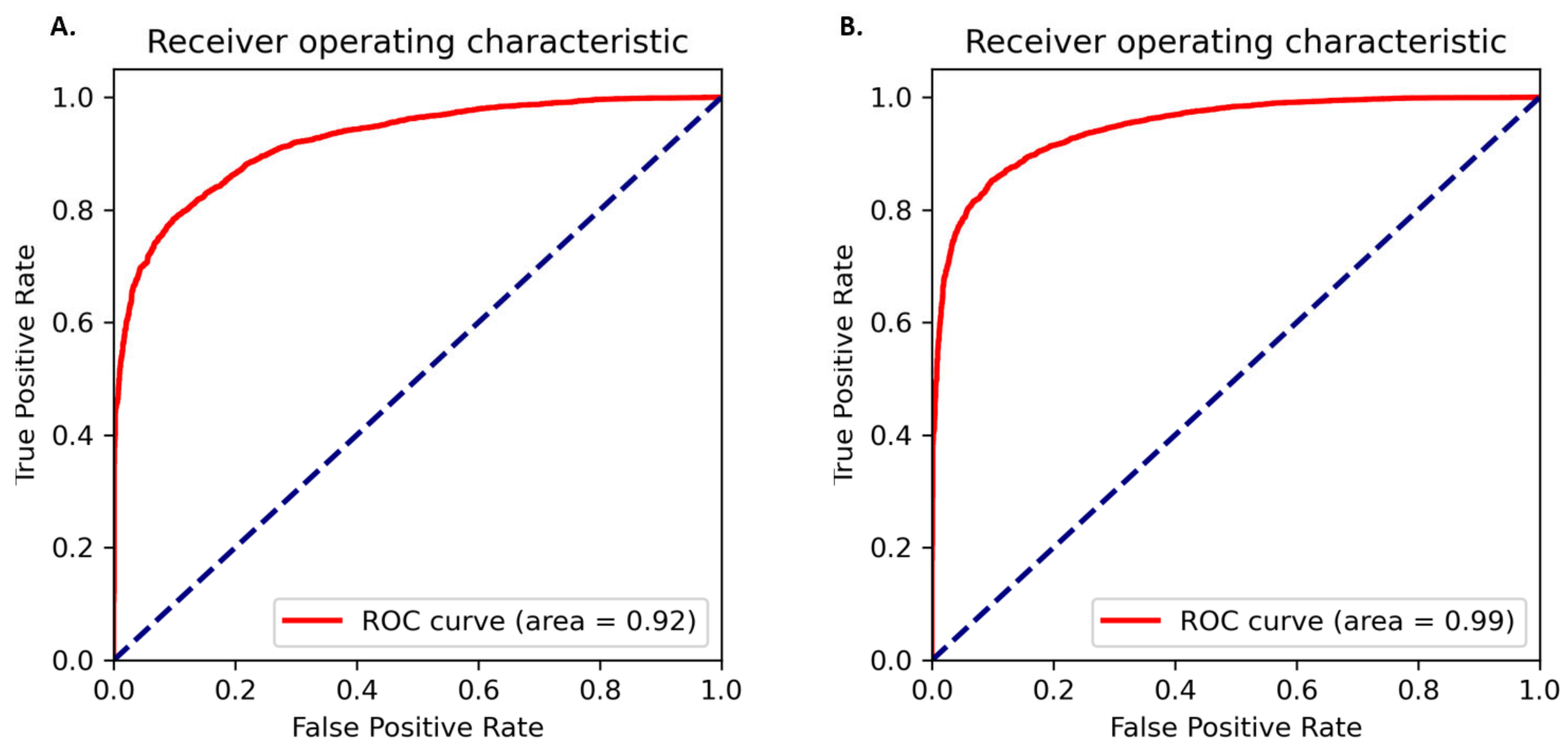
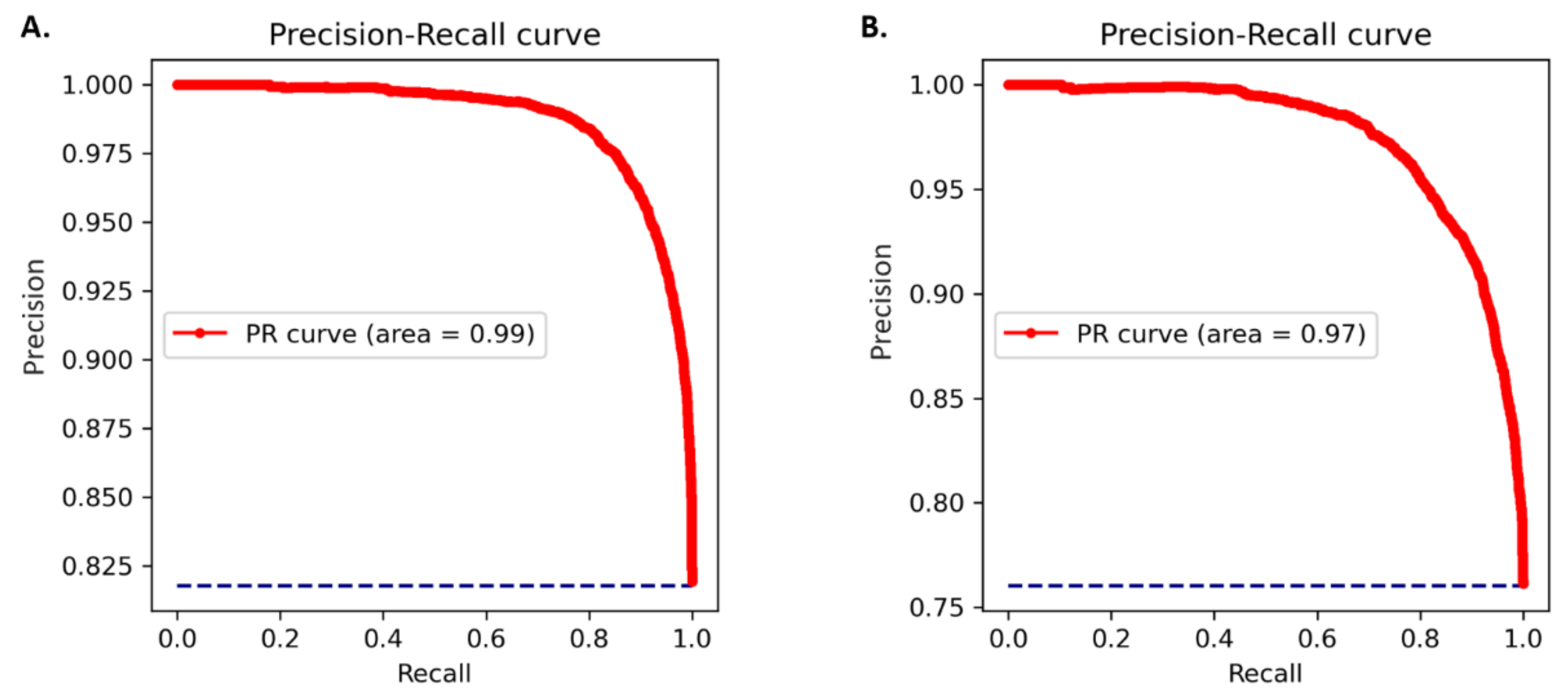
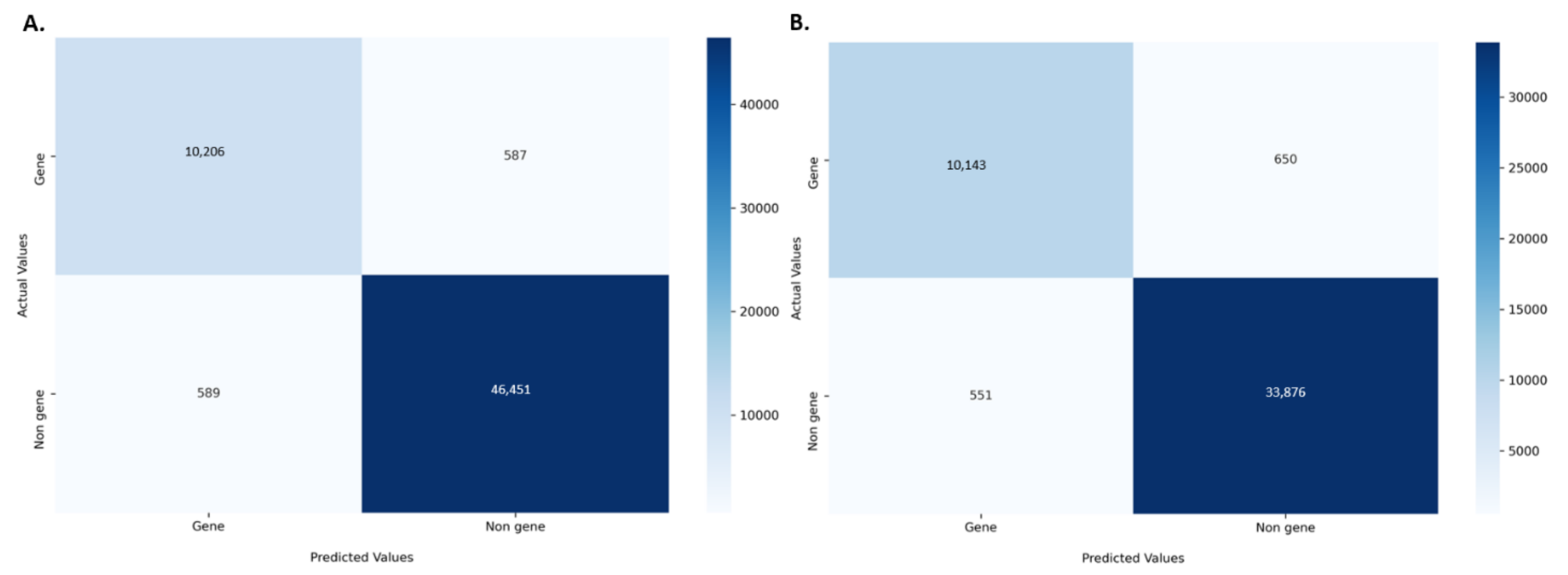
2.2. Performance Evaluation
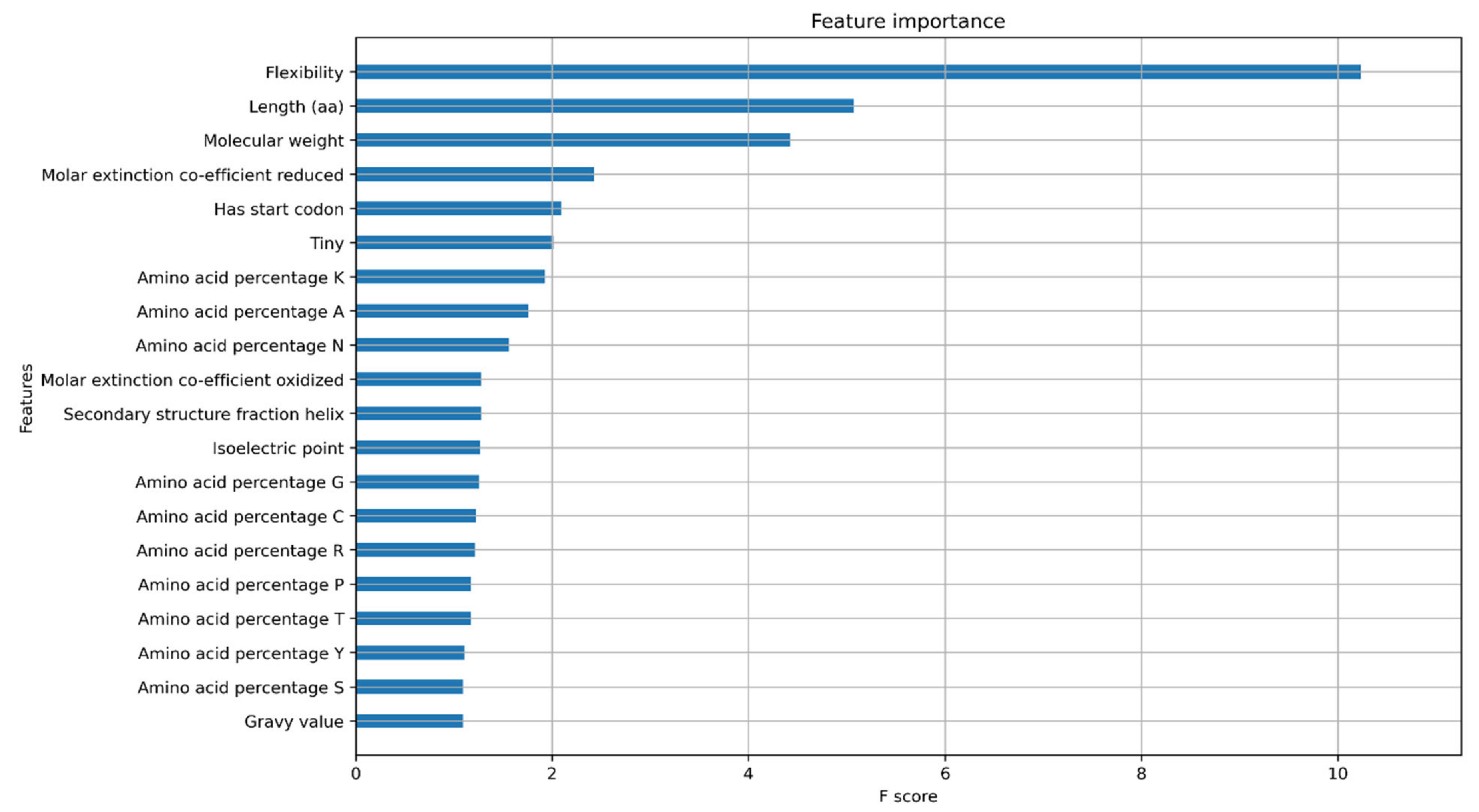
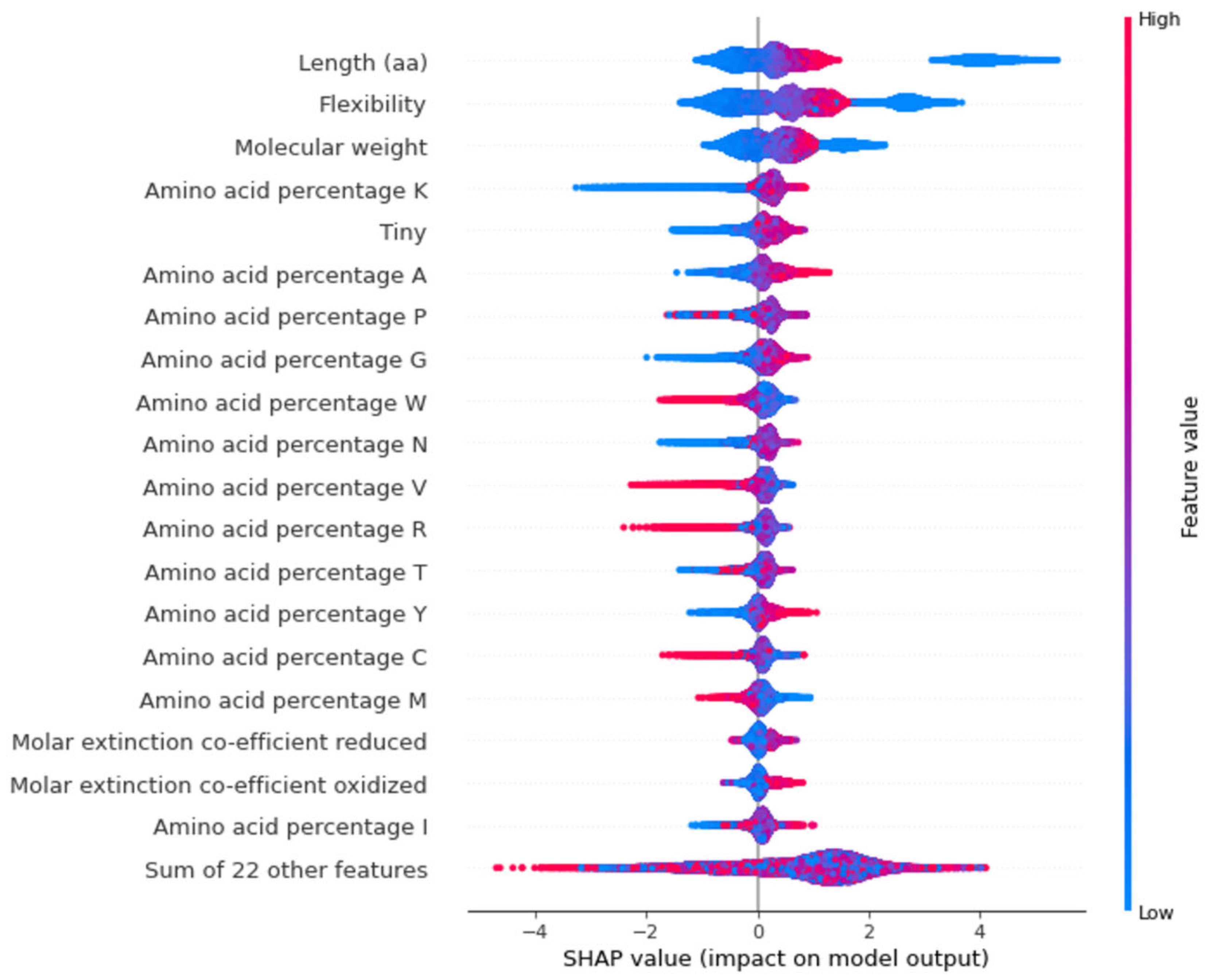
2.3. Model Explanations
3. Discussion
4. Materials and Methods
4.1. Dataset
4.2. Model Training
4.3. Model Evaluation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Marks, R.A.; Hotaling, S.; Frandsen, P.B.; VanBuren, R. Representation and participation across 20 years of plant genome sequencing. Nat. Plants 2021, 7, 1571–1578. [Google Scholar] [CrossRef] [PubMed]
- Bayer, P.E.; Golicz, A.A.; Scheben, A.; Batley, J.; Edwards, D. Plant pan-genomes are the new reference. Nat. Plants 2020, 6, 914–920. [Google Scholar] [CrossRef] [PubMed]
- Schnable, J.C. Genes and gene models, an important distinction. New Phytol. 2020, 228, 50–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerstein, M.B.; Bruce, C.; Rozowsky, J.S.; Zheng, D.; Du, J.; Korbel, J.O.; Emanuelsson, O.; Zhang, Z.D.; Weissman, S.; Snyder, M. What is a gene, post-ENCODE? History and updated definition. Genome Res. 2007, 17, 669–681. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, S.; Buell, C.R. The TIGR Plant Repeat Databases: A collective resource for the identification of repetitive sequences in plants. Nucleic Acids Res. 2004, 32, D360–D363. [Google Scholar] [CrossRef] [Green Version]
- Vaattovaara, A.; Leppälä, J.; Salojärvi, J.; Wrzaczek, M. High-throughput sequencing data and the impact of plant gene annotation quality. J. Exp. Bot. 2018, 70, 1069–1076. [Google Scholar] [CrossRef]
- Schnoes, A.M.; Brown, S.D.; Dodevski, I.; Babbitt, P.C. Annotation error in public databases: Misannotation of molecular function in enzyme superfamilies. PLoS Comput. Biol. 2009, 5, e1000605. [Google Scholar] [CrossRef]
- Golicz, A.A.; Bayer, P.E.; Barker, G.C.; Edger, P.P.; Kim, H.; Martinez, P.A.; Chan, C.K.K.; Severn-Ellis, A.; McCombie, W.R.; Parkin, I.A.P.; et al. The pangenome of an agronomically important crop plant Brassica oleracea. Nat. Commun. 2016, 7, 13390. [Google Scholar] [CrossRef] [Green Version]
- Rana, D.; van den Boogaart, T.; O’Neill, C.M.; Hynes, L.; Bent, E.; Macpherson, L.; Park, J.Y.; Lim, Y.P.; Bancroft, I. Conservation of the microstructure of genome segments in Brassica napus and its diploid relatives. Plant J. 2004, 40, 725–733. [Google Scholar] [CrossRef]
- Kowalski, S.P.; Lan, T.H.; Feldmann, K.A.; Paterson, A.H. Comparative mapping of Arabidopsis thaliana and Brassica oleracea chromosomes reveals islands of conserved organization. Genetics 1994, 138, 499–510. [Google Scholar] [CrossRef]
- Moore, G.; Devos, K.M.; Wang, Z.; Gale, M.D. Cereal genome evolution. Grasses, line up and form a circle. Curr. Biol. 1995, 5, 737–739. [Google Scholar] [CrossRef] [Green Version]
- Jordan, M.I.; Mitchell, T.M. Machine learning: Trends, perspectives, and prospects. Science 2015, 349, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Sommer, M.J.; Salzberg, S.L. Balrog: A universal protein model for prokaryotic gene prediction. PLoS Comput. Biol. 2021, 17, e1008727. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, M.M.; Buske, O.J.; Wang, J.; Weng, Z.; Bilmes, J.A.; Noble, W.S. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat. Methods 2012, 9, 473–476. [Google Scholar] [CrossRef] [Green Version]
- Degroeve, S.; De Baets, B.; Van de Peer, Y.; Rouzé, P. Feature subset selection for splice site prediction. Bioinformatics 2002, 18 (Suppl. 2), S75–S83. [Google Scholar] [CrossRef] [Green Version]
- Sirén, K.; Millard, A.; Petersen, B.; Gilbert, M.; Thomas, P.; Clokie, M.R.J.; Sicheritz-Pontén, T. Rapid discovery of novel prophages using biological feature engineering and machine learning. NAR Genom. Bioinform. 2021, 3, lqaa109. [Google Scholar] [CrossRef]
- Bayer, P.E.; Scheben, A.; Golicz, A.A.; Yuan, Y.; Faure, S.; Lee, H.; Chawla, H.S.; Anderson, R.; Bancroft, I.; Raman, H.; et al. Modelling of gene loss propensity in the pangenomes of three Brassica species suggests different mechanisms between polyploids and diploids. Plant Biotechnol. J. 2021, 19, 2488–2500. [Google Scholar] [CrossRef]
- Kreplak, J.; Madoui, M.-A.; Cápal, P.; Novák, P.; Labadie, K.; Aubert, G.; Bayer, P.E.; Gali, K.K.; Syme, R.A.; Main, D.; et al. A reference genome for pea provides insight into legume genome evolution. Nat. Genet. 2019, 51, 1411–1422. [Google Scholar] [CrossRef]
- Merrick, L.; Taly, A. The Explanation Game: Explaining Machine Learning Models Using Shapley Values. In Machine Learning and Knowledge Extraction; Springer: Cham, Switzerland, 2020; pp. 17–38. [Google Scholar]
- Lundberg, S.M.; Lee, S.-I. A unified approach to interpreting model predictions. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 4768–4777. [Google Scholar]
- Chicco, D.; Jurman, G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genom. 2020, 21, 6. [Google Scholar] [CrossRef] [Green Version]
- Craveur, P.; Joseph, A.P.; Esque, J.; Narwani, T.J.; Noel, F.; Shinada, N.; Goguet, M.; Sylvain, L.; Poulain, P.; Bertrand, O.; et al. Protein flexibility in the light of structural alphabets. Front. Mol. Biosci. 2015, 2, 20. [Google Scholar] [CrossRef] [Green Version]
- Schiex, T.; Moisan, A.; Rouzé, P. Eugène: An Eukaryotic Gene Finder That Combines Several Sources of Evidence. In Computational Biology; Springer: Berlin/Heidelberg, Germany, 2001; pp. 111–125. [Google Scholar]
- Li, Y.-R.; Liu, M.-J. Prevalence of alternative AUG and non-AUG translation initiators and their regulatory effects across plants. Genome Res. 2020, 30, 1418–1433. [Google Scholar] [CrossRef]
- Schwab, S.R.; Shugart, J.A.; Horng, T.; Malarkannan, S.; Shastri, N. Unanticipated antigens: Translation initiation at CUG with leucine. PLoS Biol. 2004, 2, e366. [Google Scholar] [CrossRef]
- Depeiges, A.; Degroote, F.; Espagnol, M.C.; Picard, G. Translation initiation by non-AUG codons in Arabidopsis thaliana transgenic plants. Plant Cell Rep. 2006, 25, 55–61. [Google Scholar] [CrossRef]
- Wang, J.; Li, S.; Zhang, Y.; Zheng, H.; Xu, Z.; Ye, J.; Yu, J.; Wong, G.K.-S. Vertebrate gene predictions and the problem of large genes. Nat. Rev. Genet. 2003, 4, 741–749. [Google Scholar] [CrossRef]
- Misawa, K.; Kikuno, R.F. GeneWaltz—A new method for reducing the false positives of gene finding. BioData Min. 2010, 3, 6. [Google Scholar] [CrossRef] [Green Version]
- Storz, G.; Wolf, Y.I.; Ramamurthi, K.S. Small proteins can no longer be ignored. Annu. Rev. Biochem. 2014, 83, 753–777. [Google Scholar] [CrossRef] [Green Version]
- Bowman, M.J.; Pulman, J.A.; Liu, T.L.; Childs, K.L. A modified GC-specific MAKER gene annotation method reveals improved and novel gene predictions of high and low GC content in Oryza sativa. BMC Bioinform. 2017, 18, 522. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Ming, R.; Yu, Q. Comparative Analysis of GC Content Variations in Plant Genomes. Trop. Plant Biol. 2016, 9, 136–149. [Google Scholar] [CrossRef]
- Lukashin, A.V.; Borodovsky, M. GeneMark.hmm: New solutions for gene finding. Nucleic Acids Res. 1998, 26, 1107–1115. [Google Scholar] [CrossRef] [Green Version]
- Khandelwal, G.; Bhyravabhotla, J. A Phenomenological Model for Predicting Melting Temperatures of DNA Sequences. PLoS ONE 2010, 5, e12433. [Google Scholar] [CrossRef] [Green Version]
- Dineen, D.G.; Wilm, A.; Cunningham, P.; Higgins, D.G. High DNA melting temperature predicts transcription start site location in human and mouse. Nucleic Acids Res. 2009, 37, 7360–7367. [Google Scholar] [CrossRef] [Green Version]
- Gailly, J.-l.; Adler, M. Zlib Compression Library. 2004. Available online: https://www.dspace.cam.ac.uk/handle/1810/3486 (accessed on 25 October 2021).
- Dash, S.; Campbell, J.D.; Cannon, E.K.S.; Cleary, A.M.; Huang, W.; Kalberer, S.R.; Karingula, V.; Rice, A.G.; Singh, J.; Umale, P.E.; et al. Legume information system (LegumeInfo.org): A key component of a set of federated data resources for the legume family. Nucleic Acids Res. 2015, 44, D1181–D1188. [Google Scholar] [CrossRef] [Green Version]
- Cock, P.J.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2012, 12, 2825–2830. [Google Scholar]
- McKinney, W. Data Structures for Statistical Computing in Python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010; pp. 56–61. [Google Scholar]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]


| Feature 1 | Feature 2 | Correlation Co-Efficient (R) |
|---|---|---|
| Length | Flexibility | 0.99 |
| Length | Molecular weight | 0.99 |
| Length | Molar extinction coefficient reduced | 0.81 |
| Length | Molar extinction coefficient oxidised | 0.81 |
| Aliphaticity | Aliphatic index | 0.93 |
| Gravy Value | Non-polar amino acids | 0.86 |
| Tiny amino acids | Amino acid percentage G | 0.57 |
| Iso-electric point | Acidic amino acids | −0.58 |
| Tiny amino acids | Amino acid percentage A | 0.48 |
| Feature 1 | Feature 2 | Correlation Co-Efficient (R) |
|---|---|---|
| Length | Molecular weight | 0.99 |
| Length | Entropy | 0.83 |
| Length | Melting temperature | 0.17 |
| Length | Zlib compression ratio | −0.68 |
| GC content | Melting temperature | 0.92 |
| GC content | GC at position 3 | 0.72 |
| GC content | GC at position 2 | 0.71 |
| GC content | GC at position 1 | 0.58 |
| Molecular weight | Entropy | 0.83 |
| Evaluation Metric | Protein Model | Nucleotide Model |
|---|---|---|
| Prediction Accuracy | 89.92% | 86.72% |
| 10-fold cross validation | 88.66% (±0.65%) | 85.38% (±0.40%) |
| F1_score | 0.94 | 0.91 |
| Average precision score | 0.93 | 0.90 |
| MCC | 0.93 | 0.93 |
| AUC value | 0.94 | 0.92 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Upadhyaya, S.R.; Bayer, P.E.; Tay Fernandez, C.G.; Petereit, J.; Batley, J.; Bennamoun, M.; Boussaid, F.; Edwards, D. Evaluating Plant Gene Models Using Machine Learning. Plants 2022, 11, 1619. https://doi.org/10.3390/plants11121619
Upadhyaya SR, Bayer PE, Tay Fernandez CG, Petereit J, Batley J, Bennamoun M, Boussaid F, Edwards D. Evaluating Plant Gene Models Using Machine Learning. Plants. 2022; 11(12):1619. https://doi.org/10.3390/plants11121619
Chicago/Turabian StyleUpadhyaya, Shriprabha R., Philipp E. Bayer, Cassandria G. Tay Fernandez, Jakob Petereit, Jacqueline Batley, Mohammed Bennamoun, Farid Boussaid, and David Edwards. 2022. "Evaluating Plant Gene Models Using Machine Learning" Plants 11, no. 12: 1619. https://doi.org/10.3390/plants11121619
APA StyleUpadhyaya, S. R., Bayer, P. E., Tay Fernandez, C. G., Petereit, J., Batley, J., Bennamoun, M., Boussaid, F., & Edwards, D. (2022). Evaluating Plant Gene Models Using Machine Learning. Plants, 11(12), 1619. https://doi.org/10.3390/plants11121619










