Resistance and Not Plant Fruit Traits Determine Root-Associated Bacterial Community Composition along a Domestication Gradient in Tomato
Abstract
:1. Introduction
2. Results
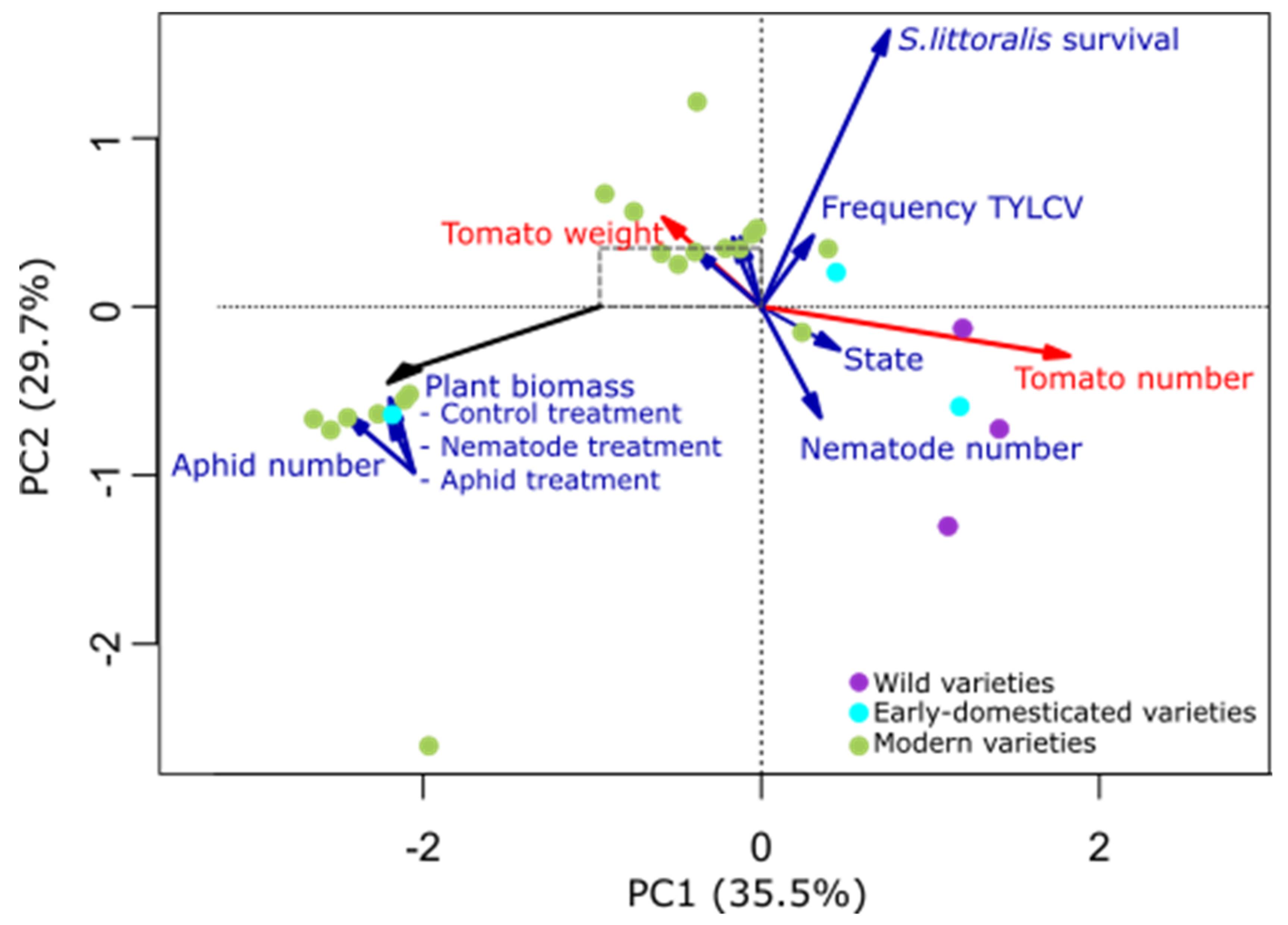
2.1. Plant Traits Affected by Domestication
2.2. Plant Domestication Influence Soil Bacterial Communities
2.3. Plant Traits and Soil Characteristics Influence Soil Bacterial Communities
3. Discussion
4. Materials and Methods
4.1. Field Experiment
4.2. Resistance to Pests Data
4.3. Soil Chemical Characterisation
4.4. Molecular Analyses of Soil Bacteria
4.5. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Plenchette, C.; Clermont-Dauphin, C.; Meynard, J.M.; Fortin, J.A. Managing arbuscular mycorrhizal fungi in cropping systems. Can. J. Plant Sci. 2005, 85, 31–40. [Google Scholar] [CrossRef]
- Lareen, A.; Burton, F.; Schäfer, P. Plant root-microbe communication in shaping root microbiomes. Plant Mol. Biol. 2016, 90, 575–587. [Google Scholar] [CrossRef] [Green Version]
- Pieterse, C.M.J.; Zamioudis, C.; Does, D.V.; Van Wees, S.C.M. Signalling networks involved in induced resistance. In Induced Resistance for Plant Defense; Walters, D.R., Newton, A.C., Lyon, G.D., Eds.; John Wiley & Sons, Ltd.: Chichester, UK, 2014; pp. 58–80. [Google Scholar]
- Bulgarelli, D.; Garrido-Oter, R.; Münch, P.C.; Weiman, A.; Dröge, J.; Pan, Y.; McHardy, A.C.; Schulze-Lefert, P. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 2015, 17, 392–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preece, C.; Peñuelas, J. Rhizodeposition under drought and consequences for soil communities and ecosystem resilience. Plant Soil 2016, 409, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Jaramillo, J.E.; Mendes, R.; Raaijmakers, J.M. Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol. Biol. 2016, 90, 635–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heinen, R.; Biere, A.; Harvey, J.A.; Bezemer, T.M. Effects of Soil Organisms on Aboveground Plant-Insect Interactions in the Field: Patterns, Mechanisms and the Role of Methodology. Front. Ecol. Evol. 2018, 6, 106. [Google Scholar] [CrossRef] [Green Version]
- Olsen, K.M.; Wendel, J.F. A bountiful harvest: Genomic insights into crop domestication phenotypes. Annu. Rev. Plant Biol. 2013, 64, 47–70. [Google Scholar] [CrossRef] [Green Version]
- Milla, R.; Osborne, C.P.; Turcotte, M.M.; Violle, C. Plant domestication through an ecological lens. Trends Ecol. Evol. 2015, 30, 463–469. [Google Scholar] [CrossRef]
- Gross, B.L.; Olsen, K.M. Genetic perspectives on crop domestication. Trends Plant Sci. 2010, 15, 529–537. [Google Scholar] [CrossRef] [Green Version]
- Ferrero, V.; Baeten, L.; Blanco-Sánchez, L.; Planelló, R.; Díaz-Pendón, J.A.; Rodríguez-Echeverría, S.; Haegeman, A.; de la Peña, E. Complex patterns in tolerance and resistance to pests and diseases underpin the domestication of tomato. New Phytol. 2019, 226, 254–266. [Google Scholar] [CrossRef]
- Szymański, J.; Bocobza, S.; Panda, S.; Sonawane, P.; Cárdenas, P.D.; Lashbrooke, J.; Kamble, A.; Shahaf, N.; Meir, S.; Bovy, A.; et al. Analysis of wild tomato introgression lines elucidates the genetic basis of transcriptome and metabolome variation underlying fruit traits and pathogen response. Nat. Genet. 2020, 52, 1111–1121. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, S.R.; Turcotte, M.M.; Poveda, K. Domestication impacts on plant-herbivore interactions: A meta-analysis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372, 20160034. [Google Scholar] [CrossRef]
- Mulatu, B.; Applebaum, S.W.; Coll, M. Effect of tomato leaf traits on the potato tuber moth and its predominant larval parasitoid: A mechanism for enemy-free space. Biol. Control 2006, 37, 231–236. [Google Scholar] [CrossRef]
- Chen, Y.H.; Gols, R.; Benrey, B. Crop domestication and its impact on naturally selected trophic interactions. Annu. Rev. Entomol. 2015, 60, 35–58. [Google Scholar] [CrossRef] [Green Version]
- Mutyambai, D.M.; Bruce, T.J.A.; Midega, C.A.O.; Woodcock, C.M.; Caulfield, J.C.; Van Den Berg, J.; Pickett, J.A.; Khan, Z.R. Responses of parasitoids to volatiles induced by Chilo partellus oviposition on teosinte, a wild ancestor of maize. J. Chem. Ecol. 2015, 41, 323–329. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Saona, C.; Vorsa, N.; Singh, A.P.; Johnson-Cicalese, J.; Szendrei, Z.; Mescher, M.C.; Frost, C.J. Tracing the history of plant traits under domestication in cranberries: Potential consequences on anti-herbivore defences. J. Exp. Bot. 2011, 62, 2633–2644. [Google Scholar] [CrossRef]
- Doebley, J.F.; Gaut, B.S.; Smith, B.D. The molecular genetics of crop domestication. Cell 2006, 127, 1309–1321. [Google Scholar] [CrossRef] [Green Version]
- Turcotte, M.M.; Araki, H.; Karp, D.S.; Poveda, K.; Whitehead, S.R. The eco-evolutionary impacts of domestication and agricultural practices on wild species. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372, 20160033. [Google Scholar] [CrossRef] [Green Version]
- Macfadyen, S.; Bohan, D.A. Crop domestication and the disruption of species interactions. Basic Appl. Ecol. 2010, 11, 116–125. [Google Scholar] [CrossRef]
- Coolen, S.; Proietti, S.; Hickman, R.; Davila Olivas, N.H.; Huang, P.-P.; Van Verk, M.C.; Van Pelt, J.A.; Wittenberg, A.H.J.; De Vos, M.; Prins, M.; et al. Transcriptome dynamics of Arabidopsis during sequential biotic and abiotic stresses. Plant J. 2016, 86, 249–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cordovez, V.; Dini-Andreote, F.; Carrión, V.J.; Raaijmakers, J.M. Ecology and evolution of plant microbiomes. Annu. Rev. Microbiol. 2019, 73, 69–88. [Google Scholar] [CrossRef]
- Carrillo, J.; Ingwell, L.L.; Li, X.; Kaplan, I. Domesticated tomatoes are more vulnerable to negative plant–soil feedbacks than their wild relatives. J. Ecol. 2019, 107, 1753–1766. [Google Scholar] [CrossRef]
- Leff, J.W.; Lynch, R.C.; Kane, N.C.; Fierer, N. Plant domestication and the assembly of bacterial and fungal communities associated with strains of the common sunflower, Helianthus annuus. New Phytol. 2017, 214, 412–423. [Google Scholar] [CrossRef]
- Zhang, C.; Tanabe, K.; Tamura, F.; Itai, A.; Wang, S. Partitioning of (13)C-photosynthate from spur leaves during fruit growth of three Japanese pear (Pyrus pyrifolia) cultivars differing in maturation date. Ann. Bot. 2005, 95, 685–693. [Google Scholar] [CrossRef] [Green Version]
- Wubs, A.M.; Ma, Y.; Heuvelink, E.; Marcelis, L.F.M. Genetic differences in fruit-set patterns are determined by differences in fruit sink strength and a source: Sink threshold for fruit set. Ann. Bot. 2009, 104, 957–964. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Jaramillo, J.E.; Carrión, V.J.; de Hollander, M.; Raaijmakers, J.M. The wild side of plant microbiomes. Microbiome 2018, 6, 143. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Jaramillo, J.E.; Carrión, V.J.; Bosse, M.; Ferrão, L.F.V.; de Hollander, M.; Garcia, A.A.F.; Ramírez, C.A.; Mendes, R.; Raaijmakers, J.M. Linking rhizosphere microbiome composition of wild and domesticated Phaseolus vulgaris to genotypic and root phenotypic traits. ISME J. 2017, 11, 2244–2257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smulders, L.; Benítez, E.; Moreno, B.; López-García, Á.; Pozo, M.J.; Ferrero, V.; de la Peña, E.; Alcalá Herrera, R. Tomato Domestication Affects Potential Functional Molecular Pathways of Root-Associated Soil Bacteria. Plants 2021, 10, 1942. [Google Scholar] [CrossRef] [PubMed]
- Legendre, P.; Legendre, L. Numerical Ecology; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar]
- Bai, Y.; Lindhout, P. Domestication and breeding of tomatoes: What have we gained and what can we gain in the future? Ann. Bot. 2007, 100, 1085–1094. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Romero, E.; Aguirre-Noyola, J.L.; Taco-Taype, N.; Martínez-Romero, J.; Zuñiga-Dávila, D. Plant microbiota modified by plant domestication. Syst. Appl. Microbiol. 2020, 43, 126106. [Google Scholar] [CrossRef]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef]
- Milla, R.; García-Palacios, P.; Matesanz, S. Looking at past domestication to secure ecosystem services of future croplands. J. Ecol. 2017, 105, 885–889. [Google Scholar] [CrossRef] [Green Version]
- Weidner, S.; Koller, R.; Latz, E.; Kowalchuk, G.; Bonkowski, M.; Scheu, S.; Jousset, A. Bacterial diversity amplifies nutrient-based plant-soil feedbacks. Funct. Ecol. 2015, 29, 1341–1349. [Google Scholar] [CrossRef]
- Legay, N.; Baxendale, C.; Grigulis, K.; Krainer, U.; Kastl, E.; Schloter, M.; Bardgett, R.D.; Arnoldi, C.; Bahn, M.; Dumont, M.; et al. Contribution of above- and below-ground plant traits to the structure and function of grassland soil microbial communities. Ann. Bot. 2014, 114, 1011–1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nwachukwu, B.C.; Babalola, O.O. Perspectives for sustainable agriculture from the microbiome in plant rhizosphere. Plant Biotechnol. Rep. 2021, 15, 259–278. [Google Scholar] [CrossRef]
- Kong, X.; Han, Z.; Tai, X.; Jin, D.; Ai, S.; Zheng, X.; Bai, Z. Maize (Zea mays L. Sp.) varieties significantly influence bacterial and fungal community in bulk soil, rhizosphere soil and phyllosphere. FEMS Microbiol. Ecol. 2020, 96, fiaa020. [Google Scholar] [CrossRef]
- Essel, E.; Xie, J.; Deng, C.; Peng, Z.; Wang, J.; Shen, J.; Xie, J.; Coulter, J.A.; Li, L. Bacterial and fungal diversity in rhizosphere and bulk soil under different long-term tillage and cereal/legume rotation. Soil Tillage Res. 2019, 194, 104302. [Google Scholar] [CrossRef]
- Uroz, S.; Buée, M.; Murat, C.; Frey-Klett, P.; Martin, F. Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil. Environ. Microbiol. Rep. 2010, 2, 281–288. [Google Scholar] [CrossRef]
- Peiffer, J.A.; Spor, A.; Koren, O.; Jin, Z.; Tringe, S.G.; Dangl, J.L.; Buckler, E.S.; Ley, R.E. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 6548–6553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shenton, M.; Iwamoto, C.; Kurata, N.; Ikeo, K. Effect of wild and cultivated rice genotypes on rhizosphere bacterial community composition. Rice 2016, 9, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doornbos, R.F.; van Loon, L.C.; Bakker, P.A.H.M. Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. A review. Agron. Sustain. Dev. 2012, 32, 227–243. [Google Scholar] [CrossRef]
- Shi, S.; Tian, L.; Nasir, F.; Li, X.; Li, W.; Tran, L.-S.P.; Tian, C. Impact of domestication on the evolution of rhizomicrobiome of rice in response to the presence of Magnaporthe oryzae. Plant Physiol. Biochem. 2018, 132, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Mendes, L.W.; Raaijmakers, J.M.; de Hollander, M.; Mendes, R.; Tsai, S.M. Influence of resistance breeding in common bean on rhizosphere microbiome composition and function. ISME J. 2018, 12, 212–224. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.J.; de Jonge, R.; Berendsen, R.L. The Soil-Borne Supremacy. Trends Plant Sci. 2016, 21, 171–173. [Google Scholar] [CrossRef]
- Lebeis, S.L.; Paredes, S.H.; Lundberg, D.S.; Breakfield, N.; Gehring, J.; McDonald, M.; Malfatti, S.; Glavina del Rio, T.; Jones, C.D.; Tringe, S.G.; et al. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 2015, 349, 860–864. [Google Scholar] [CrossRef] [Green Version]
- Paudel, S.; Lin, P.-A.; Foolad, M.R.; Ali, J.G.; Rajotte, E.G.; Felton, G.W. Induced plant defenses against herbivory in cultivated and wild tomato. J. Chem. Ecol. 2019, 45, 693–707. [Google Scholar] [CrossRef]
- Rosenthal, J.P.; Dirzo, R. Effects of life history, domestication and agronomic selection on plant defence against insects: Evidence from maizes and wild relatives. Evol. Ecol. 1997, 11, 337–355. [Google Scholar] [CrossRef]
- Brunel, C.; Pouteau, R.; Dawson, W.; Pester, M.; Ramirez, K.S.; van Kleunen, M. Towards unraveling macroecological patterns in rhizosphere microbiomes. Trends Plant Sci. 2020, 25, 1017–1029. [Google Scholar] [CrossRef] [PubMed]
- FAO; IUSS. World Reference Base for Soil Resources 2014; World Soil Resources Reports, No. 106; FAO: Rome, Italy, 2015. [Google Scholar]
- Lauber, C.L.; Zhou, N.; Gordon, J.I.; Knight, R.; Fierer, N. Effect of storage conditions on the assessment of bacterial community structure in soil and human-associated samples. FEMS Microbiol. Lett. 2010, 307, 80–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tatangelo, V.; Franzetti, A.; Gandolfi, I.; Bestetti, G.; Ambrosini, R. Effect of preservation method on the assessment of bacterial community structure in soil and water samples. FEMS Microbiol. Lett. 2014, 356, 32–38. [Google Scholar] [CrossRef]
- Wang, J.; Chapman, S.J.; Yao, H. The effect of storage on microbial activity and bacterial community structure of drained and flooded paddy soil. J. Soils Sediments 2015, 15, 880–889. [Google Scholar] [CrossRef]
- Kone, N.; Asare-Bediako, E.; Silue, S.; Kone, D.; Koita, O.; Menzel, W.; Winter, S. Influence of planting date on incidence and severity of viral disease on cucurbits under field condition. Ann. Agric. Sci. 2017, 62, 99–104. [Google Scholar] [CrossRef]
- Navas-Castillo, J.; Sánchez-Campos, S.; Díaz, J.A.; Sáez-Alonso, E.; Moriones, E. Tomato Yellow Leaf Curl Virus-Is Causes a Novel Disease of Common Bean and Severe Epidemics in Tomato in Spain. Plant Dis. 1999, 83, 29–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fortes, I.M.; Moriones, E.; Navas-Castillo, J. Tomato chlorosis virus in pepper: Prevalence in commercial crops in southeastern Spain and symptomatology under experimental conditions. Plant Pathol. 2012, 61, 994–1001. [Google Scholar] [CrossRef]
- Hartigan, J.A.; Wong, M.A. Algorithm AS 136: A K-Means Clustering Algorithm. Appl. Stat. 1979, 28, 100–108. [Google Scholar] [CrossRef]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef] [Green Version]
- Lundberg, D.S.; Yourstone, S.; Mieczkowski, P.; Jones, C.D.; Dangl, J.L. Practical innovations for high-throughput amplicon sequencing. Nat. Methods 2013, 10, 999–1002. [Google Scholar] [CrossRef]
- Vetrovský, T.; Baldrian, P.; Morais, D. SEED 2: A user-friendly platform for amplicon high-throughput sequencing data analyses. Bioinformatics 2018, 34, 2292–2294. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Chong, J.; Liu, P.; Zhou, G.; Xia, J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 2020, 15, 799–821. [Google Scholar] [CrossRef] [PubMed]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef]
- McArdle, B.H.; Anderson, M.J. Fitting multivariate models to community data: A comment on distance-based redundancy analysis. Ecology 2001, 82, 290–297. [Google Scholar] [CrossRef]
- Kembel, S.W.; Cowan, P.D.; Helmus, M.R.; Cornwell, W.K.; Morlon, H.; Ackerly, D.D.; Blomberg, S.P.; Webb, C.O. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 2010, 26, 1463–1464. [Google Scholar] [CrossRef] [Green Version]
- Laliberté, E.; Legendre, P.; Shipley, B. Measuring functional diversity (FD) from multiple traits, and other tools for functional ecology. Ecology 2014, 91, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H.; François, R.; Henry, L.; Müller, K. Dplyr: A Grammar of Data Manipulation; CRAN: 2021; Sebastopol, CA, USA, 2015. [Google Scholar]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smulders, L.; Ferrero, V.; de la Peña, E.; Pozo, M.J.; Díaz Pendón, J.A.; Benítez, E.; López-García, Á. Resistance and Not Plant Fruit Traits Determine Root-Associated Bacterial Community Composition along a Domestication Gradient in Tomato. Plants 2022, 11, 43. https://doi.org/10.3390/plants11010043
Smulders L, Ferrero V, de la Peña E, Pozo MJ, Díaz Pendón JA, Benítez E, López-García Á. Resistance and Not Plant Fruit Traits Determine Root-Associated Bacterial Community Composition along a Domestication Gradient in Tomato. Plants. 2022; 11(1):43. https://doi.org/10.3390/plants11010043
Chicago/Turabian StyleSmulders, Lisanne, Victoria Ferrero, Eduardo de la Peña, María J. Pozo, Juan Antonio Díaz Pendón, Emilio Benítez, and Álvaro López-García. 2022. "Resistance and Not Plant Fruit Traits Determine Root-Associated Bacterial Community Composition along a Domestication Gradient in Tomato" Plants 11, no. 1: 43. https://doi.org/10.3390/plants11010043
APA StyleSmulders, L., Ferrero, V., de la Peña, E., Pozo, M. J., Díaz Pendón, J. A., Benítez, E., & López-García, Á. (2022). Resistance and Not Plant Fruit Traits Determine Root-Associated Bacterial Community Composition along a Domestication Gradient in Tomato. Plants, 11(1), 43. https://doi.org/10.3390/plants11010043







