Identification and Pathogenicity of Paramyrothecium Species Associated with Leaf Spot Disease in Northern Thailand
Abstract
:1. Introduction
2. Results
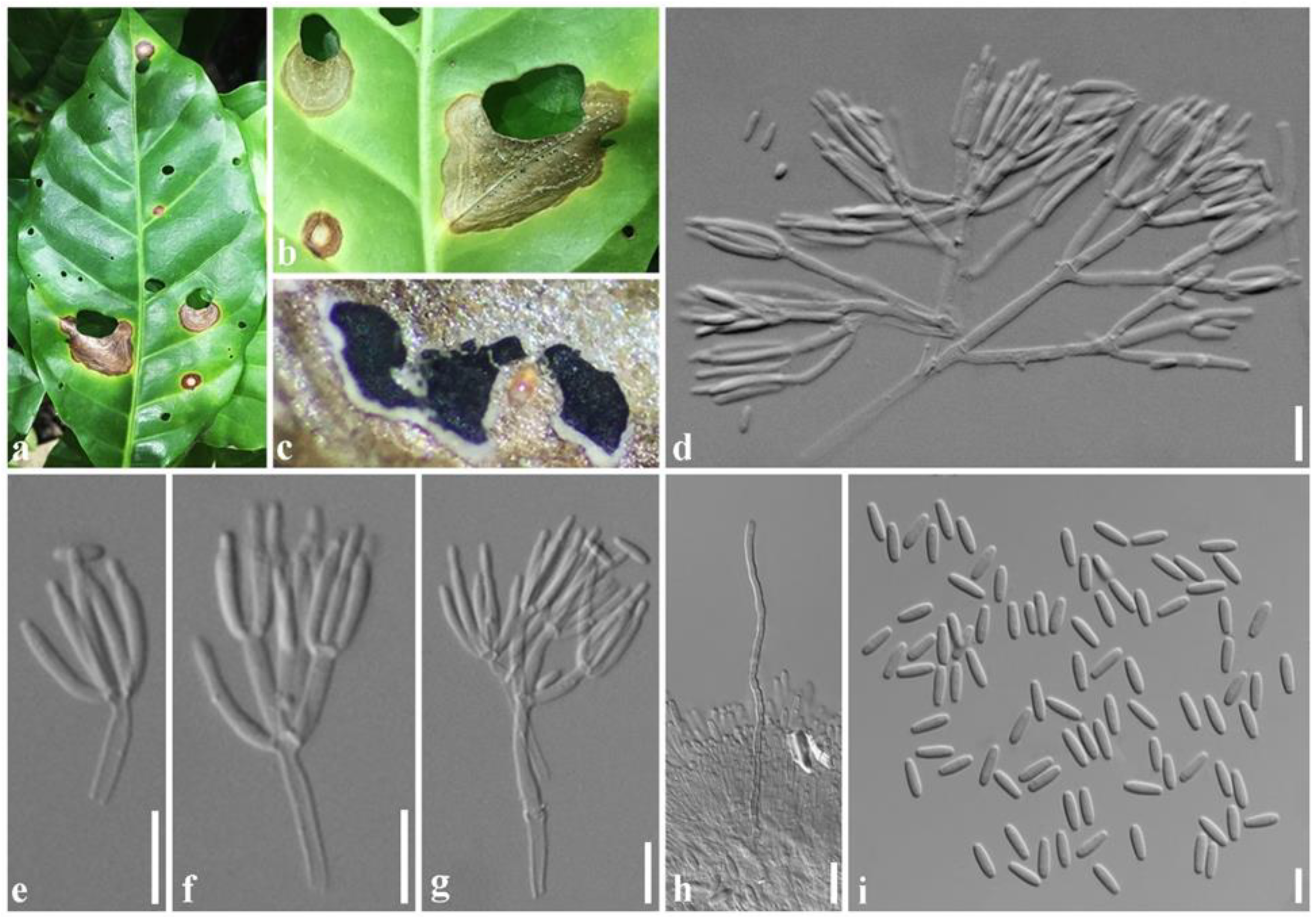
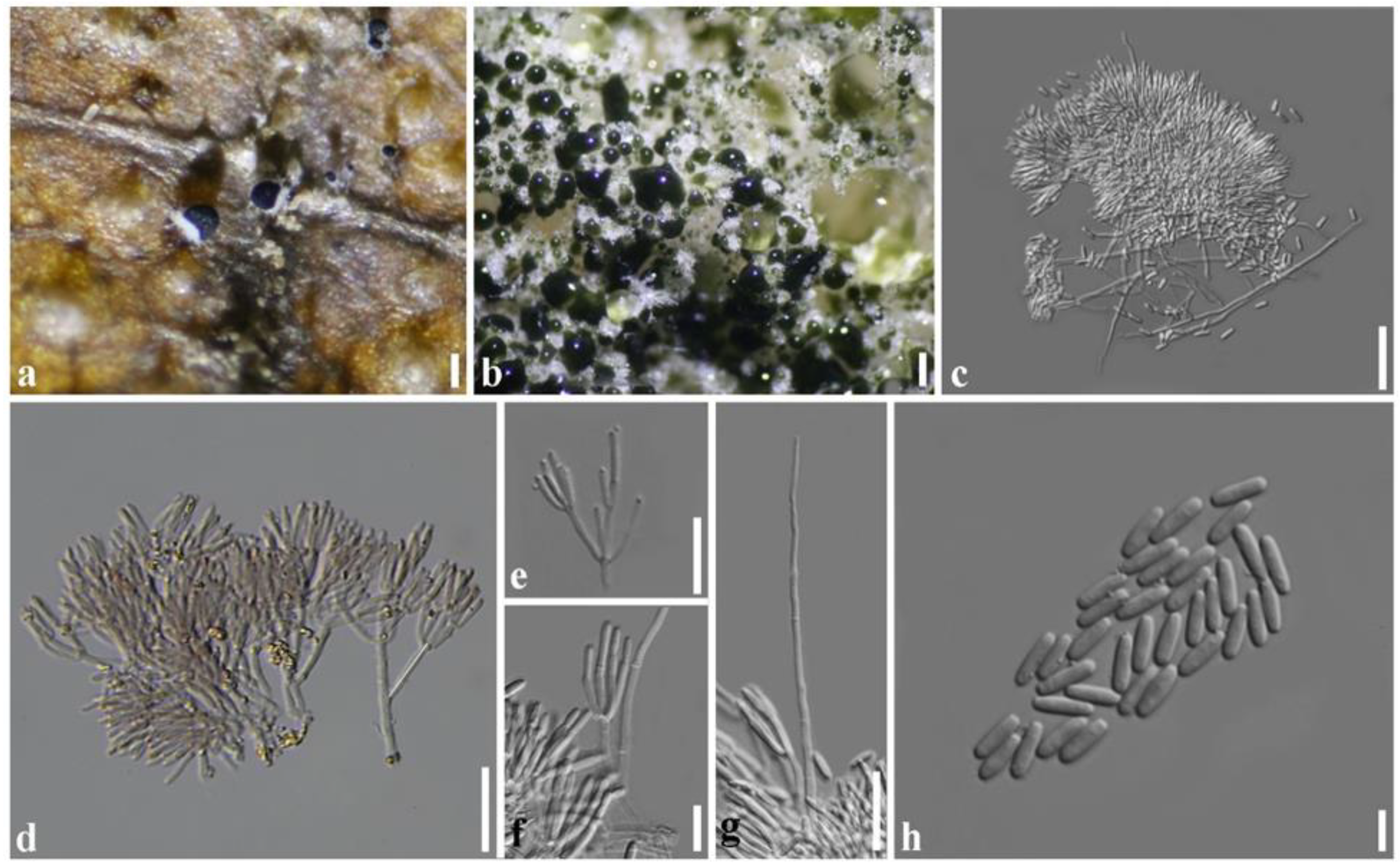
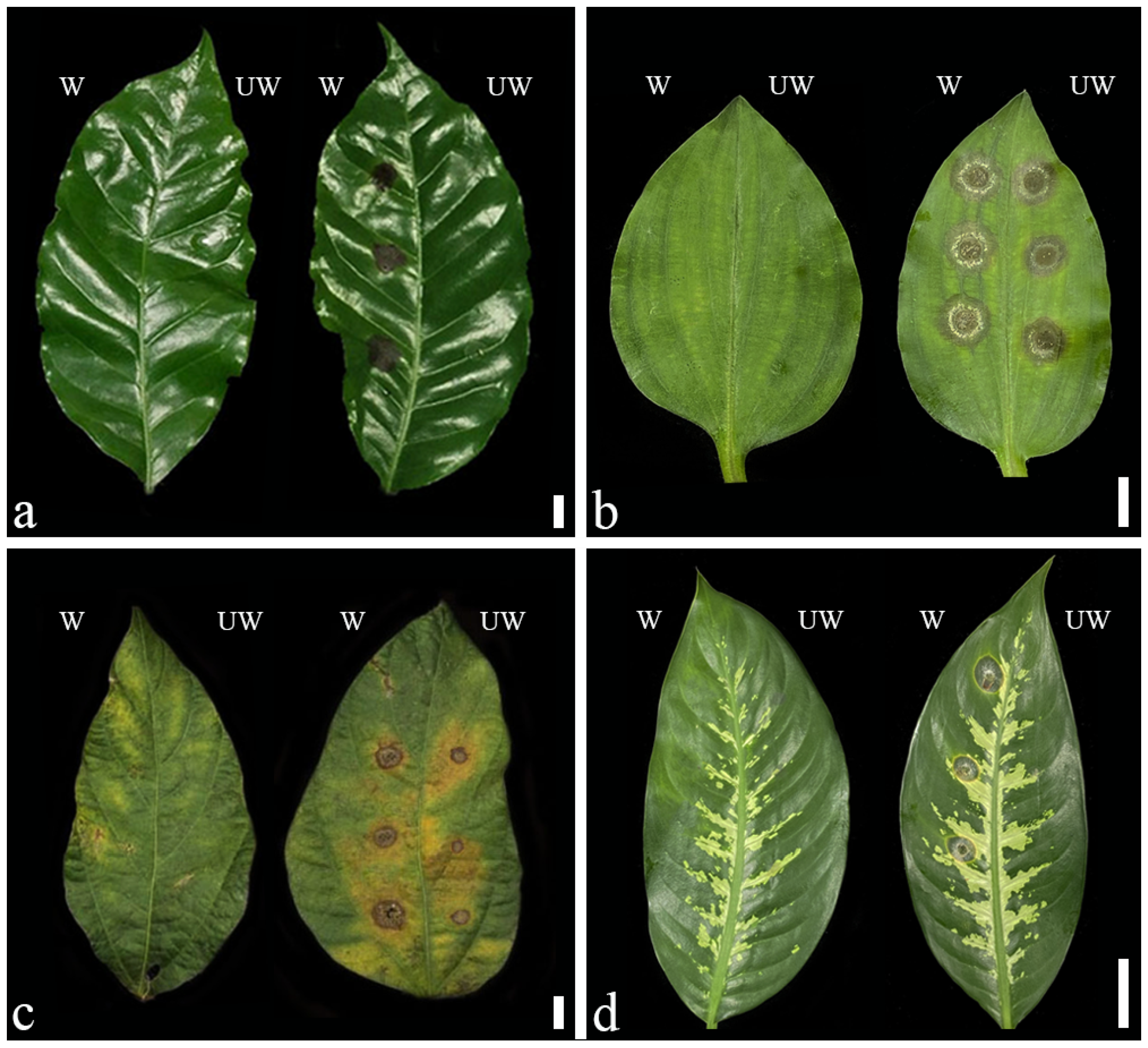
2.1. Symptoms
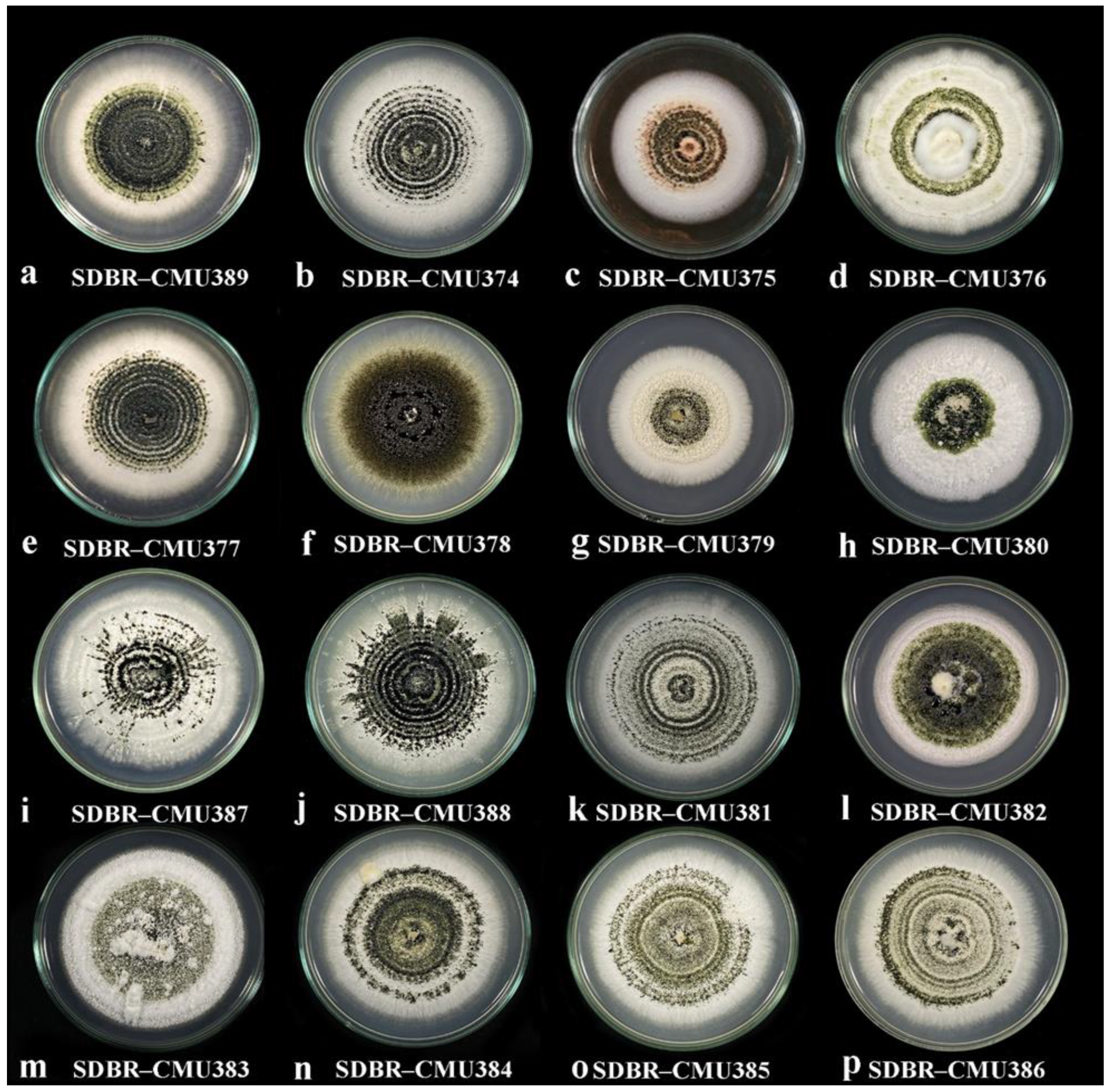
2.2. Culture Morphology
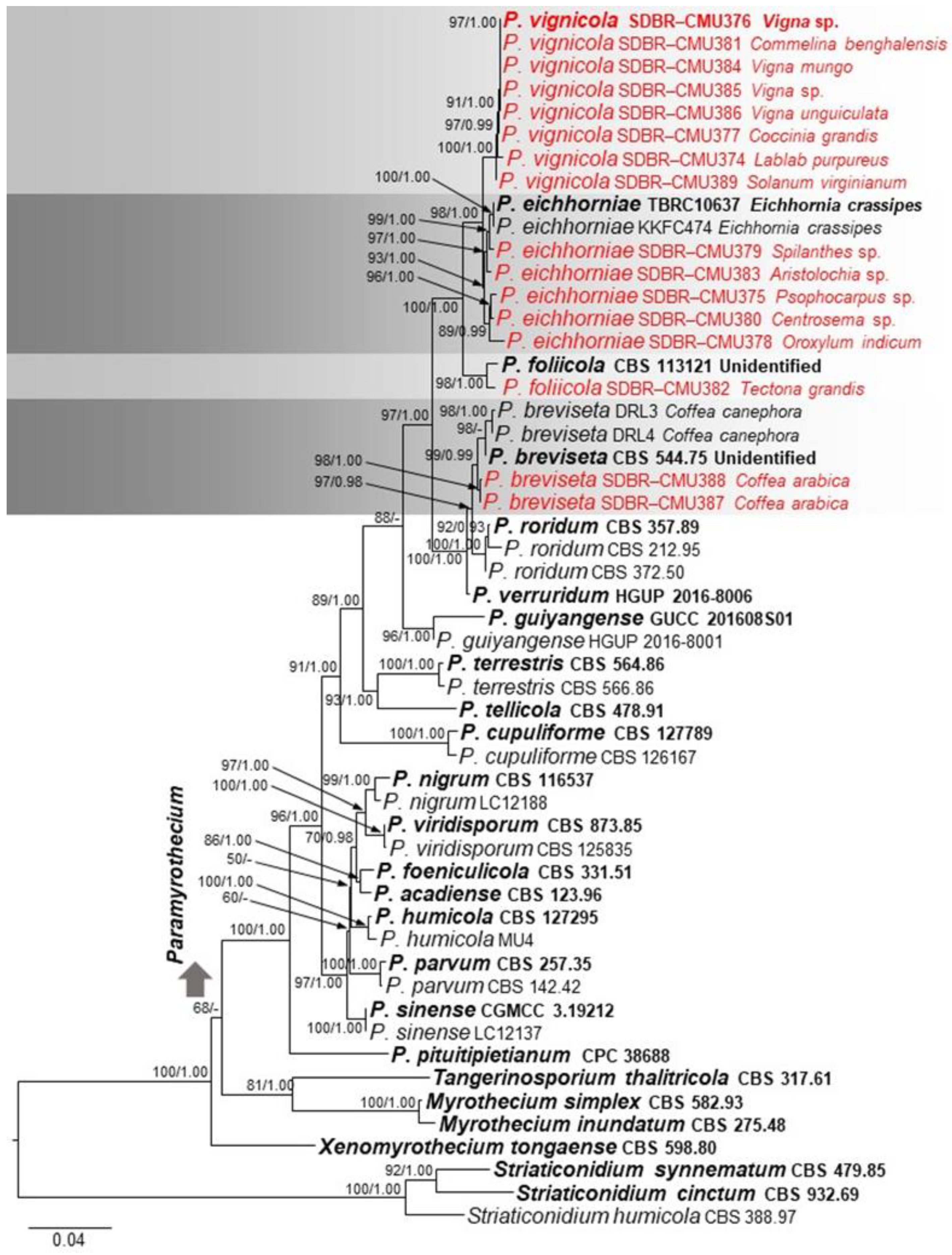
2.3. Phylogenetic Analysis
2.4. Taxonomy
2.5. Pathogenicity Test and Cross Pathogenicity
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Fungal Isolation and Taxonomic Description
4.3. DNA Extraction, Amplification, and Analyses
4.4. Pathogenicity Tests and Cross Pathogenicity
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ristaino, J.B.; Anderson, P.K.; Bebber, D.P.; Brauman, K.A.; Cunniffe, N.J.; Fedoroff, N.V.; Finegold, C.; Garrett, K.A.; Gilligan, C.A.; Jones, C.M.; et al. The persistent threat of emerging plant disease pandemics to global food security. Proc. Natl. Acad. Sci. USA 2021, 118, e2022239118. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Sarsaiya, S.; Wu, Q.; Lu, Y.; Shi, J. A review of plant leaf fungal diseases and its environment speciation. Bioengineered 2019, 10, 409–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iqbal, Z.; Khan, M.A.; Sharif, M.; Shah, J.H.; ur Rehman, M.H.; Javed, K. An automated detection and classification of citrus plant diseases using image processing techniques: A review. Comput. Electron. Agric. 2018, 153, 12–32. [Google Scholar] [CrossRef]
- Lombard, L.; Houbraken, J.; Decock, C.; Samson, R.A.; Meijer, M.; Réblová, M.; Groenewald, J.Z.; Crous, P.W. Generic hyper-diversity in Stachybotriaceae. Persoonia 2016, 36, 156–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soliman, M.S. Characterization of Paramyrothecium roridum (Basionym Myrothecium roridum) Causing Leaf Spot of Strawberry. J. Plant Prot. Res. 2020, 60, 141–149. [Google Scholar] [CrossRef]
- Farr, D.F.; Rossman, A.Y. Fungal Databases, U.S. National Fungus Collections, ARS, USDA. Available online: https://nt.ars-grin.gov/fungaldatabases/ (accessed on 25 March 2022).
- Matić, S.; Gilardi, G.; Gullino, M.L.; Garibaldi, A. Emergence of Leaf Spot Disease on Leafy Vegetable and Ornamental Crops Caused by Paramyrothecium and Albifimbria Species. Phytopathology 2019, 109, 1053–1061. [Google Scholar] [CrossRef]
- Haudenshield, J.S.; Miranda, C.; Hartman, G.L. First Report of Paramyrothecium roridum (Basionym Myrothecium roridum) Causing Myrothecium Leaf Spot on Soybean in Africa. Plant Dis. 2018, 102, 2638. [Google Scholar] [CrossRef]
- Chen, Z.D.; Li, P.L.; Chai, A.L.; Guo, W.T.; Shi, Y.X.; Xie, X.W.; Li, B.J. Crown canker caused by Paramyrothecium roridum on greenhouse muskmelon (Cucumis Melo) in China. Can. J. Plant Pathol. 2018, 40, 115–120. [Google Scholar] [CrossRef]
- Azizi, R.; Ghosta, Y.; Ahmadpour, A. New fungal canker pathogens of apple trees in Iran. J. Crop Prot. 2020, 9, 669–681. [Google Scholar]
- Rennberger, G.; Keinath, A.P. Stachybotriaceae on Cucurbits Demystified: Genetic Diversity and Pathogenicity of Ink Spot Pathogens. Plant Dis. 2020, 104, 2242–2251. [Google Scholar] [CrossRef]
- Aumentado, H.D.; Balendres, M.A. Identification of Paramyrothecium foliicola causing crater rot in eggplant and its potential hosts under controlled conditions. J. Phytopathol. 2022, 170, 148–157. [Google Scholar] [CrossRef]
- Huo, J.F.; Yao, Y.R.; Ben, H.Y.; Tian, T.; Yang, L.J.; Wang, Y.; Bai, P.H.; Hao, Y.J.; Wang, W.L. First Report of Paramyrothecium foliicola Causing Stem Canker of Cucumber (Cucumis Sativus) Seedlings in China. Plant Dis. 2021, 105, 1859. [Google Scholar] [CrossRef] [PubMed]
- Piyaboon, O.; Pawongrat, R.; Unartngam, J.; Chinawong, S.; Unartngam, A. Pathogenicity, host range and activities of a secondary metabolite and enzyme from Myrothecium roridum on water hyacinth from Thailand. Weed Biol. Manag. 2016, 16, 132–144. [Google Scholar] [CrossRef]
- Hassan, F.R.; Ghaffar, N.M.; Assaf, L.H.; Abdullah, S.K. Pathogenicity of endogenous isolate of Paramyrothecium (=Myrothecium) roridum (Tode) L. Lombard & Crous against the squash beetle Epilachna chrysomelina (F.). J. Plant Prot. Res. 2021, 61, 110–116. [Google Scholar] [CrossRef]
- Liu, H.X.; Liu, W.Z.; Chen, Y.C.; Sun, Z.H.; Tan, Y.Z.; Li, H.H.; Zhang, W.M. Cytotoxic trichothecene macrolides from the endophyte fungus Myrothecium roridum. J. Asian Nat. Prod. Res. 2016, 18, 684–689. [Google Scholar] [CrossRef]
- Basnet, B.B.; Liu, L.; Chen, B.; Suleimen, Y.M.; Yu, H.; Guo, S.; Bao, L.; Ren, J.; Liu, H. Four New Cytotoxic Arborinane–Type Triterpenes from the Endolichenic Fungus Myrothecium inundatum. Planta Med. 2019, 85, 701–707. [Google Scholar] [CrossRef]
- Elkhateeb, W.A.; Daba, G.M. Myrothecium as Promising Model for Biotechnological Applications, Potentials and Challenges. Biomed. J. Sci. Tech. Res. 2019, 16, 12126–12131. [Google Scholar] [CrossRef]
- Pinruan, U.; Unartngam, J.; Unartngam, A.; Piyaboon, O.; Sommai, S.; Khamsuntorn, P. Paramyrothecium eichhorniae sp. nov., Causing Leaf Blight Disease of Water Hyacinth from Thailand. Mycobiology 2022, 50, 12–19. [Google Scholar] [CrossRef]
- Seifert, K.A.; Louis–Seize, G.; Sampson, G. Myrothecium acadiense, a new hyphomycete isolated from the weed Tussilago farfara. Mycotaxon 2003, 87, 317–327. [Google Scholar]
- Krisai-Greilhuber, I.; Chen, Y.; Jabeen, S.; Madrid, H.; Marincowitz, S.; Kazaq, A.; Ševčiková, H.; Voglmayr, H.; Yazici, K.; Aptroot, A.; et al. Fungal Systematics and Evolution: FUSE 3. Sydowia 2017, 69, 229–264. [Google Scholar] [CrossRef]
- Crous, P.W.; Osieck, E.R.; Jurjevi, Ž.; Boers, J.; van Iperen, A.L.; Starink–Willemse, M.; Dima, B.; Balashov, S.; Bulgakov, T.S.; Johnston, P.R.; et al. Fungal Planet description sheets: 1284–1382. Persoonia 2021, 47, 178–374. [Google Scholar] [CrossRef]
- Crous, P.W.; Cowan, D.A.; Maggs–Kölling, G.; Yilmaz, N.; Larsson, E.; Angelini, C.; Brandrud, T.E.; Dearnaley, J.D.W.; Dima, B.; Dovana, F.; et al. Fungal Planet description sheets: 1112–1181. Persoonia 2020, 45, 251–409. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Cowan, D.A.; Maggs-Kölling, G.; Yilmaz, N.; Thangavel, R.; Wingfield, M.J.; Noordeloos, M.E.; Dima, B.; Brandrud, T.E.; Jansen, G.M.; et al. Fungal Planet description sheets: 1182–1283. Persoonia 2021, 46, 313–528. [Google Scholar] [CrossRef]
- Liang, J.; Li, G.; Zhou, S.; Zhao, M.; Cai, L. Myrothecium-like new species from turfgrasses and associated rhizosphere. MycoKeys 2019, 51, 29–53. [Google Scholar] [CrossRef] [PubMed]
- Potts, S.G.; Imperatriz-Fonseca, V.; Ngo, H.T.; Aizen, M.A.; Biesmeijer, J.C.; Breeze, T.D.; Dicks, L.V.; Garibaldi, L.A.; Hill, R.; Settele, J.; et al. Safeguarding pollinators and their values to human well-being. Nature 2016, 540, 220–229. [Google Scholar] [CrossRef]
- Wanasinghe, D.N.; Wijayawardene, N.N.; Xu, J.; Cheewangkoon, R.; Mortimer, P.E. Taxonomic novelties in Magnolia–associated pleosporalean fungi in the Kunming Botanical Gardens (Yunnan, China). PLoS ONE 2020, 15, e0235855. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Groenewald, J.Z.; Nakashima, C.; Nishikawa, J.; Shin, H.D.; Park, J.H.; Jama, A.N.; Groenewald, M.; Braun, U.; Crous, P.W. Species concepts in Cercospora: Spotting the weeds among the roses. Stud. Mycol. 2013, 75, 115–170. [Google Scholar] [CrossRef] [Green Version]
- O’Donnell, K.; Sarver, B.A.J.; Brandt, M.; Chang, D.C.; Noble-Wang, J.; Park, B.J.; Sutton, D.A.; Benjamin, L.; Lindsley, M.; Padhye, A.; et al. Phylogenetic Diversity and Microsphere Array-Based Genotyping of Human Pathogenic Fusaria, Including Isolates from the Multistate Contact Lens-Associated U.S. Keratitis Outbreaks of 2005 and 2006. J. Clin. Microbiol. 2007, 45, 2235–2248. [Google Scholar] [CrossRef] [Green Version]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML Web Servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES science gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [Green Version]
- Zhaxybayeva, O.; Gogarten, J.P. Bootstrap, Bayesian probability and maximum likelihood mapping: Exploring new tools for comparative genome analyses. BMC Genom. 2002, 3, 4. [Google Scholar] [CrossRef] [Green Version]
- Rannala, B.; Yang, Z. Probability distribution of molecular evolutionary trees: A new method of phylogenetic inference. J. Mol. Evol. 1996, 43, 304–311. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree. Version 1.4.3; Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, UK, 2016. [Google Scholar]



| Species | Host | Location | Conidiophores (µm) | Conidiogenous Cells (µm) | Conidia (µm) | Setae (µm) | References |
|---|---|---|---|---|---|---|---|
| Paramyrothecium acadiense | Tussilago farfara | Canada | 9–14 × 2–2.5 | – | 0–1-septate, 5.5–16.5 × 1.5–2.5 | – | [20] |
| P. breviseta | unknown | India | 6–9 × 2–4 | 6–11 × 1–2 | aseptate, 4–5 × 1–2 | 1–3-septate, 25–40 × 2–3 | [4] |
| P. cupuliforme | Soil | Namibia | 15–25 × 2–4 | 4–11 × 1–3 | aseptate, 6–8 × 1–2 | 1–3-septate, 45–90 × 2–3 | [4] |
| P. eichhorniae | Eichhornia crassipes | Thailand | 15–40 × 2–3 | (8–)11–17(–20) × 2–3 | aseptate,5– 6.5 × 1.5–2.5 | 1–3-septate, 40–120 × 2–3 | [19] |
| P. foeniculicola | Foeniculum vulgare | Netherlands | 7–17 × 2–3 | 6–16 × 1–2 | aseptate, 5–7 × 1–2 | – | [4] |
| P. foliicola | Decaying leaf | Brazil | 15–25 × 2–3 | 8–14 × 1–2 | aseptate, 5–6 × 1–2 | 1–3-septate, 60–100 × 2–3 | [4] |
| P. guiyangense | Soil | China | 10−60 × 1−3 | 8−18 × 1.6−2.7 | aseptate, 6.6−9.0 × 2−3 | 1−3-septate, 60−120 × 1−3 | [21] |
| P. humicola | Soil | USA | 12–22 × 2–3 | 8–13 × 1–3 | aseptate, 6–7 × 1–2 | 1–2-septate, 55–65 × 2–3 | [4] |
| P. lathyri | Lathyrus tuberosus | Russia | 5–10 × 2–3.5 | 5–10 × 2–3 | aseptate, (8–)9 (–10) × 2(–2.5) | 3–10-septate, up to 300 × 3–4 | [22] |
| P. nigrum | Soil | Spain | 25–45 × 2–4 | 8–13 × 1–2 | aseptate, 5–6 × 1–2 | 1–3-septate, 60–100 × 2–3 | [4] |
| P. parvum | Viola sp. | UK | 12–26 × 2–4 | 7–23 × 1–2 | aseptate, 4–5 × 1–2 | – | [4] |
| P. pituitipietianum | Grielum humifusum | South Africa | 20–35 × 3–4 | 20–35 × 3–4 | aseptate, (7–)9–10(–12) × (2–)2.5 | 7–10-septate, 100–300 × 4–5 | [23] |
| P. roridum | Gardenia sp. | Italy | 15–40 × 2–4 | 7–33 × 2–3 | aseptate, (5–)6.5–7.5(–8) × 2 | 1–3(–4)-septate, 60–100 × 2–6 | [4] |
| P. salvadorae | Salvadora persica | Namibia | 20–40 × 3–4 | 8–15 × 2–2.5 | aseptate, (8–)10–12 (–13) × 2–2.5 | 5–10- septate, 100–200 × 2.5–3 | [24] |
| P. sinense | Rhizosphere soils of Poa sp. | China | 20–30 × 2–3 | 7–16 × 1–3 | aseptate, 6–7 × 2–3 | 1–3-septate, 45–90 × 1–3 | [25] |
| P. tellicola | Soil | Turkey | 15–30 × 2–4 | 7–17 × 1–3 | aseptate, (7–)7.5–8.5 (–9) × 1–3 | 1–3-septate, 45–80 × 2–3 μm | [4] |
| P. terrestris | Soil | Turkey | 15–30 × 2–3 | 7–12 × 2–3 | aseptate, (7–)7.5–8.5 (–10) × 1–3 | 1–3-septate, 35–70 × 2–3 | [4] |
| P. verruridum | Soil | China | 20−40 × 1.5−2.5 | 12−20 × 1.7−2.7 | aseptate, 6.8−7.8 × 2−2.7 | 1−3-septate, 40−120 × 2−3 | [21] |
| P. vignicola | Vigna sp. | Thailand | 40–60 × 2–3 | 11–16 × 1–3 | aseptate, 5–7 × 1–3 µm | 3–8-septate, 80–155 × 2–3 | This study |
| P. viridisporum | Soil | Turkey | 15–35 × 2–3 | 6–12 × 3–5 | aseptate, 3–5 × 2 µm | 1–3-septate, 60–140 × 2–3 | [4] |
| Species | Isolates | Plant Hosts | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Original Host | Coffea arabica | Commelina benghalensis | Glycine max | Dieffenbachia seguine | ||||||
| w | uw | w | uw | w | uw | w | uw | |||
| P. vignicola | Vigna sp. | SDBR-CMU376 T | + | - | + | + | + | + | + | - |
| Lablab purpureus | SDBR-CMU374 | + | - | + | + | + | + | + | - | |
| Coccinia grandis | SDBR-CMU377 | + | - | + | + | + | + | + | - | |
| Commelina benghalensis | SDBR-CMU381 | + | - | + | + | + | + | + | - | |
| Vigna mungo | SDBR-CMU384 | + | - | + | + | + | + | + | - | |
| Vigna sp. | SDBR-CMU385 | + | - | + | + | + | + | + | - | |
| Vigna unguiculata | SDBR-CMU386 | + | - | + | + | + | + | + | - | |
| Solanum virginianum | SDBR-CMU389 | + | - | + | + | + | + | + | - | |
| P. brevista | Coffea arabica | SDBR-CMU387 | + | - | + | + | + | + | + | - |
| Coffea arabica | SDBR-CMU388 | + | - | + | + | + | + | + | - | |
| P. eichhorniae | Psophocarpus sp. | SDBR-CMU375 | + | - | + | + | + | + | + | - |
| Oroxylum indicum | SDBR-CMU378 | + | - | + | + | + | + | + | - | |
| Spilanthes sp. | SDBR-CMU379 | + | - | + | + | + | + | + | - | |
| Centrosema sp. | SDBR-CMU380 | + | - | + | + | + | + | + | - | |
| Aristolochia sp. | SDBR-CMU383 | + | - | - | - | + | + | + | - | |
| P. foliicola | Tectona grandis | SDBR-CMU382 | + | - | + | + | + | + | + | - |
| Gene Regions | Primers | Sequence (5′→3′) | Length (bp) | References |
|---|---|---|---|---|
| ITS | ITS5 ITS4 | GGA AGT AAA AGT CGT AAC AAG G TCC GCT TAT TGA TAT GC | ca. 600 | [28] |
| cmdA | CAL–228F CAL–737R CAL2Rd | GAG TTC AAG GAG GCC TTC TCC C CAT CTT TCT GGC CAT GG TGR TCN GCC TCD CGG ATC ATC TC | CAL–228F–CAL–737R: 470–570 CAL–228F–CAL2Rd: 680–745 | [29,30] |
| rpb2 | RPB2–5F RPB2–7cR | GAY GAY MGW GAT CAY TTY GG CCC ATR GCT TGY TTR CCC AT | ca. 1000 | [31] |
| tub2 | Bt2a Bt2b | GGT AAC CAA ATC GGT GCT TTC ACC CTC AGT GTA GTG ACC CTT GGC | ca. 320 | [32] |
| Species | Isolate No. | Substrate | Location | GenBank Accession Numbers | |||
|---|---|---|---|---|---|---|---|
| ITS | cmdA | rpb2 | tub2 | ||||
| Myrothecium inundatum | CBS 275.48 T | On decaying pileus of Russula nigricans | England | KU846452 | KU846435 | – | KU846533 |
| M. simplex | CBS 582.93 T | On decaying agaric | Japan | KU846456 | KU846439 | – | KU846537 |
| Paramyrothecium acadiense | CBS 123.96 T = DAOMC 221473 = UAMH 7653 | On leaves of Tussilago farfara | Canada | KU846288 | – | KU846350 | KU846405 |
| P. breviseta | CBS 544.75 T | unknown | India | KU846289 | KU846262 | KU846351 | KU846406 |
| DRL3 | On leaves of Coffea canephora | China | MT853067 | MT897897 | – | MT897899 | |
| DRL4 | On leaves of C. canephora | China | MT853068 | MT897898 | – | MT897900 | |
| SDBR-CMU387 | On living leaf of C. arabica | Thailand | MZ373251 | OM810407 | ON033773 | OM982450 | |
| SDBR-CMU388 | On living leaf of C. arabica | Thailand | MZ373252 | OM810408 | ON033774 | OM982451 | |
| P. cupuliforme | CBS 127789 T | On surface soil in desert | Namibia | KU846291 | KU846264 | KU846353 | KU846408 |
| CBS 126167 | On surface soil in desert | Namibia | KU846290 | KU846263 | KU846352 | KU846407 | |
| P. eichhorniae | TBRC 10637 T | On leaf of Eichhornia crassipes | Thailand | MT973996 | MT975319 | MT975317 | MT977540 |
| KKFC 474 | On leaf of E.crassipes | Thailand | MT973995 | MT975318 | MT977541 | MT975316 | |
| SDBR-CMU375 | On living leaf of Psophocarpus sp. | Thailand | MZ373241 | OM810411 | ON033781 | ON033770 | |
| SDBR-CMU378 | On living leaf of unidentified plant | Thailand | MZ373246 | OM810414 | ON033782 | ON033772 | |
| SDBR-CMU379 | On living leaf of unidentified plant | Thailand | MZ373247 | OM810415 | ON033783 | ON033768 | |
| SDBR-CMU380 | On living leaf of Centrosema sp. | Thailand | MZ373250 | OM810416 | ON033784 | ON033771 | |
| SDBR-CMU383 | On living leaf of Aristolochia sp. | Thailand | MZ373255 | OM810418 | ON033785 | ON033769 | |
| P. foeniculicola | CBS 331.51 T = IMI 140051 | On leaf sheath Foeniculum vulgare | The Netherlands | KU846292 | – | KU846354 | KU846409 |
| P. foliicola | CBS 113121 T = INIFAT C02/104 T | On rotten leaf of unknown host | Brazil | KU846294 | KU846266 | – | KU846411 |
| SDBR-CMU382 | On decaying leaf of Tectona grandis | Thailand | MZ373254 | – | ON033775 | OM982452 | |
| P. guiyangense | GUCC 201608S01 T | From soil | China | KY126418 | KY196193 | – | KY196201 |
| HGUP 2016–8001 | From soil | China | KY126417 | KY196192 | – | KY196200 | |
| P. humicola | CBS 127295 T | from tallgrass prairie soil | USA | KU846295 | – | KU846356 | KU846412 |
| MU4 | On leaf of Citrullus lanatus | USA | MN227389 | MN593629 | MN397959 | MN398054 | |
| P. nigrum | CBS 116537 T = AR 3783 | From soil | Spain | KU846296 | KU846267 | KU846357 | KU846413 |
| LC12188 | Rhizosphere soils of Poa sp. | China | MK478871 | MK500252 | MK500261 | MK500269 | |
| P. parvum | CBS 257.35 T = IMI 140049 | On Viola sp. | UK | KU846298 | – | KU846359 | KU846415 |
| CBS 142.42 = IMI 155923 = MUCL 7582 | From dune sand | France | KU846297 | KU846268 | KU846358 | KU846414 | |
| P. pituitipietianum | CPC38688 T | On stems of Grielum humifusum | South Africa | MW175358 | MW173100 | – | MW173139 |
| P. roridum | CBS 357.89 T | On Gardenia sp. | Italy | KU846300 | KU846270 | KU846361 | KU846417 |
| CBS 212.95 | From water | The Netherlands | KU846299 | KU846269 | KU846360 | KU846416 | |
| CBS 372.50 = IMI 140050 | On twig of Coffea sp. | Colombia | KU846301 | KU846271 | KU846362 | KU846418 | |
| P. sinense | CGMCC3.19212 T = LC12136 | Rhizosphere soils of Poa sp. | China | MH793296 | MH885437 | MH818824 | MH793313 |
| LC12137 | Rhizosphere soils of Poa sp. | China | MH793295 | MH885436 | MH818822 | MH793312 | |
| P. tellicola | CBS 478.91 T | From soil | Turkey | KU846302 | KU846272 | KU846363 | KU846419 |
| P. terrestris | CBS 564.86 T | From soil under Lycopersicon esculentum | Turkey | KU846303 | KU846273 | KU846364 | KU846420 |
| CBS 566.86 | From soil beneath Helianthus annuus | Turkey | KU846305 | KU846275 | KU846366 | KU846422 | |
| P. verruridum | HGUP 2016–8006 T | From soil | China | KY126422 | KY196197 | – | KY196205 |
| P. vignicola | SDBR-CMU389 | On living leaf of Solanum virginianum | Thailand | MZ373239 | OM810409 | ON033776 | ON009013 |
| SDBR-CMU374 | On living leaf of Lablab purpureus | Thailand | MZ373240 | OM810410 | ON033777 | ON009014 | |
| SDBR-CMU376T | On living leaf of Vigna sp. | Thailand | MZ373242 | OM810412 | ON033778 | ON009015 | |
| SDBR-CMU377 | On living leaf of Coccinia grandis | Thailand | MZ373244 | OM810413 | ON033779 | ON009016 | |
| SDBR-CMU381 | On living leaf of Commelina benghalensis | Thailand | MZ373253 | OM810417 | ON033780 | ON009017 | |
| SDBR-CMU384 | On living leaf of Vigna mungo | Thailand | MZ373256 | OM810419 | ON033786 | – | |
| SDBR-CMU385 | On living leaf of Vigna sp. | Thailand | MZ373257 | OM810420 | ON033787 | ON009018 | |
| SDBR-CMU386 | On living leaf of V. unguiculata | Thailand | MZ373258 | OM810421 | ON033788 | ON009019 | |
| P. viridisporum | CBS 873.85 T | From soil | Turkey | KU846308 | KU846278 | KU846369 | KU846425 |
| CBS 125835 | Rhizosphere soils of bunchgrass | USA | KU846310 | KU846280 | KU846371 | KU846427 | |
| Striaticonidium cinctum | CBS 932.69 T | From agricultural soil | The Netherlands | KU847239 | KU847216 | KU847290 | KU847329 |
| S. humicola | CBS 388.97 | From soil in tropical forest | Papua New Guinea | KU847241 | KU847217 | KU847291 | KU847331 |
| S. synnematum | CBS 479.85 T | From leaf of unknown palm | Japan | KU847242 | KU847218 | KU847292 | KU847332 |
| Tangerinosporium thalictricola | CBS 317.61 T = IMI 034815 | On Thalictrum flavum | UK | KU847243 | KU847219 | – | KU847333 |
| Xenomyrothecium tongaense | CBS 598.80 T | On dead thallus of Halimeda sp. | Tonga | KU847246 | KU847221 | KU847295 | KU847336 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Withee, P.; Haituk, S.; Senwanna, C.; Karunarathna, A.; Tamakaew, N.; Pakdeeniti, P.; Suwannarach, N.; Kumla, J.; Suttiprapan, P.; Taylor, P.W.J.; et al. Identification and Pathogenicity of Paramyrothecium Species Associated with Leaf Spot Disease in Northern Thailand. Plants 2022, 11, 1445. https://doi.org/10.3390/plants11111445
Withee P, Haituk S, Senwanna C, Karunarathna A, Tamakaew N, Pakdeeniti P, Suwannarach N, Kumla J, Suttiprapan P, Taylor PWJ, et al. Identification and Pathogenicity of Paramyrothecium Species Associated with Leaf Spot Disease in Northern Thailand. Plants. 2022; 11(11):1445. https://doi.org/10.3390/plants11111445
Chicago/Turabian StyleWithee, Patchareeya, Sukanya Haituk, Chanokned Senwanna, Anuruddha Karunarathna, Nisachon Tamakaew, Parichad Pakdeeniti, Nakarin Suwannarach, Jaturong Kumla, Piyawan Suttiprapan, Paul W. J. Taylor, and et al. 2022. "Identification and Pathogenicity of Paramyrothecium Species Associated with Leaf Spot Disease in Northern Thailand" Plants 11, no. 11: 1445. https://doi.org/10.3390/plants11111445
APA StyleWithee, P., Haituk, S., Senwanna, C., Karunarathna, A., Tamakaew, N., Pakdeeniti, P., Suwannarach, N., Kumla, J., Suttiprapan, P., Taylor, P. W. J., Samarakoon, M. C., & Cheewangkoon, R. (2022). Identification and Pathogenicity of Paramyrothecium Species Associated with Leaf Spot Disease in Northern Thailand. Plants, 11(11), 1445. https://doi.org/10.3390/plants11111445









