Phylogenomic Analysis Reconstructed the Order Matoniales from Paleopolyploidy Veil
Abstract
:1. Introduction
2. Results
2.1. Assessment of RNA-Seq Data
2.2. Evolutionary History of the Early Leptosporangiate Ferns
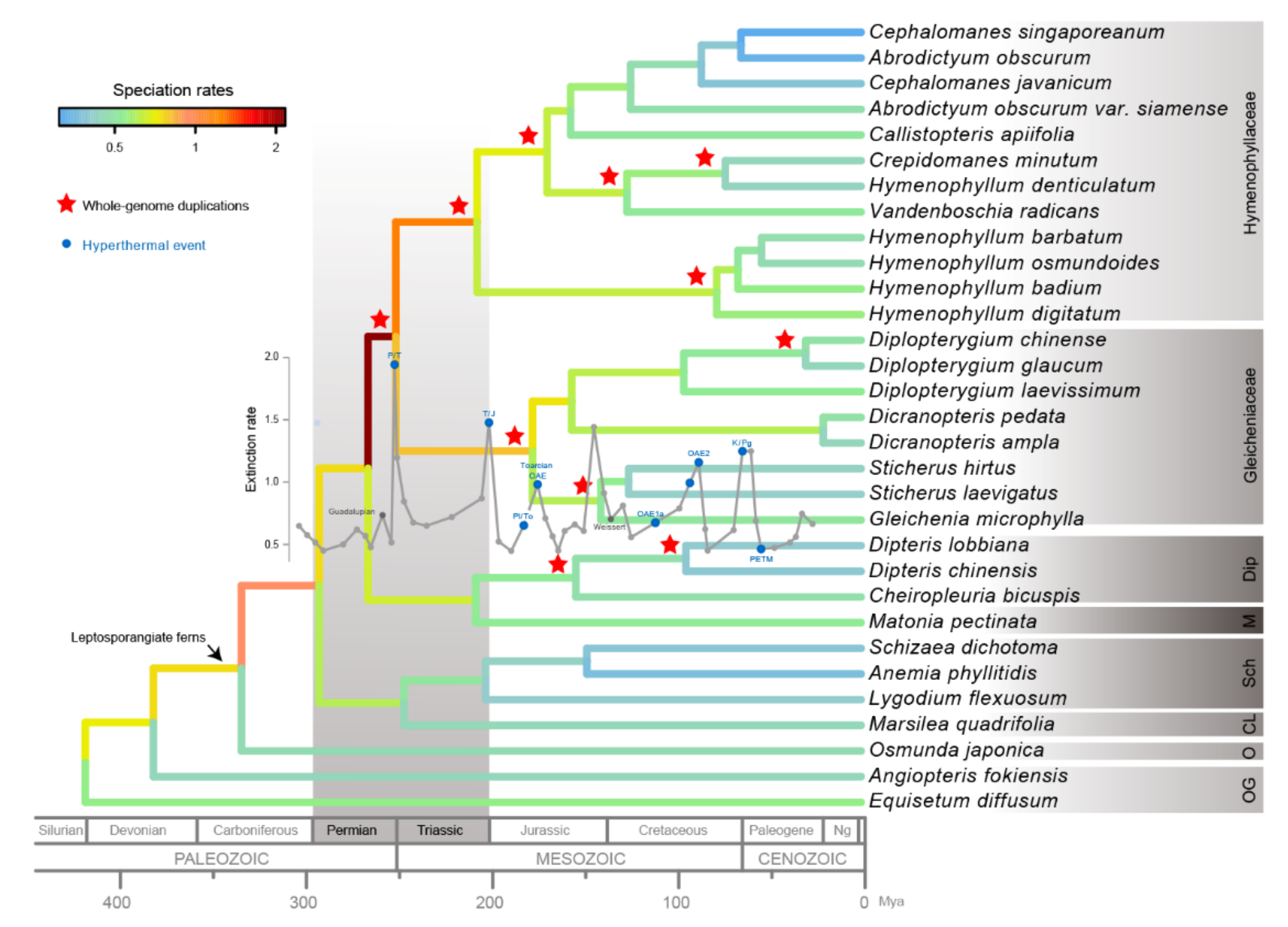
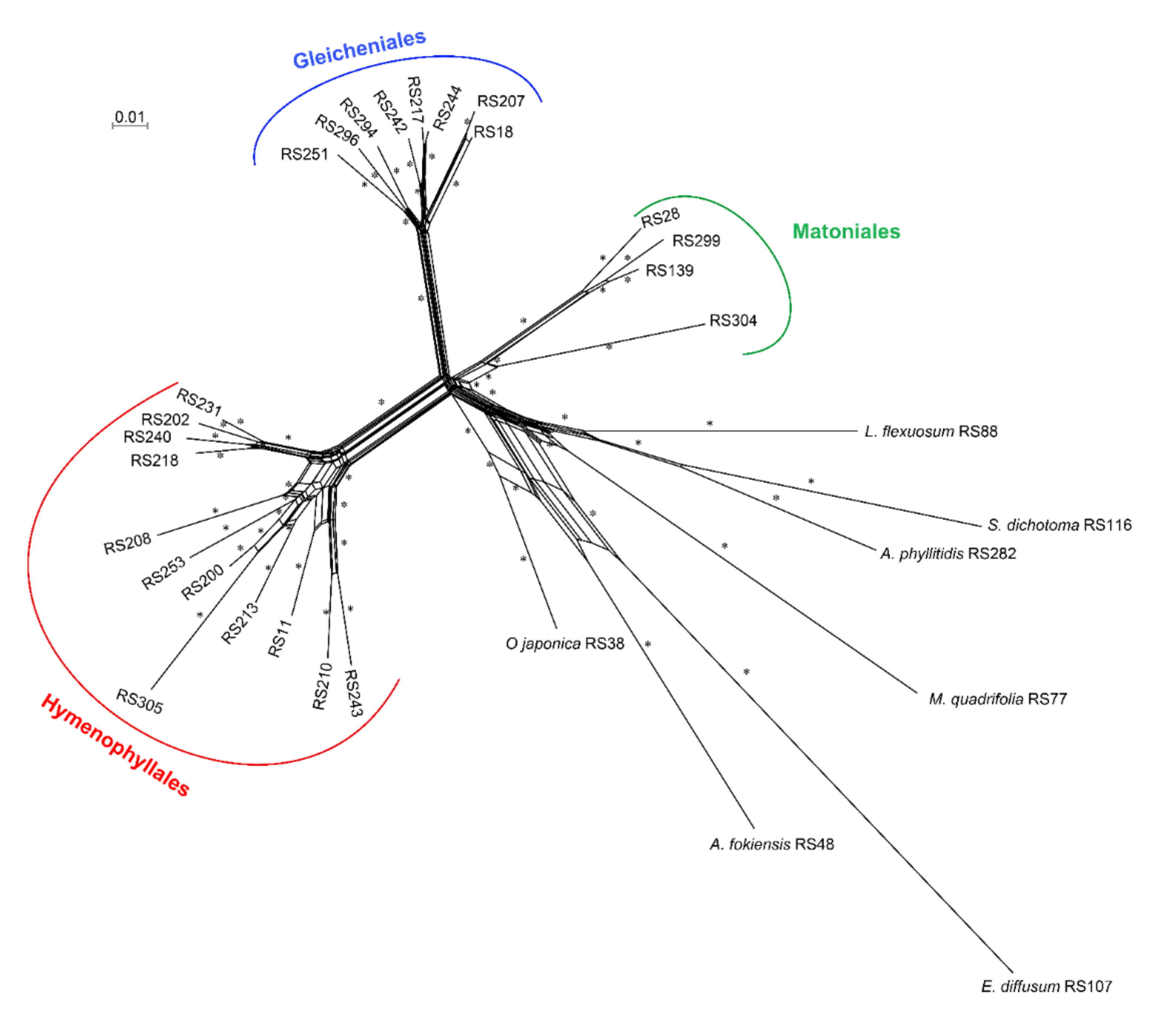
2.3. Paleopolyploidization and Reticulate Evolution in the Early Leptosporangiate Ferns
3. Discussion
3.1. Ancient Polyploidization Plays an Important Role in the Evolutionary History of Plants
3.2. Matoniales Should Be Redefined as a New Ordinal Rank
4. Materials and Methods
4.1. Taxon Sampling
4.2. Data Acquisition and Quality Assessment
4.3. Phylogenetic Tree and Network Analysis
4.4. Divergence Time Estimation
4.5. Polyploidization Inference and Localization
4.6. Species Diversification Rates
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, Z.; Rannala, B. Molecular phylogenetics: Principles and practice. Nat. Rev. Genet. 2012, 13, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Delsuc, F.; Brinkmann, H.; Philippe, H. Phylogenomics and the reconstruction of the tree of life. Nat. Rev. Genet. 2005, 6, 361–375. [Google Scholar] [CrossRef]
- Fox, G.E.; Stackebrandt, E.; Hespell, R.B.; Gibson, J.; Maniloff, J.; Dyer, T.A.; Wolfe, R.S.; Balch, W.E.; Tanner, R.S.; Magrum, L.J.; et al. The phylogeny of prokaryotes. Science 1980, 209, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Baldauf, S.L.; Roger, A.J.; Wenk-Siefert, I.; Doolittle, W.F. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 2000, 290, 972–977. [Google Scholar] [CrossRef] [PubMed]
- Dunn, C.W.; Hejnol, A.; Matus, D.Q.; Pang, K.; Browne, W.E.; Smith, S.A.; Seaver, E.; Rouse, G.W.; Obst, M.; Edgecombe, G.D.; et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 2008, 452, 745–749. [Google Scholar] [CrossRef]
- Misof, B.; Liu, S.; Meusemann, K.; Peters, R.S.; Donath, A.; Mayer, C.; Frandsen, P.B.; Ware, J.; Flouri, T.; Beutel, R.G.; et al. Phylogenomics resolves the timing and pattern of insect evolution. Science 2014, 346, 763–767. [Google Scholar] [CrossRef] [PubMed]
- James, T.Y.; Kauff, F.; Schoch, C.L.; Matheny, P.B.; Hofstetter, V.; Cox, C.J.; Celio, G.; Gueidan, C.; Fraker, E.; Miadlikowska, J.; et al. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 2006, 443, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.-L.; Lee, J.; Bernasconi-Quadroni, F.; Soltis, D.E.; Soltis, P.S.; Zanis, M.; Zimmer, E.A.; Chen, Z.; Savolainen, V.; Chase, M.W. The earliest angiosperms: Evidence from mitochondrial, plastid and nuclear genomes. Nature 1999, 402, 404–407. [Google Scholar] [CrossRef]
- Liu, Y.; Johnson, M.G.; Cox, C.J.; Medina, R.; Devos, N.; Vanderpoorten, A.; Hedenäs, L.; Bell, N.E.; Shevock, J.R.; Aguero, B.; et al. Resolution of the ordinal phylogeny of mosses using targeted exons from organellar and nuclear genomes. Nat. Commun. 2019, 10, 1485. [Google Scholar] [CrossRef]
- Leebens-Mack, J.H.; Barker, M.S.; Carpenter, E.J.; Deyholos, M.K.; Gitzendanner, M.A.; Graham, S.W.; Grosse, I.; Li, Z.; Melkonian, M.; Mirarab, S.; et al. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 2019, 574, 679–685. [Google Scholar] [CrossRef] [Green Version]
- Degnan, J.H.; Rosenberg, N.A. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol. Evol. 2009, 24, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Soltis, D.E.; Albert, V.A.; Leebens-Mack, J.; Bell, C.D.; Paterson, A.H.; Zheng, C.; Sankoff, D.; de Pamphilis, C.W.; Wall, P.K.; Soltis, P.S. Polyploidy and angiosperm diversification. Am. J. Bot. 2009, 96, 336–348. [Google Scholar] [CrossRef] [Green Version]
- Van de Peer, Y.; Mizrachi, E.; Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017, 18, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Clark, J.W.; Donoghue, P.C.J. Whole-genome duplication and plant macroevolution. Trends Plant Sci. 2018, 23, 933–945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, L.; Xi, Z.; Amorim, A.M.; Sugumaran, M.; Rest, J.S.; Liu, L.; Davis, C.C. Widespread ancient whole-genome duplications in Malpighiales coincide with Eocene global climatic upheaval. New Phytol. 2019, 221, 565–576. [Google Scholar] [CrossRef] [Green Version]
- Wood, T.E.; Takebayashi, N.; Barker, M.S.; Mayrose, I.; Greenspoon, P.B.; Rieseberg, L.H. The frequency of polyploid speciation in vascular plants. Proc. Natl. Acad. Sci. USA 2009, 106, 13875–13879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, R.; Wang, H.; Guo, C.; Zhang, N.; Zeng, L.; Chen, Y.; Ma, H.; Qi, J. Widespread whole genome duplications contribute to genome complexity and species diversity in angiosperms. Mol. Plant 2018, 11, 414–428. [Google Scholar] [CrossRef] [Green Version]
- Jiao, Y. Double the genome, double the fun: Genome duplications in angiosperms. Mol. Plant 2018, 11, 357–358. [Google Scholar] [CrossRef] [Green Version]
- Shen, H.; Jin, D.; Shu, J.-P.; Zhou, X.-L.; Lei, M.; Wei, R.; Shang, H.; Wei, H.-J.; Zhang, R.; Liu, L.; et al. Large-scale phylogenomic analysis resolves a backbone phylogeny in ferns. GigaScience 2018, 7, gix116. [Google Scholar] [CrossRef] [Green Version]
- PPGI. A community-derived classification for extant lycophytes and ferns. J. Syst. Evol. 2016, 54, 563–603. [Google Scholar] [CrossRef]
- Du, X.Y.; Lu, J.M.; Zhang, L.B.; Wen, J.; Kuo, L.Y.; Mynssen, C.M.; Schneider, H.; Li, D.Z. Simultaneous diversification of Polypodiales and angiosperms in the Mesozoic. Cladistics 2021, 37, 518–539. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Kuo, L.-Y.; Guo, C.; Li, H.; Li, Z.; Qi, J.; Wang, L.; Hu, Y.; Xiang, J.; Zhang, C.; et al. A well-resolved fern nuclear phylogeny reveals the evolution history of numerous transcription factor families. Mol. Phylogenet. Evol. 2018, 127, 961–977. [Google Scholar] [CrossRef]
- Testo, W.; Sundue, M. A 4000-species dataset provides new insight into the evolution of ferns. Mol. Phylogenet. Evol. 2016, 105, 200–211. [Google Scholar] [CrossRef] [Green Version]
- Schneider, H.; Schuettpelz, E.; Pryer, K.M.; Cranfill, R.; Magallón, S.; Lupia, R. Ferns diversified in the shadow of angiosperms. Nature 2004, 428, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Pryer, K.M.; Schneider, H.; Smith, A.R.; Cranfill, R.; Wolf, P.G.; Hunt, J.S.; Sipes, S.D. Horsetails and ferns are a monophyletic group and the closest living relatives to seed plants. Nature 2001, 409, 618–622. [Google Scholar] [CrossRef] [PubMed]
- Rothfels, C.J.; Li, F.-W.; Sigel, E.M.; Huiet, L.; Larsson, A.; Burge, D.O.; Ruhsam, M.; Deyholos, M.; Soltis, D.E.; Stewart, C.N., Jr.; et al. The evolutionary history of ferns inferred from 25 low-copy nuclear genes. Am. J. Bot. 2015, 102, 1089–1107. [Google Scholar] [CrossRef] [Green Version]
- Kuo, L.-Y.; Qi, X.; Ma, H.; Li, F.-W. Order-level fern plastome phylogenomics: New insights from Hymenophyllales. Am. J. Bot. 2018, 105, 1545–1555. [Google Scholar] [CrossRef]
- Schuettpelz, E.; Pryer, K.M. Fern phylogeny inferred from 400 leptosporangiate species and three plastid genes. Taxon 2007, 56, 1037–1050. [Google Scholar] [CrossRef] [Green Version]
- Doweld, A. Prosyllabus Tracheophytorum: Tentamen Systematis Plantarum Vascularium (Tracheophyta); Geos: Moscow, Russia, 2001. [Google Scholar]
- Reveal, J. New ordinal names for extant vascular plants. Phytologia 1993, 74, 173–177. [Google Scholar]
- Philippe, H.; Vienne, D.M.; Ranwez, V.; Roure, B.; Baurain, D.; Delsuc, F. Pitfalls in supermatrix phylogenomics. Eur. J. Taxon. 2017, 283, 1–25. [Google Scholar] [CrossRef] [Green Version]
- Jeffroy, O.; Brinkmann, H.; Delsuc, F.; Philippe, H. Phylogenomics: The beginning of incongruence? Trends Genet. 2006, 22, 225–231. [Google Scholar] [CrossRef] [Green Version]
- Heath, T.A.; Hedtke, S.M.; Hillis, D.M. Taxon sampling and the accuracy of phylogenetic analyses. J. Syst. Evol. 2008, 46, 239–257. [Google Scholar] [CrossRef]
- Pick, K.; Philippe, H.; Schreiber, F.; Erpenbeck, D.; Jackson, D.; Wrede, P.; Wiens, M.; Alié, A.; Morgenstern, B.; Manuel, M. Improved phylogenomic taxon sampling noticeably affects nonbilaterian relationships. Mol. Biol. Evol. 2010, 27, 1983–1987. [Google Scholar] [CrossRef]
- Nabhan, A.R.; Sarkar, I.N. The impact of taxon sampling on phylogenetic inference: A review of two decades of controversy. Brief Bioinform. 2012, 13, 122–134. [Google Scholar] [CrossRef] [Green Version]
- Qiu, Y.-L. Phylogeny and evolution of vascular plants. In eLS; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2018. [Google Scholar] [CrossRef]
- Haufler, C.H. Ever since Klekowski: Testing a set of radical hypotheses revives the genetics of ferns and lycophytes. Am. J. Bot. 2014, 101, 2036–2042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, J.; Hidalgo, O.; Pellicer, J.; Liu, H.; Marquardt, J.; Robert, Y.; Christenhusz, M.; Zhang, S.; Gibby, M.; Leitch, I.J. Genome evolution of ferns: Evidence for relative stasis of genome size across the fern phylogeny. New Phytol. 2016, 210, 1072–1082. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.H.; Qi, X.; Chen, D.; Qi, J.; Ma, H. Recurrent genome duplication events likely contributed to both the ancient and recent rise of ferns. J. Integr. Plant Biol. 2020, 62, 433–455. [Google Scholar] [CrossRef] [Green Version]
- Otto, S.P. The evolutionary consequences of polyploidy. Cell 2007, 131, 452–462. [Google Scholar] [CrossRef] [Green Version]
- Soltis, P.S.; Marchant, D.B.; Van de Peer, Y.; Soltis, D.E. Polyploidy and genome evolution in plants. Curr. Opin. Genet. Dev. 2015, 35, 119–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayrose, I.; Zhan, S.H.; Rothfels, C.J.; Arrigo, N.; Barker, M.S.; Rieseberg, L.H.; Otto, S.P. Methods for studying polyploid diversification and the dead end hypothesis: A reply to Soltis et al., (2014). New Phytol. 2015, 206, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Arrigo, N.; Barker, M.S. Rarely successful polyploids and their legacy in plant genomes. Curr. Opin. Plant Biol. 2012, 15, 140–146. [Google Scholar] [CrossRef]
- Mayrose, I.; Zhan, S.H.; Rothfels, C.J.; Magnuson-Ford, K.; Barker, M.S.; Rieseberg, L.H.; Otto, S.P. Recently formed polyploid plants diversify at lower rates. Science 2011, 333, 1257. [Google Scholar] [CrossRef] [Green Version]
- Schranz, M.E.; Mohammadin, S.; Edger, P.P. Ancient whole genome duplications, novelty and diversification: The WGD Radiation Lag-Time Model. Curr. Opin. Plant Biol. 2012, 15, 147–153. [Google Scholar] [CrossRef]
- Tank, D.C.; Eastman, J.M.; Pennell, M.W.; Soltis, P.S.; Soltis, D.E.; Hinchliff, C.E.; Brown, J.W.; Sessa, E.B.; Harmon, L.J. Nested radiations and the pulse of angiosperm diversification: Increased diversification rates often follow whole genome duplications. New Phytol. 2015, 207, 454–467. [Google Scholar] [CrossRef] [Green Version]
- Meier, J.I.; Marques, D.A.; Mwaiko, S.; Wagner, C.E.; Excoffier, L.; Seehausen, O. Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nat. Commun. 2017, 8, 14363. [Google Scholar] [CrossRef] [Green Version]
- Morrison, D.A. Phylogenetic networks: A review of methods to display evolutionary history. Annu. Res. Rev. Biol. 2014, 4, 1518–1543. [Google Scholar] [CrossRef]
- Chao, D.-Y.; Dilkes, B.; Luo, H.; Douglas, A.; Yakubova, E.; Lahner, B.; Salt, D.E. Polyploids exhibit higher potassium uptake and salinity tolerance in Arabidopsis. Science 2013, 341, 658–659. [Google Scholar] [CrossRef] [Green Version]
- Ramsey, J. Polyploidy and ecological adaptation in wild yarrow. Proc. Natl. Acad. Sci. USA 2011, 108, 7096–7101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diallo, A.M.; Nielsen, L.R.; Kjær, E.D.; Petersen, K.K.; Ræbild, A. Polyploidy can confer superiority to West African Acacia senegal (L.) Willd. trees. Front. Plant Sci. 2016, 7, 821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fawcett, J.A.; Maere, S.; Van De Peer, Y. Plants with double genomes might have had a better chance to survive the Cretaceous–Tertiary extinction event. Proc. Natl. Acad. Sci. USA 2009, 106, 5737–5742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanneste, K.; Baele, G.; Maere, S.; Van de Peer, Y. Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous–Paleogene boundary. Genome Res. 2014, 24, 1334–1347. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Yang, X.; Guignard, G.; Deng, S.; Tian, N.; Jiang, Z. The fossil Gleicheniaceous ferns of China: Biodiversity, systematics, spore ultrastructure and evolution. Rev. Palaeobot. Palynol. 2009, 156, 139–156. [Google Scholar] [CrossRef]
- Clapham, M.E.; Renne, P.R. Flood basalts and mass extinctions. Annu. Rev. Earth Planet. Sci. 2019, 47, 275–303. [Google Scholar] [CrossRef]
- Sun, H.; Xiao, Y.; Gao, Y.; Zhang, G.; Casey, J.F.; Shen, Y. Rapid enhancement of chemical weathering recorded by extremely light seawater lithium isotopes at the Permian–Triassic boundary. Proc. Natl. Acad. Sci. USA 2018, 115, 3782–3787. [Google Scholar] [CrossRef] [Green Version]
- Corsin, P. Paleobiogeography of the dipteridaceae and matoniaceae of the mesozoic. In Proceedings of the IV International Gondwana Symposium, Calcutta, India, 4 January 1977; pp. 51–70. [Google Scholar]
- Wang, Y. Fern ecological implications from the Lower Jurassic in western Hubei, China. Rev. Palaeobot. Palynol. 2002, 119, 125–141. [Google Scholar] [CrossRef]
- Pryer, K.M.; Schuettpelz, E.; Wolf, P.G.; Schneider, H.; Smith, A.R.; Cranfill, R. Phylogeny and evolution of ferns (monilophytes) with a focus on the early leptosporangiate divergences. Am. J. Bot. 2004, 91, 1582–1598. [Google Scholar] [CrossRef] [Green Version]
- Kramer, K.U.; Green, P.; Kubitzki, K. The Families and Genera of Vascular Plants. V. 1: Pteridophytes and Gymnosperms; Springer: Berlin/Heidelberg, Germany, 1990. [Google Scholar]
- Schuettpelz, E.; Korall, P.; Pryer, K.M. Plastid atpA data provide improved support for deep relationships among ferns. Taxon 2006, 55, 897–906. [Google Scholar] [CrossRef] [Green Version]
- Schneider, H. Vergleichende Wurzelanatomie der Farne; Shaker: Herzogenrath, Germany, 1996. [Google Scholar]
- Smith, A.R.; Pryer, K.M.; Schuettpelz, E.; Korall, P.; Schneider, H.; Wolf, P.G. A classification for extant ferns. Taxon 2006, 55, 705–731. [Google Scholar] [CrossRef]
- Bierhorst, D.W. Morphology of Vascular Plants; MacMillan Co.: New York, NY, USA, 1971. [Google Scholar]
- Tryon, A.F.; Lugardon, B. Spores of the Pteridophyta: Surface, Wall Structure, and Diversity Based on Electron Microscope Studies; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Holttum, R.E. A Revised Flora of Malaya: Ferns of Malaya; US Government Printing Office: Washington, DC, USA, 1953; Volume 2.
- Zhang, X.; Masahiro, K.; Hans, P.N. Dipteridaceae. In Flora of China; Wu, Z.Y., Raven, P.H., Hong, D.Y., Eds.; Science Press: Beijing, China; Missouri Botanical Garden Press: St. Louis, MO, USA, 2013; Volume 2–3 (Pteridophytes), pp. 116–117. [Google Scholar]
- Turland, N.J.; Wiersema, J.H.; Barrie, F.R.; Greuter, W.; Hawksworth, D.L.; Herendeen, P.S.; Knapp, S.; Kusber, W.-H.; Li, D.-Z.; Marhold, K. International Code of Nomenclature for Algae, Fungi, and Plants (Shenzhen Code) Adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017; Koeltz Botanical Books: Hessen, Germany, 2018. [Google Scholar]
- Skog, J.E. Biogeography of Mesozoic leptosporangiate ferns related to extant ferns. Brittonia 2001, 53, 236–269. [Google Scholar] [CrossRef]
- Taylor, E.L.; Taylor, T.N.; Krings, M. Paleobotany: The Biology and Evolution of Fossil Plants, 2nd ed.; Academic Press: Cambridge, MA, USA, 2009. [Google Scholar]
- Zhou, N.; Wang, Y.-D.; Li, L.-Q.; Zhang, X.-Q. Diversity variation and tempo-spatial distributions of the Dipteridaceae ferns in the Mesozoic of China. Palaeoworld 2016, 25, 263–286. [Google Scholar] [CrossRef]
- Choo, T.Y.; Escapa, I.H. Assessing the evolutionary history of the fern family Dipteridaceae (Gleicheniales) by incorporating both extant and extinct members in a combined phylogenetic study. Am. J. Bot. 2018, 105, 1315–1328. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, H. Fertile organs and in situ spores of a new dipteridaceous fern Hausmannia sinensis from the Jurassic of northern China. Proc. R. Soc. B Biol. Sci. 2010, 277, 311–320. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Li, L.; Guignard, G.; Dilcher, D.L.; Xie, X.; Tian, N.; Zhou, N.; Wang, Y. Fertile structures with in situ spores of a dipterid fern from the Triassic in southern China. J. Plant Res. 2015, 128, 445–457. [Google Scholar] [CrossRef]
- Guignard, G.; Wang, Y.; Ni, Q.; Tian, N.; Jiang, Z. A dipteridaceous fern with in situ spores from the Lower Jurassic in Hubei, China. Rev. Palaeobot. Palynol. 2009, 156, 104–115. [Google Scholar] [CrossRef]
- Chase, M.W.; Christenhusz, M.; Fay, M.; Byng, J.; Judd, W.S.; Soltis, D.; Mabberley, D.; Sennikov, A.; Soltis, P.S.; Stevens, P.F. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Klass, K.-D.; Zompro, O.; Kristensen, N.P.; Adis, J. Mantophasmatodea: A new insect order with extant members in the Afrotropics. Science 2002, 296, 1456–1459. [Google Scholar] [CrossRef] [Green Version]
- Shu, J.-P.; Shang, H.; Jin, D.; Wei, H.-J.; Zhou, X.-L.; Liu, H.-M.; Gu, Y.-F.; Wang, Y.; Wang, F.-G.; Shen, H. Re-establishment of species from synonymies based on DNA barcoding and phylogenetic analysis using Diplopterygium simulans (Gleicheniaceae) as an example. PLoS ONE 2017, 12, e0164604. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [Green Version]
- Douard, V.; Brunet, F.; Boussau, B.; Ahrens-Fath, I.; Vlaeminck-Guillem, V.; Haendler, B.; Laudet, V.; Guiguen, Y. The fate of the duplicated androgen receptor in fishes: A late neofunctionalization event? BMC Evol. Biol. 2008, 8, 336. [Google Scholar] [CrossRef] [Green Version]
- Dunn, C.W.; Howison, M.; Zapata, F. Agalma: An automated phylogenomics workflow. BMC Bioinform. 2013, 14, 330. [Google Scholar] [CrossRef] [Green Version]
- Breinholt, J.W.; Kawahara, A.Y. Phylotranscriptomics: Saturated third codon positions radically influence the estimation of trees based on next-gen data. Genome Biol. Evol. 2013, 5, 2082–2092. [Google Scholar] [CrossRef] [Green Version]
- Ran, J.-H.; Shen, T.-T.; Wang, M.-M.; Wang, X.-Q. Phylogenomics resolves the deep phylogeny of seed plants and indicates partial convergent or homoplastic evolution between Gnetales and angiosperms. Proc. R. Soc. B Biol. Sci. 2018, 285, 20181012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Mirarab, S.; Reaz, R.; Bayzid, M.S.; Zimmermann, T.; Swenson, M.S.; Warnow, T. ASTRAL: Genome-scale coalescent-based species tree estimation. Bioinformatics 2014, 30, i541–i548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, T.-K. Calculating bootstrap probabilities of phylogeny using multilocus sequence data. Mol. Biol. Evol. 2008, 25, 960–971. [Google Scholar] [CrossRef] [Green Version]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Bryant, D.; Moulton, V. Neighbor-net: An agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 2004, 21, 255–265. [Google Scholar] [CrossRef]
- Bek, J.; Pšenička, J. Senftenbergia plumosa (Artis) emend. and its spores from the Carboniferous of the Kladno and Pilsen basins, Bohemian Massif, and some related and synonymous taxa. Rev. Palaeobot. Palynol. 2001, 116, 213–232. [Google Scholar] [CrossRef]
- Axsmith, B.J.; Krings, M.; Taylor, T.N. A filmy fern from the Upper Triassic of North Carolina (USA). Am. J. Bot. 2001, 88, 1558–1567. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [Green Version]
- Tiley, G.P.; Barker, M.S.; Burleigh, J.G. Assessing the Performance of Ks Plots for Detecting Ancient Whole Genome Duplications. Genome Biol. Evol. 2018, 10, 2882–2898. [Google Scholar] [CrossRef] [PubMed]
- Conant, G.C.; Birchler, J.A.; Pires, J.C. Dosage, duplication, and diploidization: Clarifying the interplay of multiple models for duplicate gene evolution over time. Curr. Opin. Plant Biol. 2014, 19, 91–98. [Google Scholar] [CrossRef]
- Zwaenepoel, A.; Van de Peer, Y. Inference of Ancient Whole-Genome Duplications and the Evolution of Gene Duplication and Loss Rates. Mol. Biol. Evol. 2019, 36, 1384–1404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paterson, A.; Bowers, J.; Chapman, B. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. USA 2004, 101, 9903–9908. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Baniaga, A.E.; Sessa, E.B.; Scascitelli, M.; Graham, S.W.; Rieseberg, L.H.; Barker, M.S. Early genome duplications in conifers and other seed plants. Sci. Adv. 2015, 1, e1501084. [Google Scholar] [CrossRef] [Green Version]
- Maliet, O.; Hartig, F.; Morlon, H. A model with many small shifts for estimating species-specific diversification rates. Nat. Ecol. Evol. 2019, 3, 1086–1092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stadler, T. Species-specific diversification. Nat. Ecol. Evol. 2019, 3, 1003–1004. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shu, J.-P.; Wang, H.; Shen, H.; Wang, R.-J.; Fu, Q.; Wang, Y.-D.; Jiao, Y.-N.; Yan, Y.-H. Phylogenomic Analysis Reconstructed the Order Matoniales from Paleopolyploidy Veil. Plants 2022, 11, 1529. https://doi.org/10.3390/plants11121529
Shu J-P, Wang H, Shen H, Wang R-J, Fu Q, Wang Y-D, Jiao Y-N, Yan Y-H. Phylogenomic Analysis Reconstructed the Order Matoniales from Paleopolyploidy Veil. Plants. 2022; 11(12):1529. https://doi.org/10.3390/plants11121529
Chicago/Turabian StyleShu, Jiang-Ping, Hao Wang, Hui Shen, Rui-Jiang Wang, Qiang Fu, Yong-Dong Wang, Yuan-Nian Jiao, and Yue-Hong Yan. 2022. "Phylogenomic Analysis Reconstructed the Order Matoniales from Paleopolyploidy Veil" Plants 11, no. 12: 1529. https://doi.org/10.3390/plants11121529
APA StyleShu, J.-P., Wang, H., Shen, H., Wang, R.-J., Fu, Q., Wang, Y.-D., Jiao, Y.-N., & Yan, Y.-H. (2022). Phylogenomic Analysis Reconstructed the Order Matoniales from Paleopolyploidy Veil. Plants, 11(12), 1529. https://doi.org/10.3390/plants11121529





