Corn Starch: Quality and Quantity Improvement for Industrial Uses
Abstract
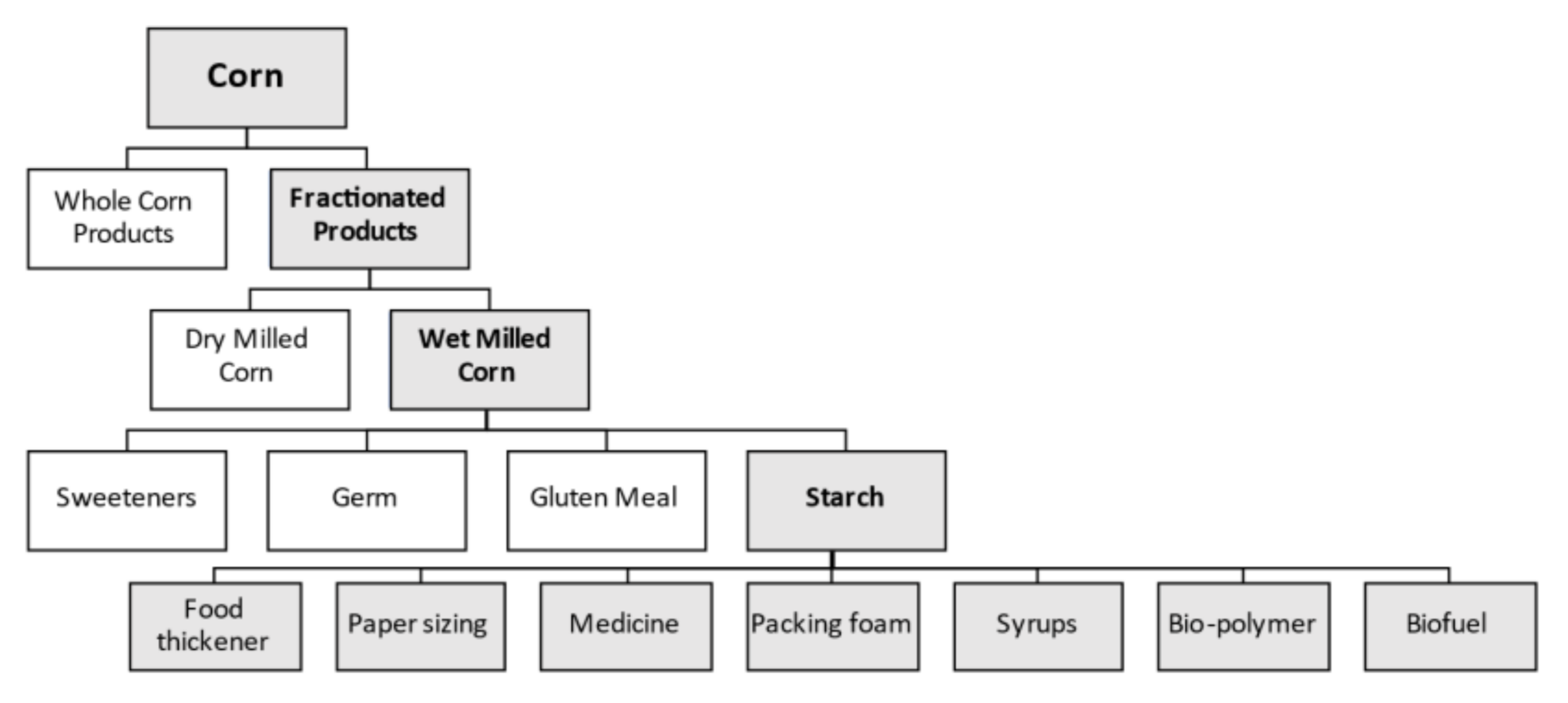
:1. Introduction
2. Wet Milling Process
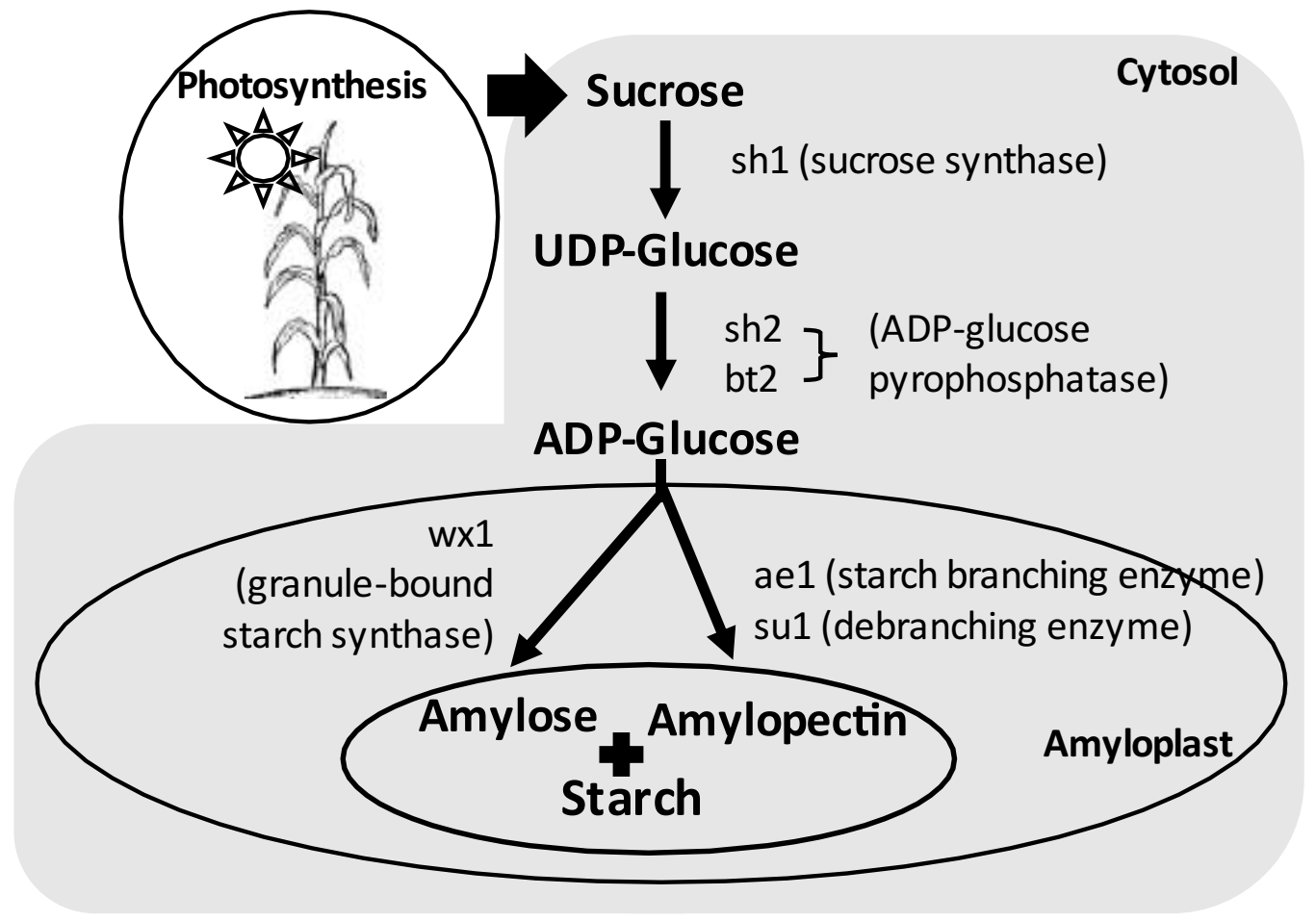
3. Starch Biosynthesis
4. Starch Quality Improvement: High Amylopectin (Waxy Corn and Glutinous Corn)
5. Starch Quality Improvement: High Amylose (Amylomaize)
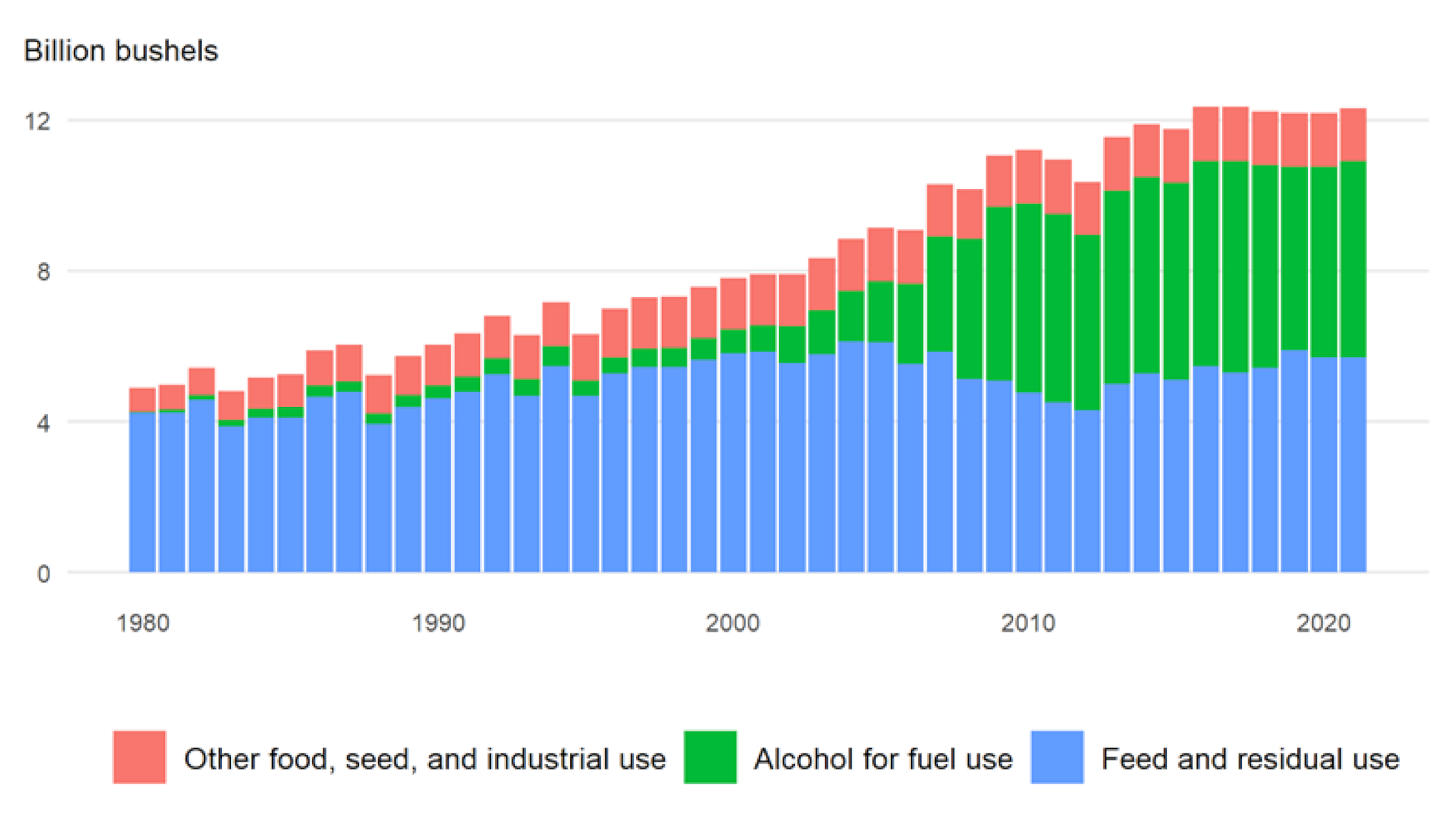
6. Starch Quantity Improvement
7. Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pollak, L.M.; Scott, M.P. Breeding for grain quality traits. Maydica 2005, 50, 247–257. [Google Scholar]
- Burrell, M.M. Starch: The need for improved quality or quantity—An overview. J. Exp. Bot. 2003, 54, 451–456. [Google Scholar] [CrossRef] [Green Version]
- Kaur, B.; Ariffin, F.; Bhat, R.; Karim, A.A. Progress in starch modification in the last decade. Food Hydrocoll. 2012, 26, 398–404. [Google Scholar] [CrossRef]
- Pajic, Z.; Radosavljevic, M.; Filipovic, M.; Todorovic, G.; Srdic, J.; Pavlov, M. Breeding of specialty maize for industrial purposes. Genetika 2010, 42, 57–66. [Google Scholar] [CrossRef]
- Zhang, R.; Ma, S.; Li, L.; Zhang, M.; Tian, S.; Wang, D.; Liu, K.; Liu, H.; Zhu, W.; Wang, X. Comprehensive utilization of corn starch processing by-products—A review. Grain Oil Sci. Technol. 2021, 4, 89–107. [Google Scholar] [CrossRef]
- Somavat, P.; Liu, W.; Singh, V. Wet milling characteristics of corn mutants using modified processes and improving starch yields from high amylose corn. Food Bioprod. Process. 2021, 126, 104–112. [Google Scholar] [CrossRef]
- Ramírez, E.C.; Johnston, D.B.; McAloon, A.J.; Singh, V. Enzymatic corn wet milling: Engineering process and cost model. Biotechnol. Biofuels 2009, 2, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Yu, X.; Wang, Y.; Fang, G.; Liu, Y. A Cleaner approach for corn starch production by ultrasound-assisted laboratory scale wet-milling. Food Sci. Technol. Res. 2020, 4, 469–478. [Google Scholar] [CrossRef]
- Jeon, J.S.; Ryoo, N.; Hahn, T.R.; Walia, H.; Nakamura, Y. Starch biosynthesis in cereal endosperm. Plant Physiol. Biochem. 2010, 48, 383–392. [Google Scholar] [CrossRef] [PubMed]
- James, M.G.; Denyer, K.; Myers, A.M. Starch synthesis in the cereal endosperm. Curr. Opin. Plant Biol. 2003, 6, 215–222. [Google Scholar] [CrossRef]
- Tetlow, I.J. Starch biosynthesis in developing seeds. Seed Sci. Res. 2011, 21, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Nelson, O.; Pan, D. Starch synthesis in maize endosperms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1995, 46, 475–496. [Google Scholar] [CrossRef]
- Tayade, R.; Kulkarni, K.P.; Jo, H.; Song, J.T.; Lee, J.-D. Insight into the prospects for the improvement of seed starch in legume—A Review. Front. Plant Sci. 2019, 10, 1213. [Google Scholar] [CrossRef]
- James, M.G.; Robertson, D.S.; Myers, A.M. Characterization of the maize gene sugary1, a determinant of starch composition in kernels. Plant Cell 1995, 7, 417–429. [Google Scholar]
- Zheng, H.; Wang, H.; Yang, H.; Wu, J.; Shi, B.; Cai, R.; Xu, Y.; Wu, A.; Luo, L. Genetic diversity and molecular evolution of Chinese waxy maize germplasm. PLoS ONE 2013, 8, e66606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Huang, Y.; Huang, R.; Wu, Y.; Wang, W. The genetic architecture of amylose biosyntheses in maize kernel. Plant Biotechnol. J. 2018, 16, 688–695. [Google Scholar] [CrossRef]
- Liu, J.; Rong, T.; Li, W. Mutation loci and intragenic selection marker of the granule-bound starch synthase gene in waxy maize. Mol. Breed. 2007, 20, 93–102. [Google Scholar] [CrossRef]
- Qi, X.; Wu, H.; Jiang, H.; Zhu, J.; Huang, C.; Zhang, X.; Liu, C.; Cheng, B. Conversion of a normal maize hybrid into a waxy version using in vivo CRISPR/Cas9 targeted mutation activity. Crop J. 2020, 8, 440–448. [Google Scholar] [CrossRef]
- Gao, H.; Gadlage, M.J.; Lafitte, H.R.; Lenderts, B.; Yang, M.; Schroder, M.; Farrell, J.; Snopek, K.; Peterson, D.; Feigenbutz, L.; et al. Superior field performance of waxy corn engineered using CRISPR-Cas9. Nat. Biotechnol. 2020, 38, 579–581. [Google Scholar] [CrossRef]
- Raigond, P.; Ezekiel, R.; Raigond, B. Resistant starch in food: A review. J. Sci. Food Agric. 2015, 95, 1968–1978. [Google Scholar] [CrossRef]
- Li, L.; Jiang, H.; Campbell, M.; Blanco, M.; Jane, J. Characterization of maize amylose-extender (ae) mutant starches. Part I: Relationship between resistant starch contents and molecular structures. Carbohydr. Polym. 2008, 74, 396–404. [Google Scholar] [CrossRef]
- Cai, C.; Lin, L.; Man, J.; Zhao, L.; Wang, Z.; Wei, C. Different structural properties of high-amylose maize starch fractions varying in granule size. J. Agric. Food Chem. 2014, 62, 11711–11721. [Google Scholar] [CrossRef]
- Wang, J.; Hu, P.; Chen, Z.; Liu, Q.; Wei, C. Progress in high-amylose cereal crops through inactivation of starch branching enzymes. Front. Plant Sci. 2017, 8, 469. [Google Scholar] [CrossRef] [Green Version]
- He, W.; Liu, X.; Lin, L.; Xu, A.; Hao, D.; Wei, C. The defective effect of starch branching enzyme IIb from weak to strong induces the formation of biphasic starch granules in amylose-extender maize endosperm. Plant Mol. Biol. 2020, 103, 355–371. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.D.; Gao, X.C.; Li, Z.W.; Xu, L.C.; Li, Y.B.; Zhang, R.H.; Xu, J.Q.; Guo, D.W. The effect of amylose on kernel phenotypic characteristics, starch-related gene expression and amylose inheritance in naturally mutated high-amylose maize. J. Integr. Agric. 2020, 19, 1554–1564. [Google Scholar] [CrossRef]
- Vineyard, M.L.; Bear, R.P.; MacMasters, M.M.; Deatherage, W.L. Development of “Amylomaize”—Corn hybrids with high amylose starch: I. Genetic considerations. Agron. J. 1958, 50, 595–598. [Google Scholar] [CrossRef]
- Campbell, M.R.; Jane, J.L.; Pollak, L.; Blanco, M.; O'Brien, A. Registration of maize germplasm line GEMS-0067. J. Plant Regist. 2007, 1, 60–61. [Google Scholar] [CrossRef]
- Wu, Y.; Campbell, M.; Yen, Y.; Wicks, Z.; Ibrahim, A. Genetic analysis of high amylose content in maize (Zea mays L.) using a triploid endosperm model. Euphytica 2009, 166, 155–164. [Google Scholar] [CrossRef]
- Han, N.; Li, W.; Xie, C.; Fu, F. Breeding potential of maize germplasm line GEMS-0067 for high amylose proportion. Preprints 2021, 2021050560. [Google Scholar] [CrossRef]
- Liu, D.; Wellner, N.; Parker, M.L.; Morris, V.J.; Cheng, F. In situ mapping of the effect of additional mutations on starch granule structure in amylose-extender (ae) maize kernels. Carbohydr. Polym. 2015, 118, 199–208. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, N.; Li, B.; Li, Z.; Xie, G.; Zhang, J. Reduced expression of starch branching enzyme IIa and IIb in maize endosperm by RNAi constructs greatly increases the amylose content in kernel with nearly normal morphology. Planta 2015, 241, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Vermerris, W.; Saballos, A.; Ejeta, G.; Mosier, N.; Ladisch, M.; Carpita, N. Molecular breeding to enhance ethanol production from corn and sorghum stover. Crop. Sci. 2007, 47, S142–S153. [Google Scholar] [CrossRef]
- Zeeman, S.C.; Kossmann, J.; Smith, A.M. Starch: Its metabolism, evolution, and biotechnological modification in plants. Annu. Rev. Plant Biol. 2010, 61, 209–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, N.; Zhang, S.; Zhao, Y.; Li, B.; Zhang, J. Over-expression of AGPase genes enhances seed weight and starch content in transgenic maize. Planta 2011, 233, 241–250. [Google Scholar] [CrossRef]
- Qi, X.; Li, S.; Zhu, Y.; Zhao, Q.; Zhu, D.; Yu, J. ZmDof3, a maize endosperm-specific Dof protein gene, regulates starch accumulation and aleurone development in maize endosperm. Plant Mol. Biol. 2016, 93, 7–20. [Google Scholar] [CrossRef]
- Jiang, L.; Yu, X.; Qi, X.; Yu, Q.; Deng, S.; Bai, B.; Li, N.; Zhang, A.; Zhu, C.; Liu, B.; et al. Multigene engineering of starch biosynthesis in maize endosperm increases the total starch content and the proportion of amylose. Transgenic Res. 2013, 22, e1133–e1142. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Xue, Y.; Guo, Z.; Li, W.; Tang, J. Genome-wide association study identifies candidate genes for starch content regulation in maize kernels. Front. Plant Sci. 2016, 7, 1046. [Google Scholar] [CrossRef] [PubMed]
- Bahaji, A.; Li, J.; Sánchez-López, A.M.; Baroja-Fernández, E.; Muñoz, F.J.; Ovecka, M.; Almagro, G.; Montero, M.; Ezquer, I.; Etxeberria, E.; et al. Starch biosynthesis, its regulation and biotechnological approaches to improve crop yields. Biotechnol. Adv. 2014, 32, 87–106. [Google Scholar] [CrossRef]
- Niu, L.; Ding, H.; Zhang, J.; Wang, W. Proteomic analysis of starch biosynthesis in maize seeds. Starch 2019, 71, 1800294. [Google Scholar] [CrossRef]
- Wang, T.; Wang, M.; Hu, S.; Xiao, Y.; Tong, H.; Pan, Q.; Xue, J.; Yan, J.; Li, J.; Yang, X. Genetic basis of maize kernel starch content revealed by high-density single nucleotide polymorphism markers in a recombinant inbred line population. BMC Plant Biol. 2015, 15, 288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varshney, R.K.; Bohra, A.; Yu, J.; Graner, A.; Zhang, Q.; Sorrells, M.E. Designing future crops: Genomics-assisted breeding comes of age. Trends Plant Sci. 2021, 26, 631–649. [Google Scholar] [CrossRef]
- Thitisaksakul, M.; Jimenez, R.C.; Arias, M.C.; Beckles, D.M. Effects of environmental factors on cereal starch biosynthesis and composition. J. Cereal Sci. 2012, 56, 67–80. [Google Scholar] [CrossRef]
- Beckles, D.M.; Thitisaksakul, M. How environmental stress affects starch composition and functionality in cereal endosperm. Starch-Starke 2014, 66, 58–71. [Google Scholar] [CrossRef] [Green Version]
- Van Dijk, A.D.J.; Kootstra, G.; Kruijer, W.; de Ridder, D. Machine learning in plant science and plant breeding. iScience 2020, 24, 101890. [Google Scholar] [CrossRef]
- Tong, H.; Nikoloski, Z. Machine learning approaches for crop improvement: Leveraging phenotypic and genotypic big data. J. Plant Physiol. 2021, 257, 153354. [Google Scholar] [CrossRef] [PubMed]

| Mutant | Gene | Enzyme | Starch Change | |
|---|---|---|---|---|
| Quality | Quantity | |||
| shrunken2 (sh2) | AGPLSU | AGP large subunit | Yes | |
| brittle2 (bt2) | AGPSSU | AGP small subunit | Yes | |
| waxy (wx1) | GBSSI | Granule-bound starch synthase | Low amylose | |
| amylose extender (ae) | SBEIIb | Starch branching enzyme | High amylose | |
| sugary1 (su1) | ISA | Isoamylase-type debranching enzyme | Granule number and form | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, J.-K.; Moon, Y.-S. Corn Starch: Quality and Quantity Improvement for Industrial Uses. Plants 2022, 11, 92. https://doi.org/10.3390/plants11010092
Yu J-K, Moon Y-S. Corn Starch: Quality and Quantity Improvement for Industrial Uses. Plants. 2022; 11(1):92. https://doi.org/10.3390/plants11010092
Chicago/Turabian StyleYu, Ju-Kyung, and Yong-Sun Moon. 2022. "Corn Starch: Quality and Quantity Improvement for Industrial Uses" Plants 11, no. 1: 92. https://doi.org/10.3390/plants11010092
APA StyleYu, J.-K., & Moon, Y.-S. (2022). Corn Starch: Quality and Quantity Improvement for Industrial Uses. Plants, 11(1), 92. https://doi.org/10.3390/plants11010092






