Unraveling NPR-like Family Genes in Fragaria spp. Facilitated to Identify Putative NPR1 and NPR3/4 Orthologues Participating in Strawberry-Colletotrichum fructicola Interaction
Abstract
:1. Introduction
2. Results
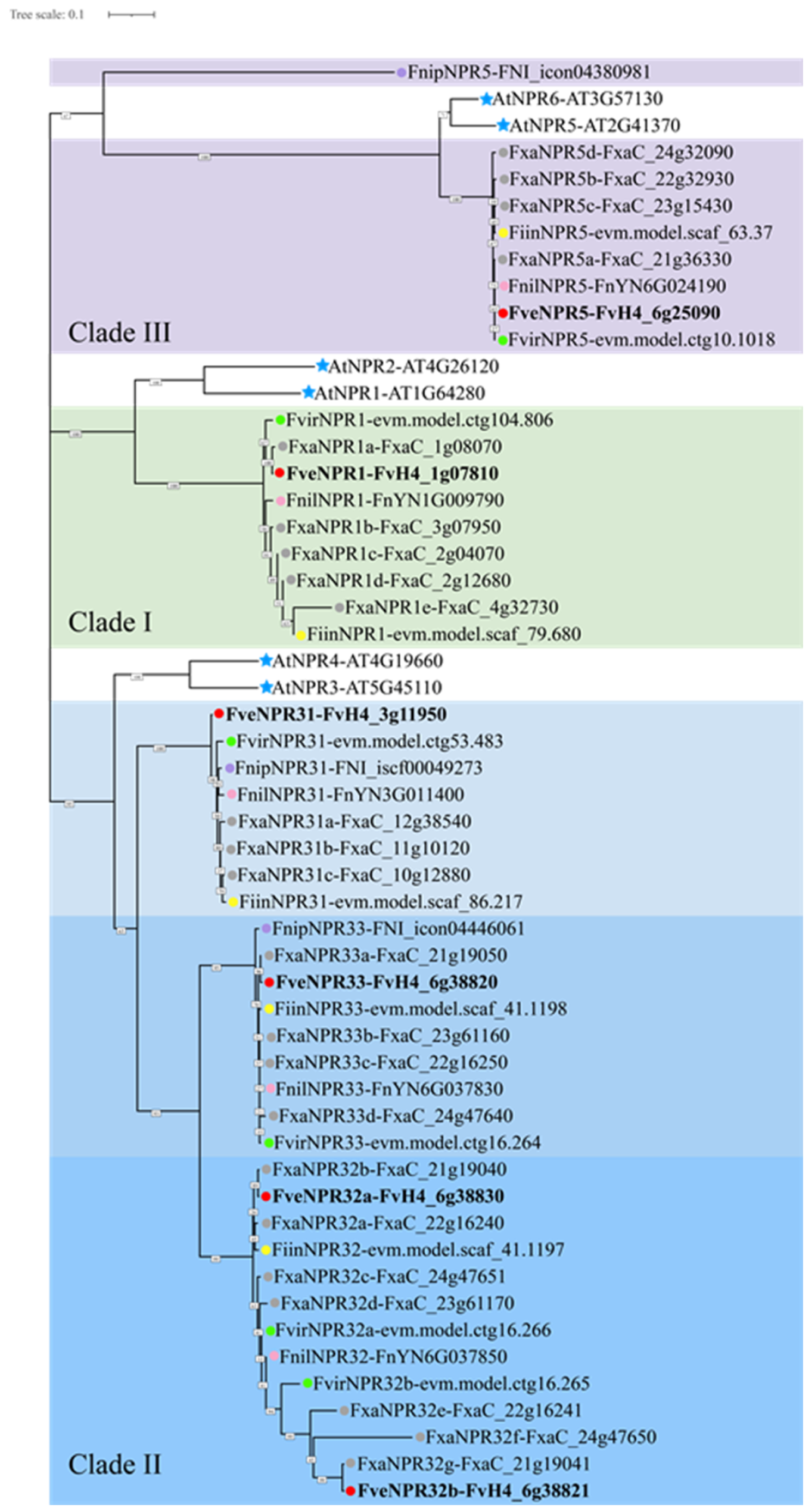
2.1. Diversity and Phylogeny of NPR-like Members in Fragaria Species
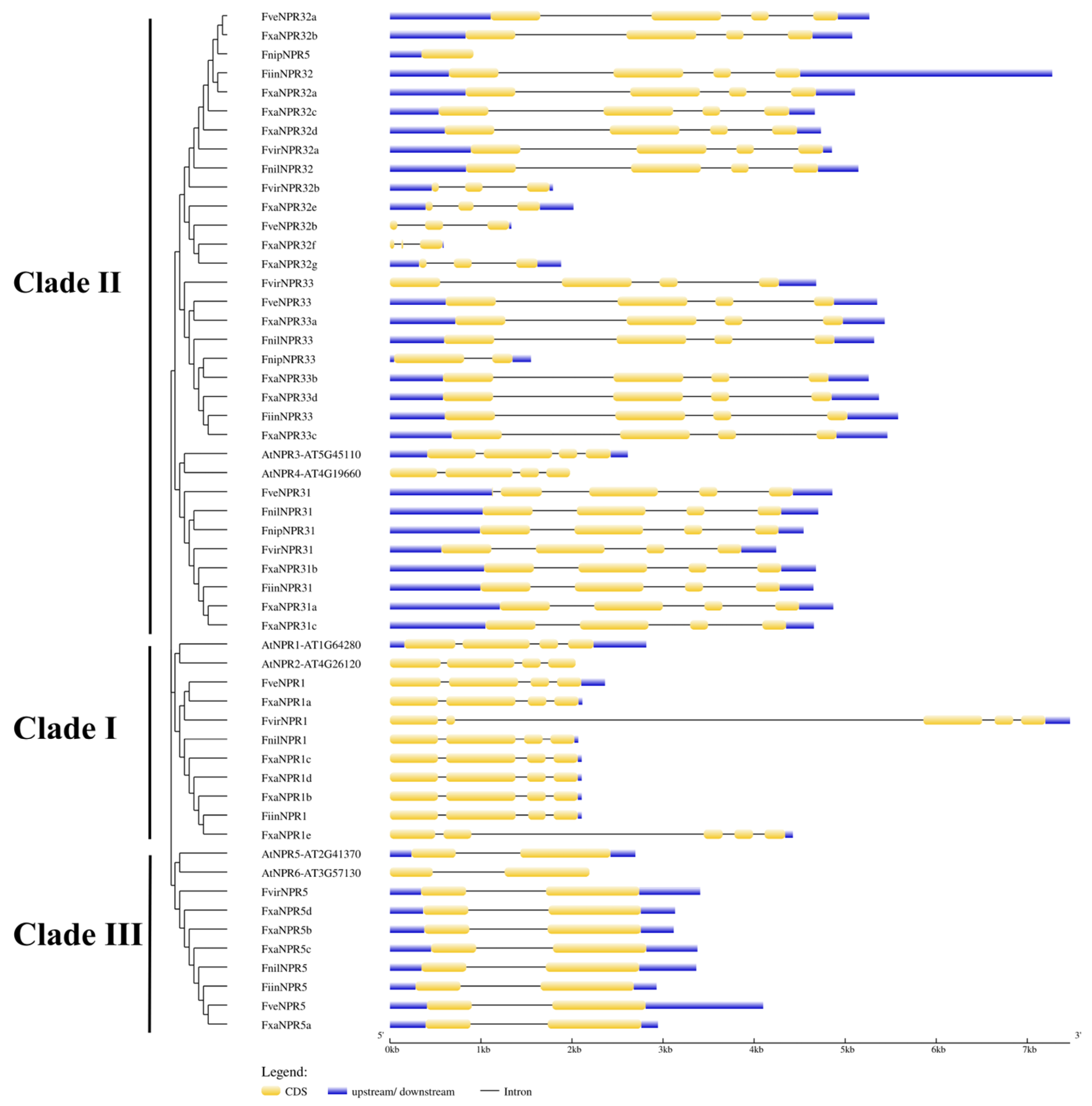
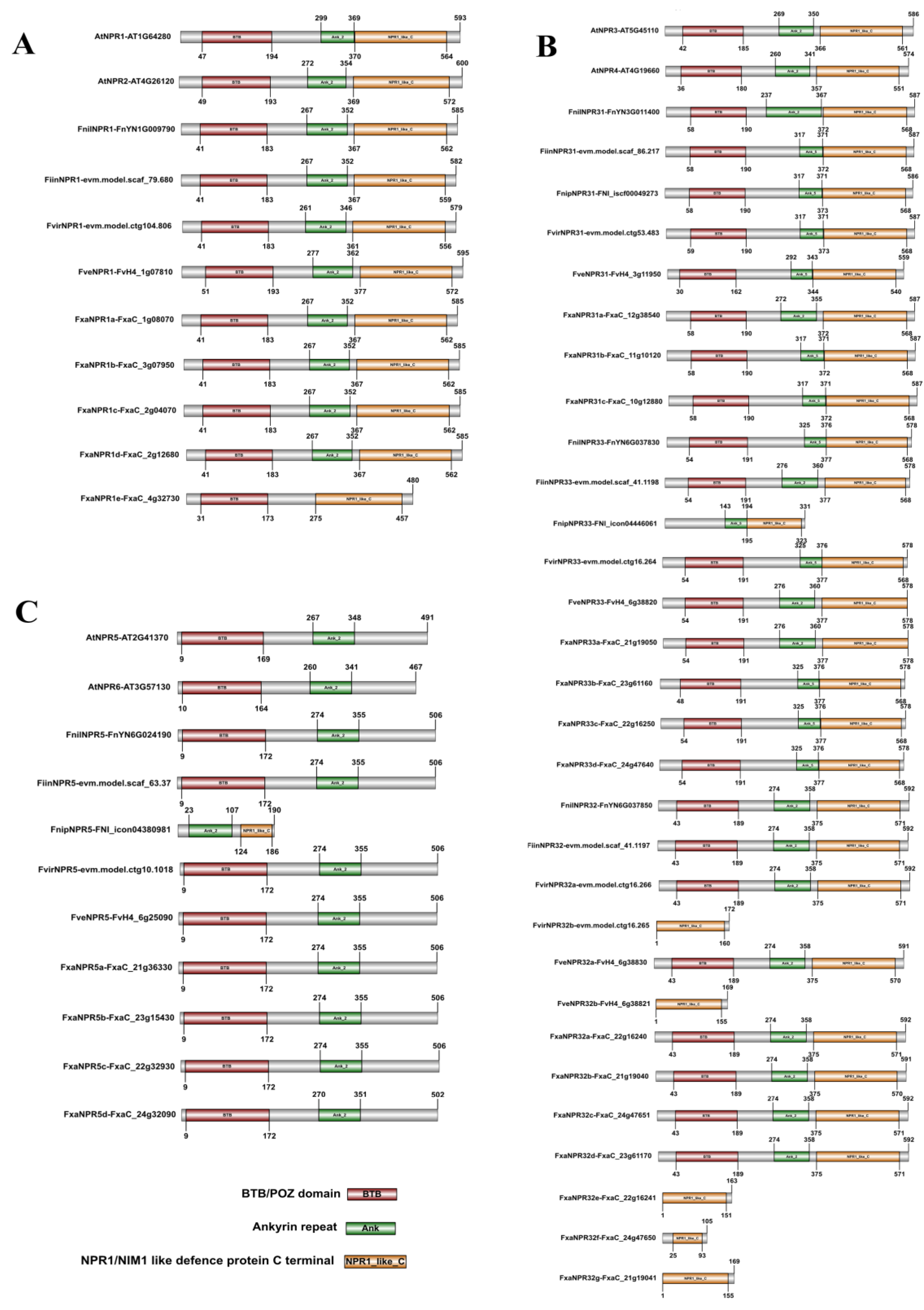
2.2. Structural Features of Fragaria NPR-like Members in DNA and Amino Acid Sequences
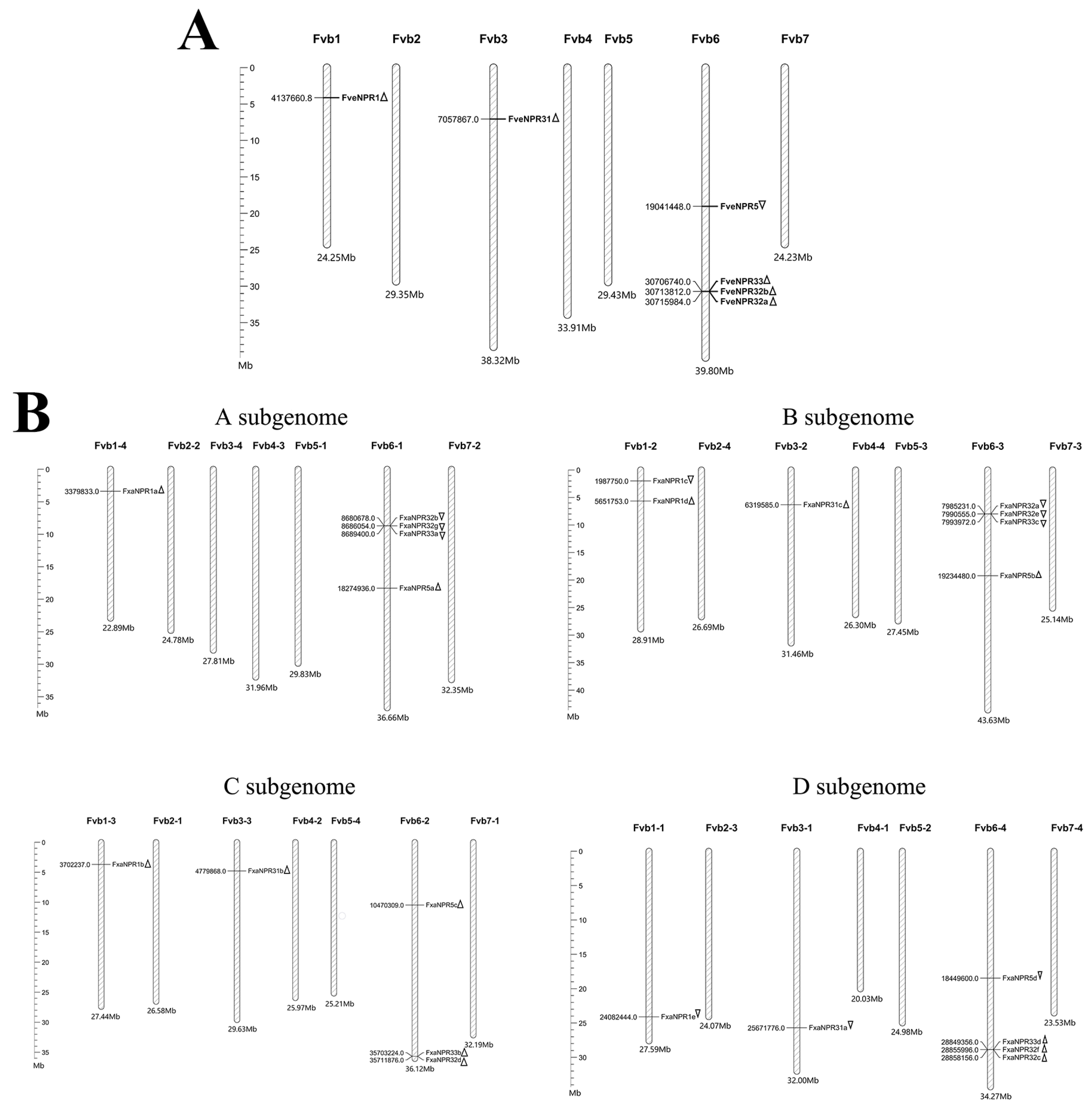
2.3. Physical Distribution of NPR1-like Loci in the Genomes of F. vesca and F. × ananassa
2.4. Cis-Elements in the Promoter Regions of NPR-like Genes from F. vesca and F. × ananassa
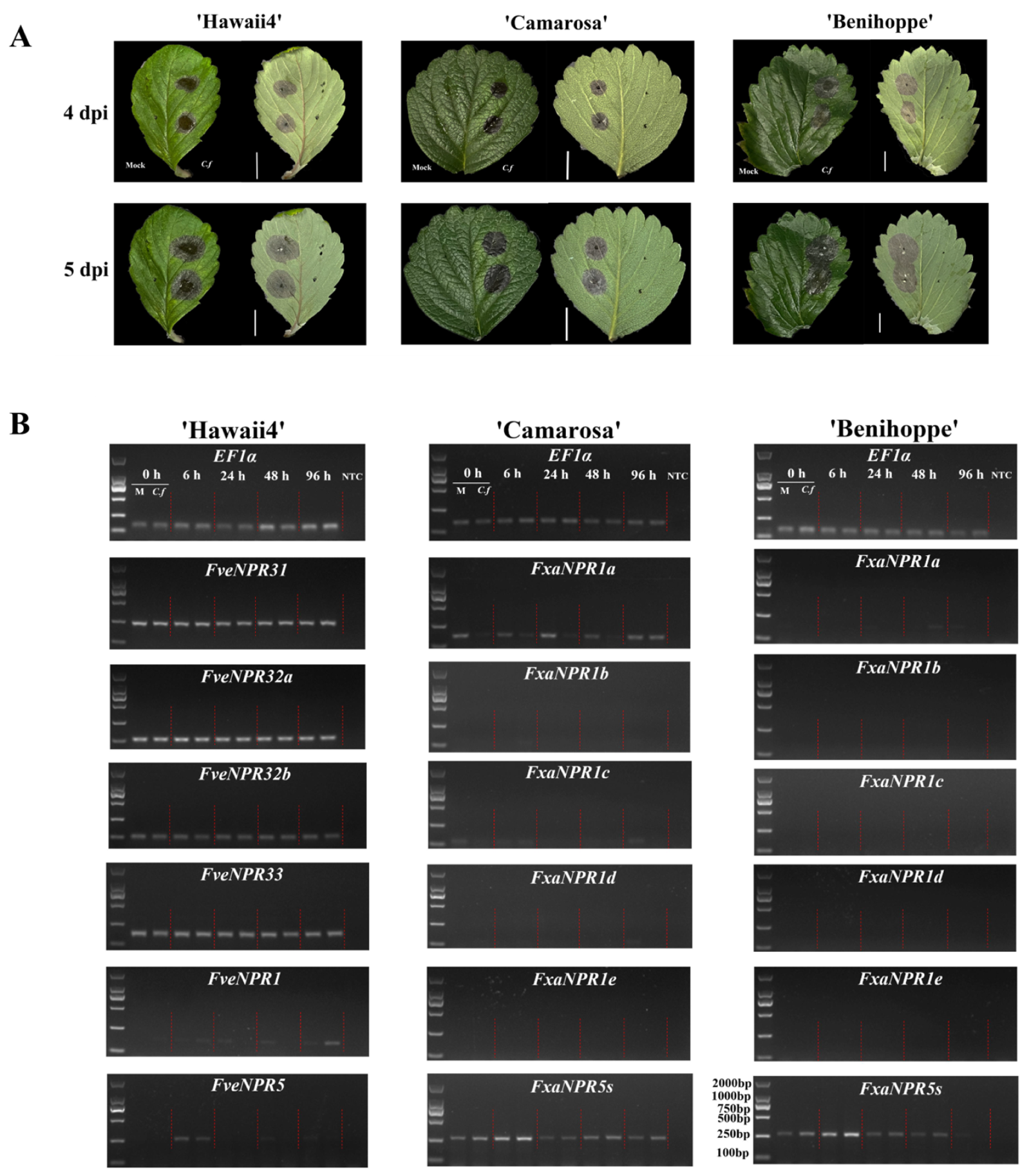
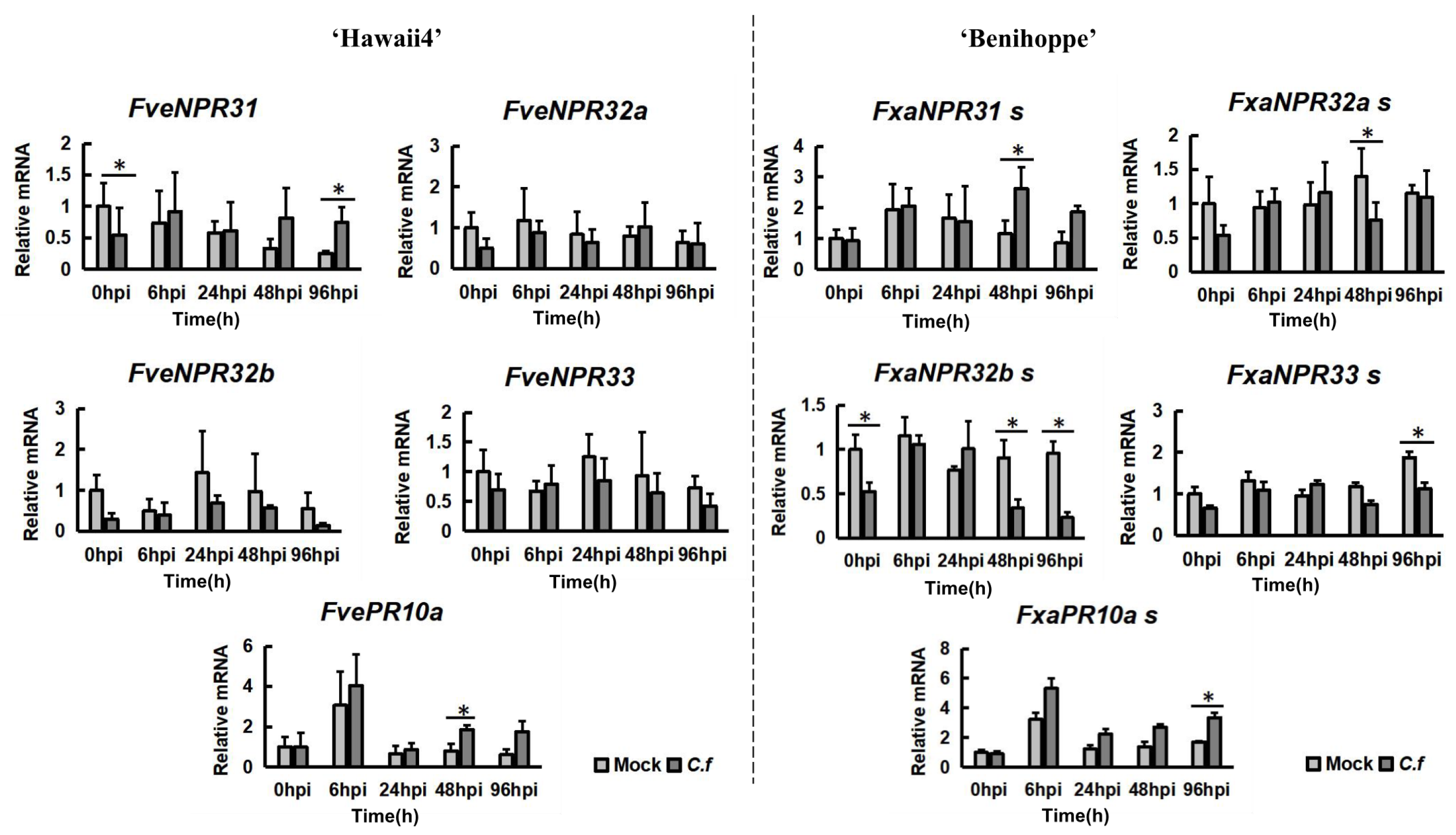
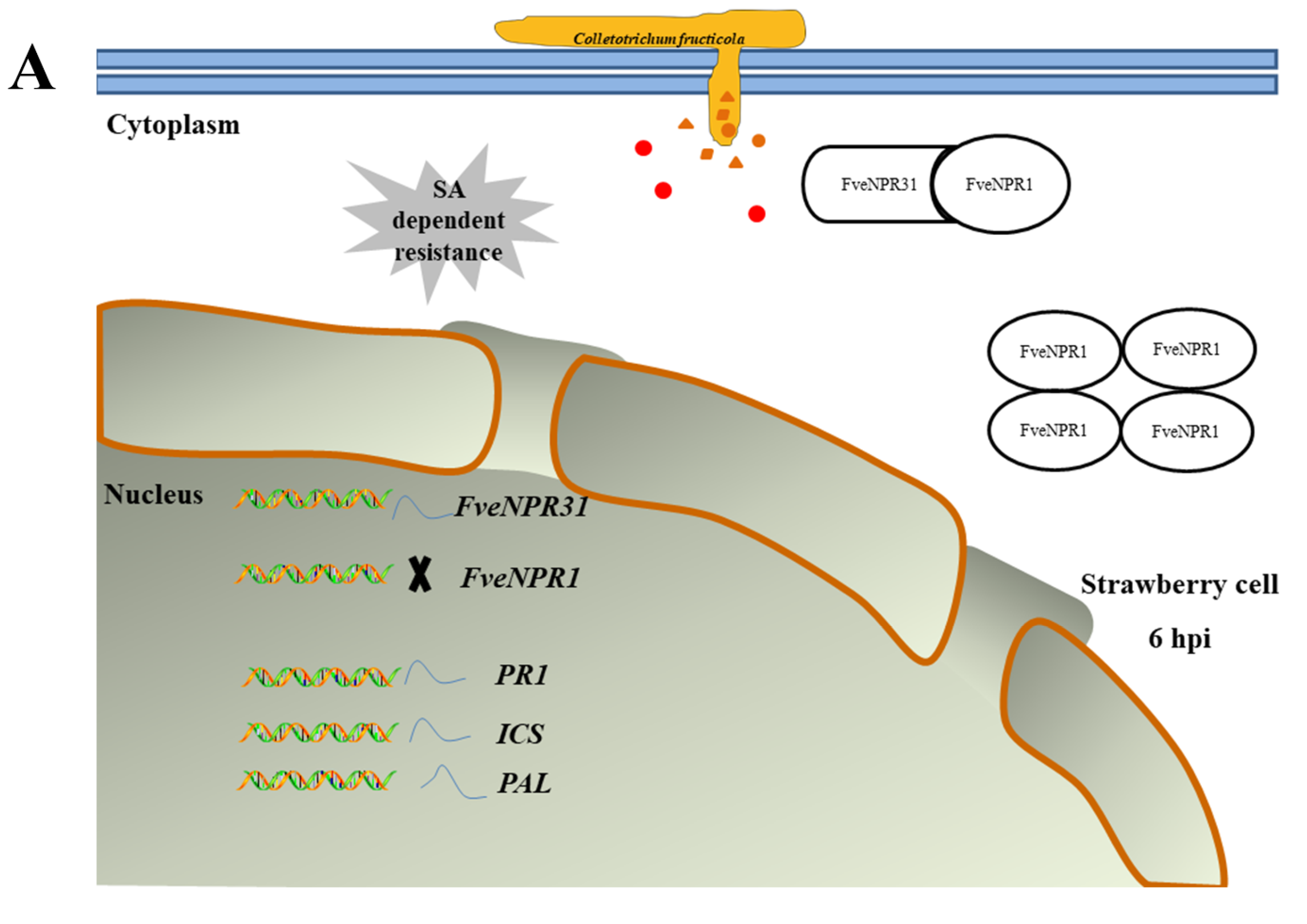
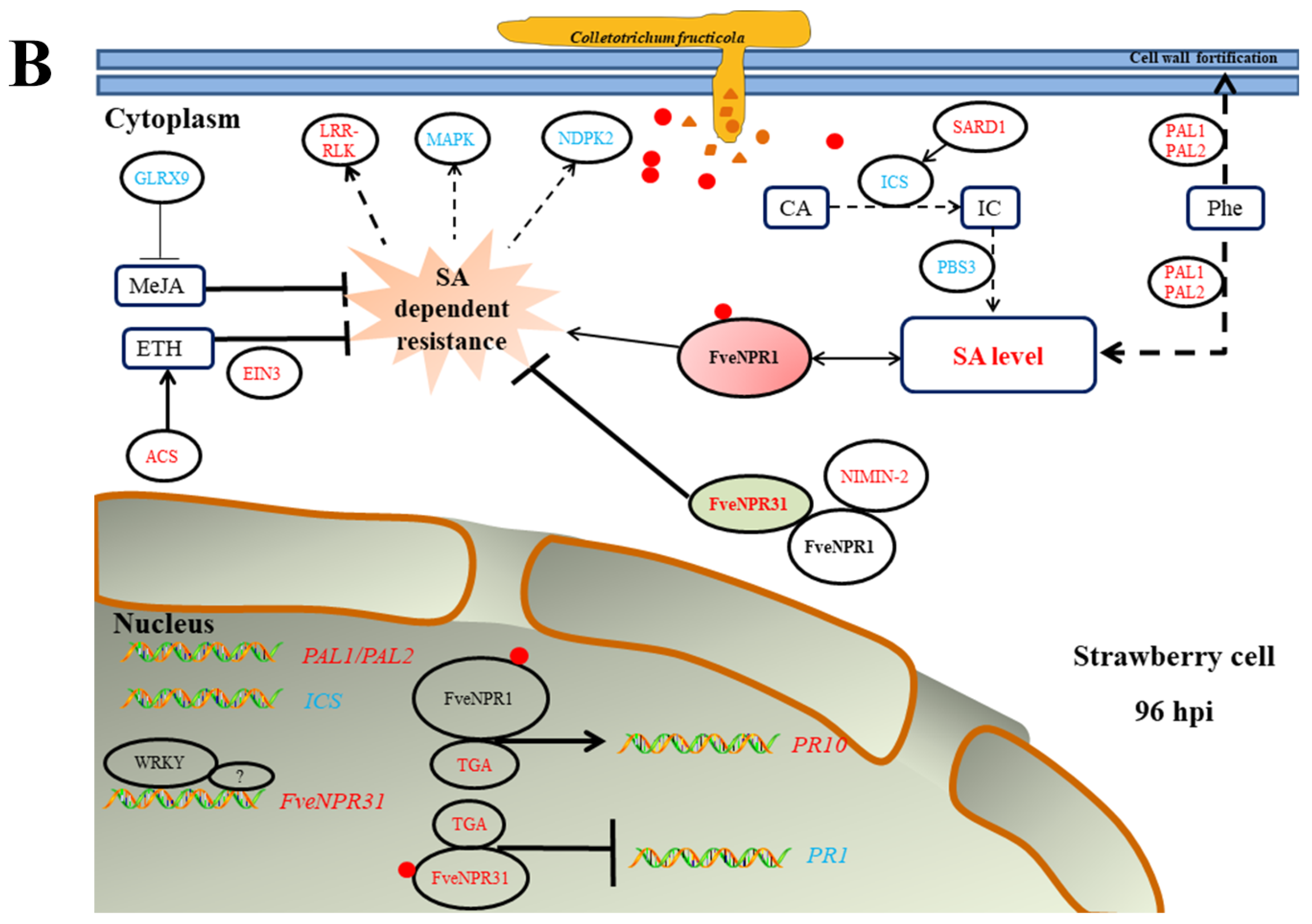
2.5. Transcriptional Responses of Strawberry NPR-like Genes to C. fructicola
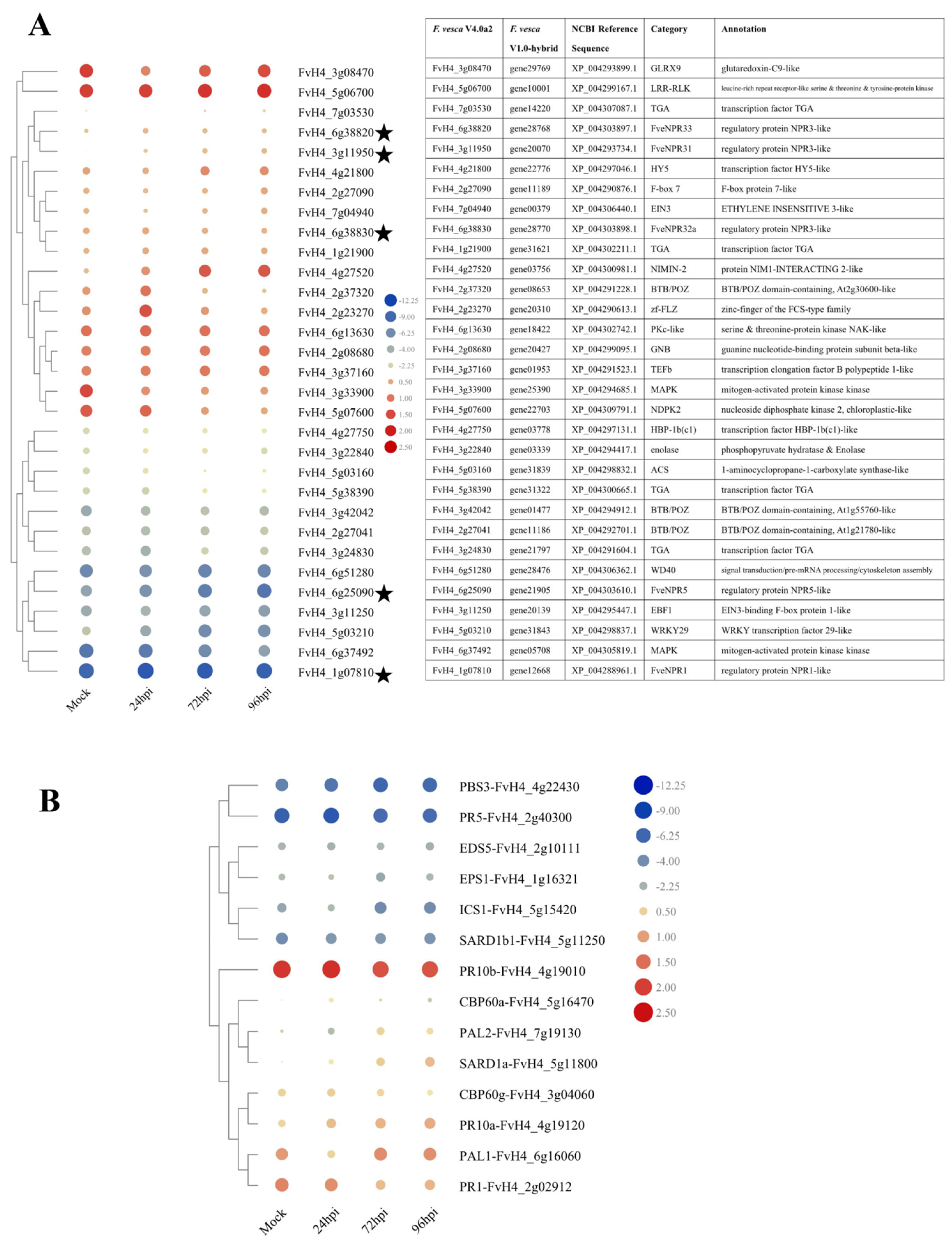
2.6. Transcriptome Profile of NPR-like and Related Genes in Strawberry Infected with C. fructicola
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and Inoculation with C. fructicola
4.2. Identification and Phylogenic Analysis of NPR-like Genes in Fragaria Species
4.3. Exon-Intron Structure in NPR-like Genes and Domain Organization in Deduced Proteins
4.4. Chromosomal Locations of NPR-like Genes in F. vesca and F. × ananassa
4.5. Cis-Elements in Promoters of NPR-like Loci in F. vesca and F. × ananassa
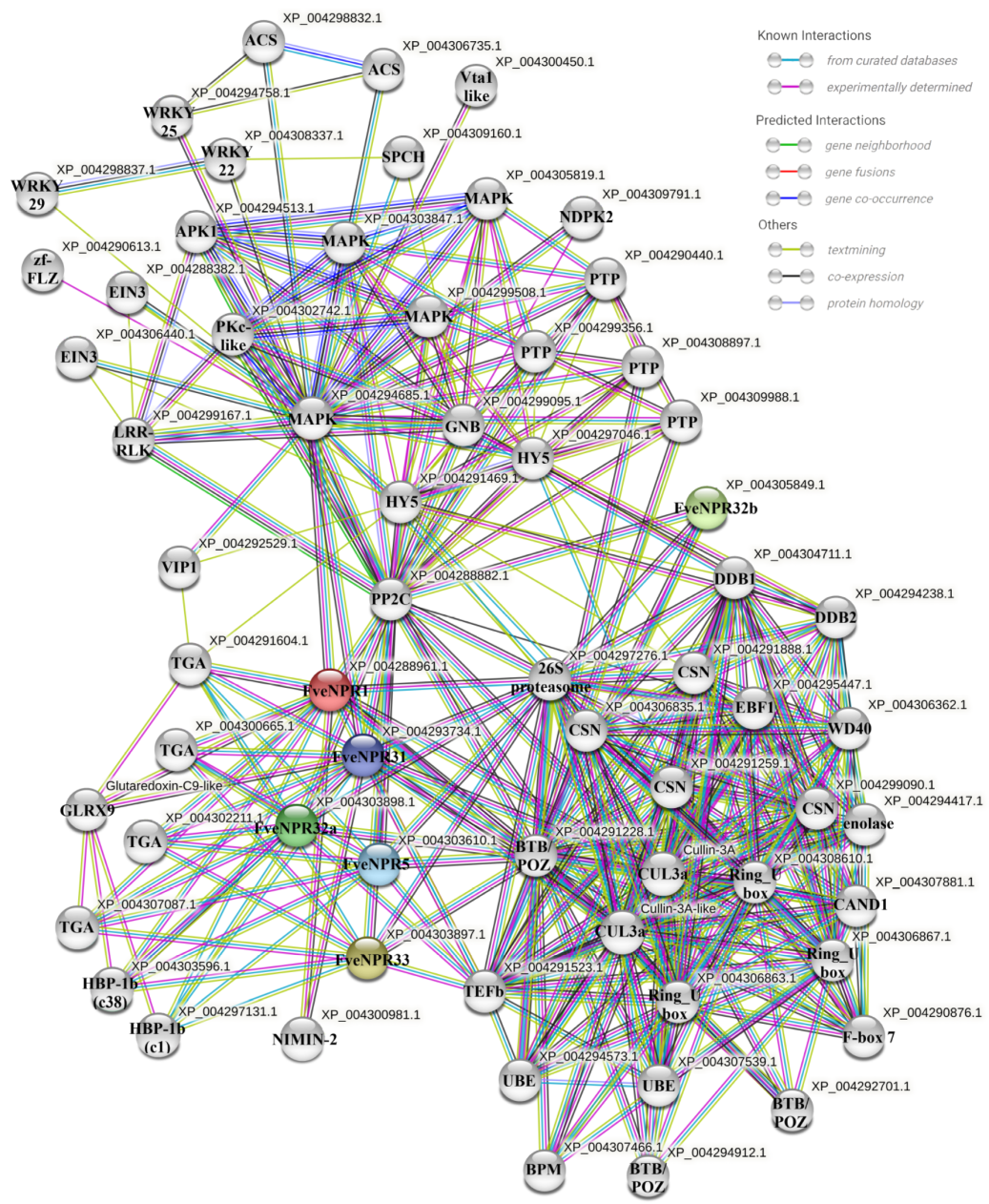
4.6. Protein-Protein Interaction Prediction for Strawberry NPR-like Proteins
4.7. RNA Purification, RT-PCR, and RNA Sequencing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cao, H.; Bowling, S.A.; Gordon, A.S.; Dong, X. Characterization of an Arabidopsis Mutant That Is Nonresponsive to Inducers of Systemic Acquired Resistance. Plant Cell 1994, 6, 1583–1592. [Google Scholar] [CrossRef] [PubMed]
- Kinkema, M.; Fan, W.; Dong, X. Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 2000, 12, 2339–2350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mou, Z.; Fan, W.; Dong, X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 2003, 113, 935–944. [Google Scholar] [CrossRef] [Green Version]
- Fu, Z.Q.; Yan, S.; Saleh, A.; Wang, W.; Ruble, J.; Oka, N.; Mohan, R.; Spoel, S.H.; Tada, Y.; Zheng, N.; et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 2012, 486, 228–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saleh, A.; Withers, J.; Mohan, R.; Marqués, J.; Gu, Y.; Yan, S.; Zavaliev, R.; Nomoto, M.; Tada, Y.; Dong, X. Posttranslational Modifications of the Master Transcriptional Regulator NPR1 Enable Dynamic but Tight Control of Plant Immune Responses. Cell Host Microbe 2015, 18, 169–182. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Sun, T.; Sun, Y.; Zhang, Y.; Radojičić, A.; Ding, Y.; Tian, H.; Huang, X.; Lan, J.; Chen, S.; et al. Diverse Roles of the Salicylic Acid Receptors NPR1 and NPR3/NPR4 in Plant Immunity. Plant Cell 2020, 32, 4002–4016. [Google Scholar] [CrossRef]
- Ding, P.; Ding, Y. Stories of Salicylic Acid: A Plant Defense Hormone. Trends Plant Sci. 2020, 25, 549–565. [Google Scholar] [CrossRef]
- Paeng, S.K.; Chi, Y.H.; Kang, C.H.; Chae, H.B.; Lee, E.S.; Park, J.H.; Wi, S.D.; Bae, S.B.; Phan, K.A.T.; Lee, S.Y. Chaperone function of Arabidopsis NPR1. Plant Biotechnol. Rep. 2020, 14, 227–233. [Google Scholar] [CrossRef]
- Wang, W.; Withers, J.; Li, H.; Zwack, P.J.; Rusnac, D.V.; Shi, H.; Liu, L.; Yan, S.; Hinds, T.R.; Guttman, M.; et al. Structural basis of salicylic acid perception by Arabidopsis NPR proteins. Nature 2020, 586, 311–316. [Google Scholar] [CrossRef]
- Zhang, Y.; Cheng, Y.T.; Qu, N.; Zhao, Q.; Bi, D.; Li, X. Negative regulation of defense responses in Arabidopsis by two NPR1 paralogs. Plant J. 2006, 48, 647–656. [Google Scholar] [CrossRef]
- Endah, R.; Beyene, G.; Kiggundu, A.; van den Berg, N.; Schluter, U.; Kunert, K.; Chikwamba, R. Elicitor and Fusarium-induced expression of NPR1-like genes in banana. Plant Physiol. Biochem. 2008, 46, 1007–1014. [Google Scholar] [CrossRef]
- Bergeault, K.; Bertsch, C.; Merdinoglu, D.; Walter, B. Low level of polymorphism in two putative NPR1 homologs in the Vitaceae family. Biol. Direct 2010, 5, 9. [Google Scholar] [CrossRef] [Green Version]
- Backer, R.; Mahomed, W.; Reeksting, B.J.; Engelbrecht, J.; Ibarra-Laclette, E.; van den Berg, N. Phylogenetic and expression analysis of the NPR1-like gene family from Persea americana (Mill.). Front. Plant. Sci. 2015, 6, 300. [Google Scholar] [CrossRef] [Green Version]
- Shu, L.J.; Liao, J.Y.; Lin, N.C.; Chung, C.L. Identification of a strawberry NPR-like gene involved in negative regulation of the salicylic acid-mediated defense pathway. PLoS ONE 2018, 13, e0205790. [Google Scholar] [CrossRef]
- Liu, X.; Liu, Z.; Niu, X.; Xu, Q.; Yang, L. Genome-Wide Identification and Analysis of the NPR1-Like Gene Family in Bread Wheat and Its Relatives. Int. J. Mol. Sci. 2019, 20, 5974. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, N.; Srivastava, R.; Verma, A.; Rai, K.M.; Singh, B.; Verma, P.C. Unravelling Cotton Nonexpressor of Pathogenesis-Related 1(NPR1)-Like Genes Family: Evolutionary Analysis and Putative Role in Fiber Development and Defense Pathway. Plants 2020, 9, 999. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, L.; Liu, D.; Ma, S.; Dai, Y.; Zhang, X.; Wang, Y.; Hu, T.; Xiao, M.; Zhou, Y.; et al. Identification and Expression of the Multidrug and Toxic Compound Extrusion (MATE) Gene Family in Capsicum annuum and Solanum tuberosum. Plants 2020, 9, 1448. [Google Scholar] [CrossRef]
- Li, R.; Li, Y.; Zhang, Y.; Sheng, J.; Zhu, H.; Shen, L. Transcriptome analysis reveals that SlNPR1 mediates tomato fruit resistance against Botrytis cinerea by modulating phenylpropanoid metabolism and balancing ROS homeostasis. Postharvest Biol. Technol. 2021, 172, 111382. [Google Scholar] [CrossRef]
- Wang, P.; Zhao, Z.; Zhang, Z.; Cai, Z.; Liao, J.; Tan, Q.; Xiang, M.; Chang, L.; Xu, D.; Tian, Q.; et al. Genome-wide identification and analysis of NPR family genes in Brassica juncea var. tumida. Gene 2021, 769, 145210. [Google Scholar] [CrossRef]
- Wei, Y.; Zhao, S.; Liu, N.; Zhang, Y. Genome-wide identification, evolution, and expression analysis of the NPR1-like gene family in pears. PeerJ 2021, 9, e12617. [Google Scholar] [CrossRef]
- Backer, R.; Naidoo, S.; van den Berg, N. The NONEXPRESSOR of PATHOGENESIS-RELATED GENES 1 (NPR1) and RELATED FAMILY: Mechanistic insights in plant disease resistance. Front. Plant Sci. 2019, 10, 102. [Google Scholar] [CrossRef] [Green Version]
- Darrow, G.M. The Strawberry History, Breeding and Physiology; Mickey, G.H., Ed.; Holt, Rinehart and Winston: New York, NY, USA, 1966. [Google Scholar]
- Edger, P.P.; Poorten, T.J.; VanBuren, R.; Hardigan, M.A.; Colle, M.; McKain, M.R.; Smith, R.D.; Teresi, S.J.; Nelson, A.D.L.; Wai, C.M.; et al. Origin and evolution of the octoploid strawberry genome. Nat. Genet. 2019, 51, 541–547. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Wang, H.; Cheng, X.; Su, X.; Zhao, Y.; Jiang, T.; Jin, Q.; Lin, Y.; Cai, Y. Comparative genomic analysis of the PAL genes in five Rosaceae species and functional identification of Chinese white pear. PeerJ 2019, 7, e8064. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Li, M.; Liu, Z.; Ai, X.; Li, Y. Reannotation of the cultivated strawberry genome and establishment of a strawberry genome database. Hortic. Res. 2021, 8, 41. [Google Scholar] [CrossRef]
- Qiao, Q.; Edger, P.P.; Xue, L.; Qiong, L.; Lu, J.; Zhang, Y.; Cao, Q.; Yocca, A.E.; Platts, A.E.; Knapp, S.J.; et al. Evolutionary history and pan-genome dynamics of strawberry (Fragaria spp.). Proc. Natl. Acad. Sci. USA 2021, 118, e2105431118. [Google Scholar] [CrossRef]
- Liston, A.; Wei, N.; Tennessen, J.A.; Li, J.; Dong, M.; Ashman, T.-L. Revisiting the origin of octoploid strawberry. Nat. Genet. 2020, 52, 2–4. [Google Scholar] [CrossRef]
- Feng, C.; Wang, J.; Harris, A.J.; Folta, K.M.; Zhao, M.; Kang, M. Tracing the Diploid Ancestry of the Cultivated Octoploid Strawberry. Mol. Biol. Evol. 2021, 38, 478–485. [Google Scholar] [CrossRef]
- Hardigan, M.A.; Lorant, A.; Pincot, D.D.A.; Feldmann, M.J.; Famula, R.A.; Acharya, C.B.; Lee, S.; Verma, S.; Whitaker, V.M.; Bassil, N.; et al. Unraveling the Complex Hybrid Ancestry and Domestication History of Cultivated Strawberry. Mol. Biol. Evol. 2021, 38, 2285–2305. [Google Scholar] [CrossRef]
- Wang, L.; Guo, Z.; Zhang, Y.; Wang, Y.; Yang, G.; Yang, L.; Wang, L.; Wang, R.; Xie, Z. Overexpression of LhSorNPR1, a NPR1-like gene from the oriental hybrid lily ‘Sorbonne’, conferred enhanced resistance to Pseudomonas syringae pv. tomato DC3000 in Arabidopsis. Physiol. Mol. Biol. Plants 2017, 23, 793–808. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, W.-H.; Ma, L.-Y.; Li, X.; Zhao, F.-Y.; Tan, X.-L. Overexpression of Brassica napus NPR1 enhances resistance to Sclerotinia sclerotiorum in oilseed rape. Physiol. Mol. Plant Pathol. 2020, 110, 101460. [Google Scholar] [CrossRef]
- Peng, A.; Zou, X.; He, Y.; Chen, S.; Liu, X.; Zhang, J.; Zhang, Q.; Xie, Z.; Long, J.; Zhao, X. Overexpressing a NPR1-like gene from Citrus paradisi enhanced Huanglongbing resistance in C. sinensis. Plant Cell Rep. 2021, 40, 529–541. [Google Scholar] [CrossRef] [PubMed]
- Silva, K.J.P.; Brunings, A.; Peres, N.A.; Mou, Z.; Folta, K.M. The Arabidopsis NPR1 gene confers broad-spectrum disease resistance in strawberry. Transgenic Res. 2015, 24, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Amil-Ruiz, F.; Garrido-Gala, J.; Gadea, J.; Blanco-Portales, R.; Muñoz-Mérida, A.; Trelles, O.; de Los Santos, B.; Arroyo, F.T.; Aguado-Puig, A.; Romero, F.; et al. Partial Activation of SA- and JA-Defensive Pathways in Strawberry upon Colletotrichum acutatum Interaction. Front. Plant Sci. 2016, 7, 1036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Huang, X.; He, C.; Zhang, Q.Y.; Zou, X.; Duan, K.; Gao, Q. Novel Fungal Pathogenicity and Leaf Defense Strategies Are Revealed by Simultaneous Transcriptome Analysis of Colletotrichum fructicola and Strawberry Infected by This Fungus. Front Plant Sci. 2018, 9, 434. [Google Scholar] [CrossRef] [Green Version]
- He, C.; Duan, K.; Zhang, L.; Zhang, L.; Song, L.; Yang, J.; Zou, X.; Wang, Y.; Gao, Q. Fast Quenching the Burst of Host Salicylic Acid Is Common in Early Strawberry/Colletotrichum fructicola Interaction. Phytopathology 2019, 109, 531–541. [Google Scholar] [CrossRef] [Green Version]
- Feng, J.; Cheng, Y.; Zheng, C. Expression patterns of octoploid strawberry TGA genes reveal a potential role in response to Podosphaera aphanis infection. Plant Biotechnol. Rep. 2019, 14, 55–67. [Google Scholar] [CrossRef]
- Feng, J.; Zhang, M.; Yang, K.N.; Zheng, C.X. Salicylic acid-primed defence response in octoploid strawberry ‘Benihoppe’ leaves induces resistance against Podosphaera aphanis through enhanced accumulation of proanthocyanidins and upregulation of pathogenesis-related genes. BMC Plant Biol. 2020, 20, 149. [Google Scholar] [CrossRef] [Green Version]
- Maier, F.; Zwicker, S.; Huckelhoven, A.; Meissner, M.; Funk, J.; Pfitzner, A.J.; Pfitzner, U.M. nonexpressor of pathogenesis-related proteins1 (NPR1) and some NPR1-related proteins are sensitive to salicylic acid. Mol. Plant Pathol. 2011, 12, 73–91. [Google Scholar] [CrossRef]
- Germain, H.; Lachance, D.; Pelletier, G.; Fossdal, C.G.; Solheim, H.; Séguin, A. The expression pattern of the Picea glauca Defensin 1 promoter is maintained in Arabidopsis thaliana, indicating the conservation of signalling pathways between angiosperms and gymnosperms. J. Exp. Bot. 2012, 63, 785–795. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, Z. Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol. 2002, 129, 706–716. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Chen, C.; Fan, B.; Chen, Z. Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 2006, 18, 1310–1326. [Google Scholar] [CrossRef] [Green Version]
- Goldsbrough, A.P.; Albrecht, H.; Stratford, R. Salicylic acid-inducible binding of a tobacco nuclear protein to a 10 bp sequence which is highly conserved amongst stress-inducible genes. Plant J. 1993, 3, 563–571. [Google Scholar] [CrossRef]
- Salazar, M.; González, E.; Casaretto, J.A.; Casacuberta, J.M.; Ruiz-Lara, S. The promoter of the TLC1.1 retrotransposon from Solanum chilense is activated by multiple stress-related signaling molecules. Plant Cell Rep. 2007, 26, 1861–1868. [Google Scholar] [CrossRef]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T. The ocs element in the soybean GH2/4 promoter is activated by both active and inactive auxin and salicylic acid analogues. Plant Mol. Biol. 1994, 26, 1055–1064. [Google Scholar] [CrossRef]
- Xiang, C.; Miao, Z.H.; Lam, E. Coordinated activation of as-1-type elements and a tobacco glutathione S-transferase gene by auxins, salicylic acid, methyl-jasmonate and hydrogen peroxide. Plant Mol. Biol. 1996, 32, 415–426. [Google Scholar] [CrossRef]
- Krawczyk, S.; Thurow, C.; Niggeweg, R.; Gatz, C. Analysis of the spacing between the two palindromes of activation sequence-1 with respect to binding to different TGA factors and transcriptional activation potential. Nucleic Acids Res. 2002, 30, 775–781. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Zhang, J.; Kong, M.; Freeman, A.; Chen, H.; Liu, F. More stories to tell: NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1, a salicylic acid receptor. Plant Cell Environ. 2021, 44, 1716–1727. [Google Scholar] [CrossRef]
- Amil-Ruiz, F.; Garrido-Gala, J.; Blanco-Portales, R.; Folta, K.M.; Muñoz-Blanco, J.; Caballero, J.L. Identification and validation of reference genes for transcript normalization in strawberry (Fragaria × ananassa) defense responses. PLoS ONE 2013, 8, e70603. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Holub, E.B.; Alonso, J.M.; Ecker, J.R.; Fobert, P.R. An Arabidopsis NPR1-like gene, NPR4, is required for disease resistance. Plant J. 2005, 41, 304–318. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhong, S.; Li, Q.; Zhu, Z.; Lou, Y.; Wang, L.; Wang, J.; Wang, M.; Li, Q.; Yang, D.; et al. Functional analysis of rice NPR1-like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnol. J. 2007, 5, 313–324. [Google Scholar] [CrossRef]
- Wang, Z.; Ma, L.Y.; Li, X.; Zhao, F.Y.; Sarwar, R.; Cao, J.; Li, Y.L.; Ding, L.N.; Zhu, K.M.; Yang, Y.H.; et al. Genome-wide identification of the NPR1-like gene family in Brassica napus and functional characterization of BnaNPR1 in resistance to Sclerotinia sclerotiorum. Plant Cell Rep. 2020, 39, 709–722. [Google Scholar] [CrossRef]
- Amil-Ruiz, F. Molecular Mechanisms of Strawberry Plant Defence against Colletotrichum acutatum. Ph.D. Thesis, Universidad de Córdoba, Córdoba, Spain, 2013. [Google Scholar]
- Tada, Y.; Spoel, S.H.; Pajerowska-Mukhtar, K.; Mou, Z.; Song, J.; Wang, C.; Zuo, J.; Dong, X. Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science 2008, 321, 952–956. [Google Scholar] [CrossRef] [Green Version]
- Ding, Y.; Sun, T.; Ao, K.; Peng, Y.; Zhang, Y.; Li, X.; Zhang, Y. Opposite Roles of Salicylic Acid Receptors NPR1 and NPR3/NPR4 in Transcriptional Regulation of Plant Immunity. Cell 2018, 173, 1454–1467.e15. [Google Scholar] [CrossRef]
- Ha, C.M.; Jun, J.H.; Nam, H.G.; Fletcher, J.C. BLADE-ON-PETIOLE1 Encodes a BTB/POZ Domain Protein Required for Leaf Morphogenesis in Arabidopsis thaliana. Plant Cell Physiol. 2004, 45, 1361–1370. [Google Scholar] [CrossRef]
- Wang, Y.; Salasini, B.C.; Khan, M.; Devi, B.; Bush, M.; Subramaniam, R.; Hepworth, S.R. Clade I TGACG-Motif Binding Basic Leucine Zipper Transcription Factors Mediate BLADE-ON-PETIOLE-Dependent Regulation of Development. Plant Physiol. 2019, 180, 937–951. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.; Yang, J.; Li, X.; Zhang, Y. Salicylic Acid: Biosynthesis and Signaling. Annu. Rev. Plant Biol. 2021, 72, 761–791. [Google Scholar] [CrossRef]
- Kang, C.; Darwish, O.; Geretz, A.; Shahan, R.; Alkharouf, N.; Liu, Z. Genome-scale transcriptomic insights into early-stage fruit development in woodland strawberry Fragaria vesca. Plant Cell 2013, 25, 1960–1978. [Google Scholar] [CrossRef] [Green Version]
- Xu, G.; Yuan, M.; Ai, C.; Liu, L.; Zhuang, E.; Karapetyan, S.; Wang, S.; Dong, X. uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature 2017, 545, 491–494. [Google Scholar] [CrossRef]
- Ren, X.; Li, Y.; Lu, J.; Yang, B.; Dai, F. Identification of Colletotrichum species from strawberry in Shanghai. Acta Phytopathol. Sin. 2008, 3, 325–328. (In Chinese) [Google Scholar]
- Chung, P.C.; Wu, H.Y.; Wang, Y.W.; Ariyawansa, H.A.; Hu, H.P.; Hung, T.H.; Tzean, S.S.; Chung, C.L. Diversity and pathogenicity of Colletotrichum species causing strawberry anthracnose in Taiwan and description of a new species, Colletotrichum miaoliense sp. nov. Sci. Rep. 2020, 10, 14664. [Google Scholar] [CrossRef]
- Jung, S.; Lee, T.; Cheng, C.-H.; Buble, K.; Zheng, P.; Yu, J.; Humann, J.; Ficklin, S.P.; Gasic, K.; Scott, K.; et al. 15 years of GDR: New data and functionality in the Genome Database for Rosaceae. Nucleic Acids Res. 2019, 47, D1137–D1145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Xie, Y.; Ma, J.; Luo, X.; Nie, P.; Zuo, Z.; Lahrmann, U.; Zhao, Q.; Zheng, Y.; Zhao, Y.; et al. IBS: An illustrator for the presentation and visualization of biological sequences. Bioinformatics 2015, 31, 3359–3361. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [Green Version]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [Green Version]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–2. [Google Scholar] [CrossRef] [Green Version]

| GENE NAME | Locus ID | Chromosomal Location | DNA (bp) | mRNA (bp) | Protein (aa) | LOC a | MW b (kDa) | Pi c |
|---|---|---|---|---|---|---|---|---|
| FnilNPR1 | FnYN1G009790 | GWHABKC00000007:1g6682576-6684604 | 2029 | 1758 | 585 | Nuc:Ct_Nuc (6.5:6) | 64.7 | 6.74 |
| FnilNPR31 | FnYN3G011400 | GWHABKC00000002:3g8012487-8017100 | 4614 | 3096 | 587 | Ct:ER (4:3) | 65.3 | 6.39 |
| FnilNPR32 | FnYN6G037850 | GWHABKC00000001:6g39083311-39088355 | 5045 | 2958 | 592 | Ct | 66.3 | 5.87 |
| FnilNPR33 | FnYN6G037830 | GWHABKC00000001:6g39072614-39077826 | 5213 | 2660 | 578 | Nuc | 64.7 | 5.77 |
| FnilNPR5 | FnYN6G024190 | GWHABKC00000001:6g24090462-24093763 | 3302 | 2430 | 506 | CL:ER(4:3) | 55.1 | 6.30 |
| FiinNPR1 | evm.model.scaf_79.680 | Chr1:3535351-3537414 | 2064 | 1749 | 582 | Ct:Ct_Nuc (6.5:6.5) | 64.6 | 6.34 |
| FiinNPR31 | evm.model.scaf_86.217 | Chr3:6321505-6326064 | 4560 | 3037 | 587 | CL:Ct:ER(3:3:3) | 65.3 | 6.15 |
| FiinNPR32 | evm.model.scaf_41.1197 | Chr6:34174583-34181716 | 7134 | 5053 | 592 | Nuc | 66.1 | 5.88 |
| FiinNPR33 | evm.model.scaf_41.1198 | Chr6:34168193-34173663 | 5471 | 2786 | 578 | Nuc | 64.8 | 5.71 |
| FiinNPR5 | evm.model.scaf_63.37 | Chr6:22982676-22985547 | 2872 | 1994 | 506 | CL:ER(4:3) | 55.2 | 6.25 |
| FnipNPR31 | FNI_iscf00049273 | FNI_iscf00049273.1:1606-6149 | 4544 | 3026 | 586 | CL:Ct:ER(3:3:3) | 65.2 | 6.48 |
| FnipNPR33 | FNI_icon04446061 | FNI_icon04446061.1:1..1551 | 1551 | 1244 | 331 | CL:Ct (5:4) | 37.5 | 5.93 |
| FnipNPR5 | FNI_icon04380981 | FNI_icon04380981.1:1-919 | 919 | 919 | 190 | Ct | 21.3 | 5.44 |
| FvirNPR1 | evm.model.ctg104.806 | Fvir1_FvScbg_v1.0:4416036-4423359 | 7324 | 1830 | 579 | Nuc:Ct_Nuc (6.5:6) | 64.2 | 6.39 |
| FvirNPR31 | evm.model.ctg53.483 | Fvir3_FvScbg_v1.0:7054130-7058288 | 4159 | 2628 | 587 | CL:Ct:ER(3:3:3) | 65.3 | 6.46 |
| FvirNPR32a | evm.model.ctg16.266 | Fvir6_FvScbg_v1.0:28680195-28684954 | 4760 | 2667 | 592 | Ct | 66.3 | 5.84 |
| FvirNPR32b | evm.model.ctg16.265 | Fvir6_FvScbg_v1.0:28677873-28679626 | 1754 | 978 | 172 | Nuc:Nuc_Pm (7: 5.5) | 19.3 | 5.47 |
| FvirNPR33 | evm.model.ctg16.264 | Fvir6_FvScbg_v1.0:28671131-28675723 | 4593 | 2056 | 578 | Nuc | 64.7 | 5.71 |
| FvirNPR5 | evm.model.ctg10.1018 | Fvir6_FvScbg_v1.0:17711496-17714838 | 3343 | 2466 | 506 | CL:ER(4:3) | 55.1 | 6.30 |
| FveNPR1 | FvH4_1g07810 | Fvb1_v4.0.a1:1g4137661-4139975 | 2315 | 2001 | 595 | Ct_Nuc:Nuc:Ct (5:4.5:4.5) | 66.1 | 6.56 |
| FveNPR31 | FvH4_3g11950 | Fvb3_v4.0.a1:3g7057867-7062282 | 4763 | 3137 | 559 | Nuc:Ct_Nuc (6.5:5.5) | 62.5 | 6.62 |
| FveNPR32a | FvH4_6g38830 | Fvb6_v4.0.a1:6g30715984-30720865 | 5163 | 3125 | 591 | Nuc | 65.9 | 6.21 |
| FveNPR32b | FvH4_6g38821 | Fvb6_v4.0.a1:6g30713812-30715119 | 1308 | 510 | 169 | Ct | 18.9 | 6.52 |
| FveNPR33 | FvH4_6g38820 | Fvb6_v4.0.a1:6g30706741-30711990 | 5250 | 2720 | 578 | Nuc | 64.6 | 5.77 |
| FveNPR5 | FvH4_6g25090 | Fvb6_v4.0.a1:6g19041447-19045465 | 4019 | 3135 | 506 | CL:ER(4:3) | 55.1 | 6.30 |
| FxaNPR1a | FxaC_1g08070 | Fvb1-4:1g3379833-3381904 | 2072 | 1758 | 585 | Nuc:Ct_Nuc (6.5:6.5) | 65.0 | 6.31 |
| FxaNPR1b | FxaC_3g07950 | Fvb1-3:3g3702237-3704301 | 2065 | 1758 | 585 | Nuc:Ct_Nuc:Ct (6.5:6.5:5.5) | 64.8 | 6.34 |
| FxaNPR1c | FxaC_2g04070 | Fvb1-2:2g1987750-1989813 | 2064 | 1758 | 585 | Nuc:Ct_Nuc:Ct (6.5:6.5:5.5) | 64.8 | 6.31 |
| FxaNPR1d | FxaC_2g12680 | Fvb1-2:2g5651753-5653817 | 2065 | 1758 | 585 | Nuc:Ct_Nuc:Ct (6.5:6.5:5.5) | 64.9 | 6.74 |
| FxaNPR1e | FxaC_4g32730 | Fvb1-1:4g24082443-24086784 | 4342 | 1443 | 480 | CL:Ct_Nuc(6:4.5) | 53.1 | 6.17 |
| FxaNPR31a | FxaC_12g38540 | Fvb3-1:12g25671777-25676553 | 4777 | 3253 | 587 | CL:Ct:Vc(3:3:3) | 65.4 | 6.69 |
| FxaNPR31b | FxaC_11g10120 | Fvb3-3:11g4779868-4784455 | 4588 | 3087 | 587 | Ct:Nuc(4:3) | 65.3 | 6.39 |
| FxaNPR31c | FxaC_10g12880 | Fvb3-2:10g6319585-6324148 | 4564 | 3026 | 587 | CL:Ct:ER(3:3:3) | 65.2 | 6.33 |
| FxaNPR32a | FxaC_22g16240 | Fvb6-3:22g7985231-7990237 | 5007 | 2939 | 592 | Nuc | 66.1 | 6.10 |
| FxaNPR32b | FxaC_21g19040 | Fvb6-1:21g8680678-8685655 | 4978 | 2943 | 591 | Nuc | 65.9 | 6.24 |
| FxaNPR32c | FxaC_24g47651 | Fvb6-4:24g28858157-28862733 | 4577 | 2505 | 592 | Ct | 66.1 | 6.07 |
| FxaNPR32d | FxaC_23g61170 | Fvb6-2:23g35711876-35716520 | 4645 | 2552 | 592 | Ct | 66.3 | 6.94 |
| FxaNPR32e | FxaC_22g16241 | Fvb6-3:22g7990555-7992532 | 1978 | 1213 | 163 | Nuc:Nuc_Pm(6:5) | 18.0 | 6.31 |
| FxaNPR32f | FxaC_24g47650 | Fvb6-4:24g28855998-28856575 | 578 | 318 | 105 | CL | 12.1 | 5.29 |
| FxaNPR32g | FxaC_21g19041 | Fvb6-1:21g8686054-8687896 | 1843 | 1051 | 169 | Ct | 18.8 | 6.21 |
| FxaNPR33a | FxaC_21g19050 | Fvb6-1:21g8689400-8694727 | 5328 | 2804 | 578 | Nuc | 64.6 | 5.83 |
| FxaNPR33b | FxaC_23g61160 | Fvb6-2:23g35703222-35708378 | 5157 | 2657 | 578 | Nuc | 64.7 | 5.71 |
| FxaNPR33c | FxaC_22g16250 | Fvb6-3:22g7993972-7999331 | 5360 | 2868 | 578 | Nuc | 64.8 | 5.77 |
| FxaNPR33d | FxaC_24g47640 | Fvb6-4:24g28849358-28854622 | 5265 | 2733 | 578 | Nuc | 64.9 | 5.55 |
| FxaNPR5a | FxaC_21g36330 | Fvb6-1:21g18274937-18277824 | 2888 | 2040 | 506 | CL:ER(4:3) | 55.1 | 6.28 |
| FxaNPR5b | FxaC_22g32930 | Fvb6-3:22g19234481-19237536 | 3056 | 2197 | 506 | CL:ER(4:3) | 55.3 | 6.27 |
| FxaNPR5c | FxaC_23g15430 | Fvb6-2:23g10470309-10473618 | 3310 | 2467 | 506 | CL:ER(4:3) | 55.2 | 6.20 |
| FxaNPR5d | FxaC_24g32090 | Fvb6-4:24g18449601-18452673 | 3073 | 2192 | 502 | CL:ER(4:3) | 54.7 | 6.23 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, Y.; Li, Z.; Zhu, J.; Chen, S.; Dong, C.; Gao, Q.; Duan, K. Unraveling NPR-like Family Genes in Fragaria spp. Facilitated to Identify Putative NPR1 and NPR3/4 Orthologues Participating in Strawberry-Colletotrichum fructicola Interaction. Plants 2022, 11, 1589. https://doi.org/10.3390/plants11121589
Bai Y, Li Z, Zhu J, Chen S, Dong C, Gao Q, Duan K. Unraveling NPR-like Family Genes in Fragaria spp. Facilitated to Identify Putative NPR1 and NPR3/4 Orthologues Participating in Strawberry-Colletotrichum fructicola Interaction. Plants. 2022; 11(12):1589. https://doi.org/10.3390/plants11121589
Chicago/Turabian StyleBai, Yun, Ziyi Li, Jiajun Zhu, Siyu Chen, Chao Dong, Qinghua Gao, and Ke Duan. 2022. "Unraveling NPR-like Family Genes in Fragaria spp. Facilitated to Identify Putative NPR1 and NPR3/4 Orthologues Participating in Strawberry-Colletotrichum fructicola Interaction" Plants 11, no. 12: 1589. https://doi.org/10.3390/plants11121589
APA StyleBai, Y., Li, Z., Zhu, J., Chen, S., Dong, C., Gao, Q., & Duan, K. (2022). Unraveling NPR-like Family Genes in Fragaria spp. Facilitated to Identify Putative NPR1 and NPR3/4 Orthologues Participating in Strawberry-Colletotrichum fructicola Interaction. Plants, 11(12), 1589. https://doi.org/10.3390/plants11121589







