Eco-Geographical, Morphological and Molecular Characterization of a Collection of the Perennial Endemic Species Medicago tunetana (Murb.) A.W. Hill (Fabaceae) from Tunisia
Abstract
1. Introduction
2. Results
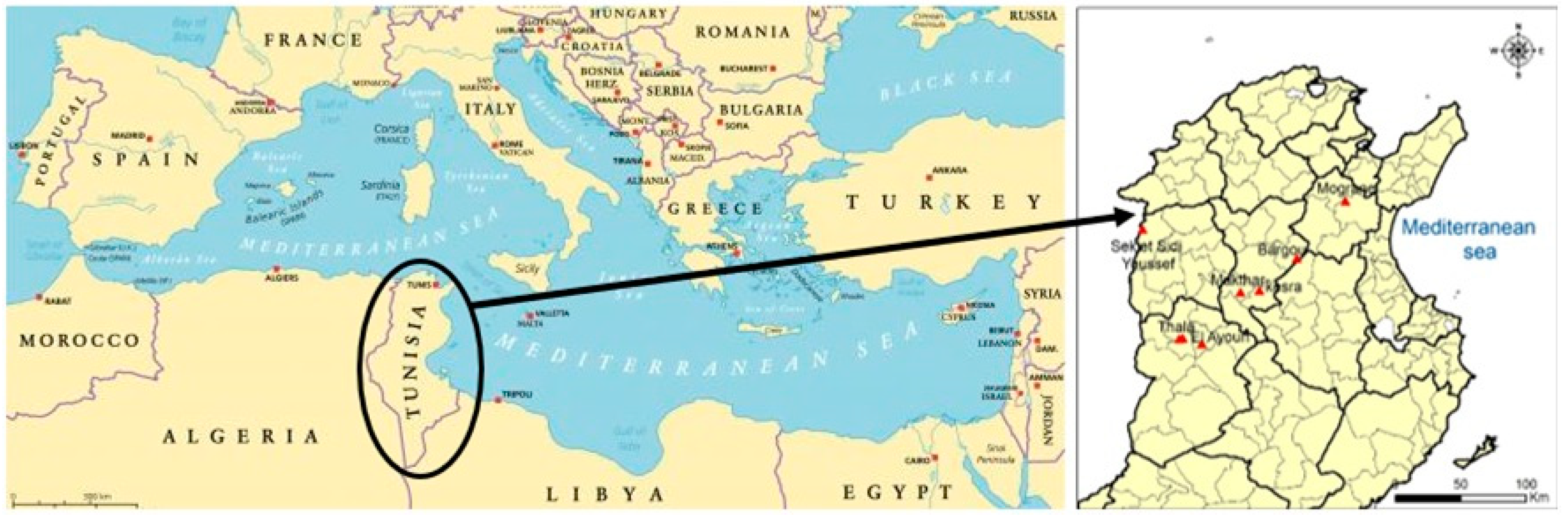
2.1. Ecology of M. tunetana Sites
2.2. Morphological Variation
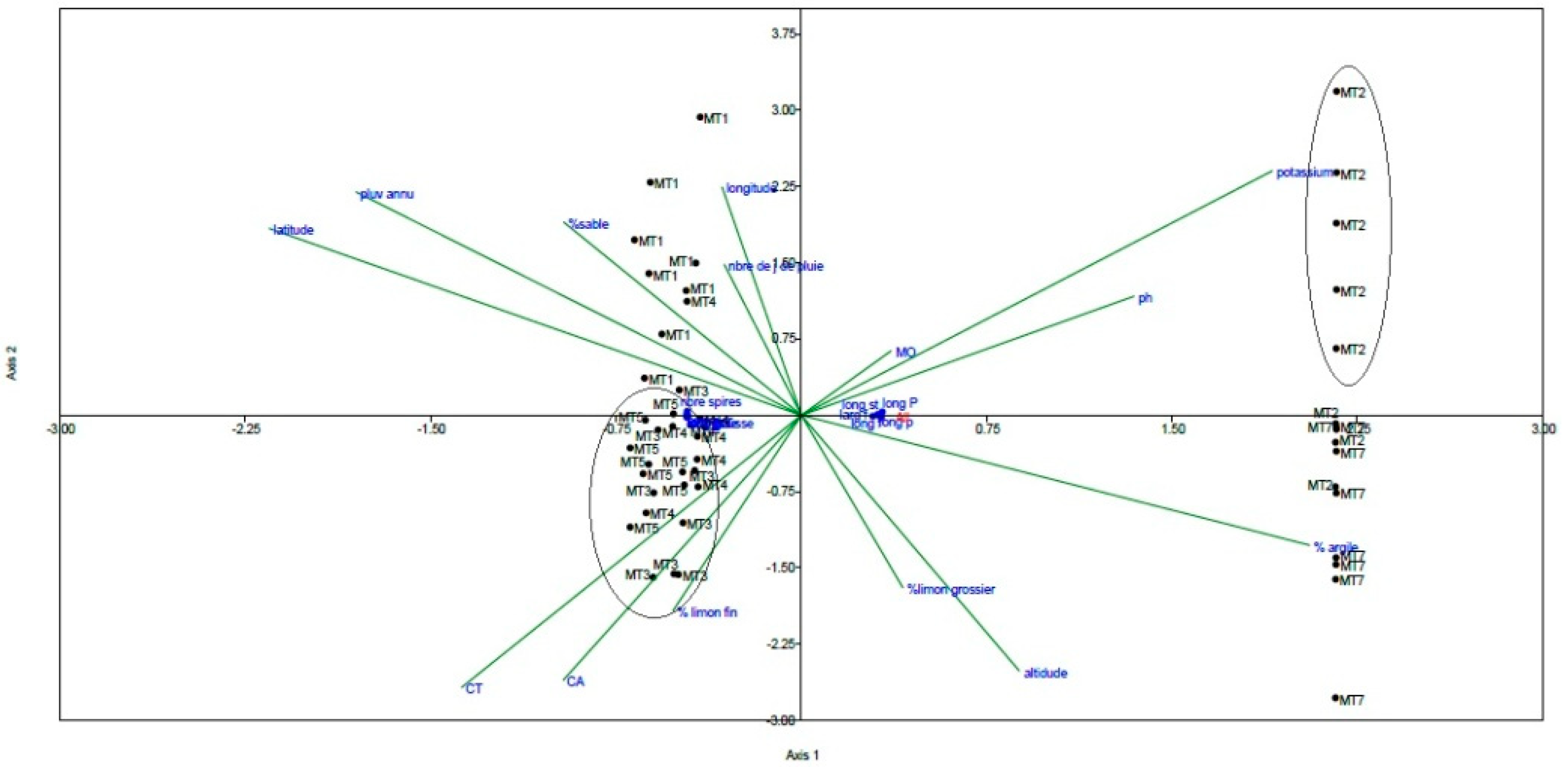
2.3. Canonical Correspondence Analysis
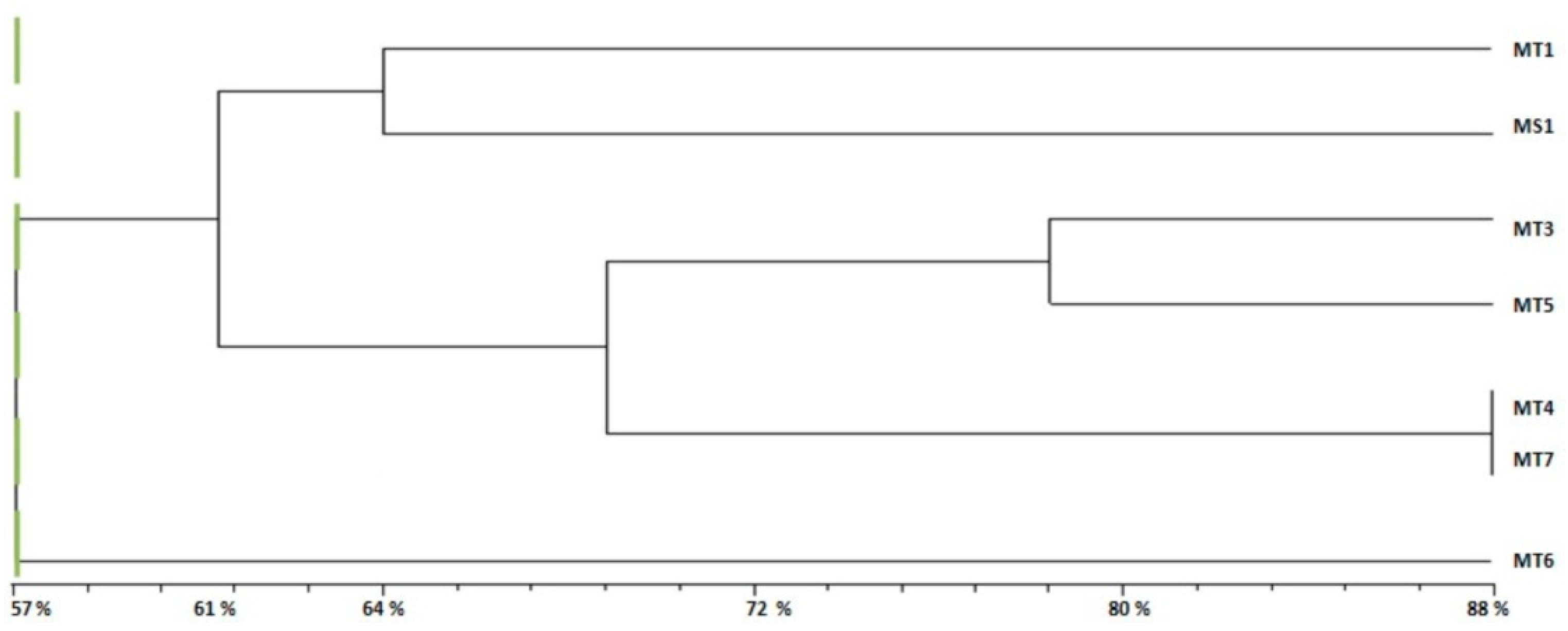
2.4. Genetic Diversity Analysis
- Group 1 (G1): Formed by two subgroups at 61% of genetic similarity with other accessions.
- −
- Subgroup 1: Formed by MT1 from Bargou and MS1, the variety of M. sativa.
- −
- Subgroup 2: Included four accessions of M. tunetana; MT3, MT5, MT4 and MT7 from Kesra, Thala, Sekiet Sidi Youssef and El Ayoun respectively.
- Group 2 (G2): Formed by MT6 from Thala with a 57% of similarity to other accessions of M. tunetana and the reference variety of M. sativa.
3. Discussion
4. Material and Methods
4.1. Origin of Plant Material Collection
4.2. Sample Size
4.3. Ecological Analyses
4.4. Morphological Traits
4.5. Molecular Analysis
4.6. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abdelguerfi, A.; Abdelguerfi-Laouar, M. Les ressources génétiques d’intérêt fourrager et/ou pastoral: Diversité, collecte et valorisation au niveau méditerranéen. Cah. Options Méditerranéennes 2004, 62, 29–41. [Google Scholar]
- Boussaid, M.; Ben Fadhel, N.; Zouali, Y.; Ben, S.A.; Abdelkefi, A. Plantes pastorales en milieux arides de l’Afrique du Nord. Cah. Options Méditerranéennes 2004, 62, 55–59. [Google Scholar]
- Robert, P.; Thiebeau, P.; Coulmier, D.; Larbre, D. Luzerne et Eau: Mieux Vaut Prévenir Que Guérir; COOP de France Désydratation France: Paris, France, 2010. [Google Scholar]
- El Makki-Ben Brahim, N.; Chaabani, A.; Toumi, L.; Sebei, H. Les plantes rares de la Tunisie Septentrionale et central. Ann. l’INRAT 2014, 87, 128–145. [Google Scholar]
- Chakroun, M.; Zouaghi, M. Conservation et valorisation des ressources génétiques fourragères et pastorales du Nord Tunisien. PGINA 2004, 123, 46–51. [Google Scholar]
- Wang, Z.; Yu, G.; Shi, B.; Wang, X.; Qiang, H.; Gao, H. Development and characterization of simple sequence repeat (SSR) markers based on RNA-Sequencing of Medicago sativa and in silico mapping onto the M. truncatula genome. PLoS ONE 2014, 9, e92029. [Google Scholar] [CrossRef]
- Benabderrahim, M.A.; Haddad, M.; Ferchichi, A. Essai d’adaptation de 16 cultivars de luzerne pérenne (Medicago sativa L.) dans un système oasien du sud tunisien: Gabès (local) et 15 cultivars étrangers. Cah. Option Méditerranéenne 2008, 79, 419–422. [Google Scholar]
- Gebahrd, C.A.; Buchi, L.; Liebisch, F.; Sinaj, S.; Ramseier, H.; Charles, R. Screening de légumineuses pour couverts végétaux: Azote et adventices. Rech. Agron. Suisse 2013, 4, 384–393. [Google Scholar]
- Tlahig, S.; Ben Khaled, A.; Ben Hmed, L.; Loumerem, M. Evaluation des rendements fourragers et semenciers des progéniteurs d’un polycross des génotypes de luzerne (Medicago sativa L. ssp. sativa) sélectionnées pour les régions arides tunisiennes. Rev. Régions Arid. 2017, 43, 81–88. [Google Scholar]
- Tlahig, S.; Yahia, H.; Loumerem, M. Agro-morphological homogeneity of Lucerne (Medicago sativa L. subsp. sativa) half-sib progenies bred for outside oases conditions of southern Tunisia. J. New Sci. 2017, 37, 2031–2034. [Google Scholar]
- Aridhi, F.; Sghaier, H.; Gaitanoros, A.; Khadri, A.; Aschi-Smiti, S.; Brouquisse, R. Nitric oxide production is involved in maintaining energy state in Alfalfa (Medicago sativa L.) nodulated roots under both salinity and flooding. Planta 2020, 252, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Le Floc’h, E.; Boulos, L.; Vela, E. Catalogue Synonymique Commenté de la FLORE DE TUNISIE; République Tunisienne Ministère de l’Environnement et du Développement Durable Banque Nationale de Gènes: Tunis, Tunisie, 2010. [Google Scholar]
- Quiros, C.F.; Bauchan, G.R. The genus Medicago and the origin of the Medicago sativa complex. In Alfalfa and alfalfa improvement. Alfalfa AlfalfaImprov. 1988, 29, 93–124. [Google Scholar] [CrossRef]
- Small, E.; Jomphe, M. A synopsis of the genus Medicago (Leguminosae). Can. J. Bot. 1989, 67, 3260–3294. [Google Scholar] [CrossRef]
- Pottier-Alapetite, G. Flore de la Tunisie Angiospermes-Dialypétales; Imprimerie Officielle de la République Tunisienne: Tunis, Tunisie, 1979. [Google Scholar]
- Catalogue Synonymique Commenté De La Flore De Tunisie. 2010. Available online: https://www.researchgate.net/profile/Errol-Vela/publication/224023795_Catalogue_Synonymique_Commente_De_La_Flore_De_Tunisie/links/0922b4f8c55746ec48000000/Catalogue-Synonymique-Commente-De-La-Flore-De-Tunisie.pdf (accessed on 15 July 2021).
- Lesins, K.A.; Lesins, I. Genus Medicago (Leguminosae): A Taxogenetic Study; W. Junk: The Hague, The Netherlands, 1979; 228p. [Google Scholar]
- The Plant List (TPL) Site. Available online: http://www.theplantlist.org/tpl1.1/record/ild-8549 (accessed on 15 July 2021).
- Tharachand, C.; Immanuel Selvarej, C.; Abraham, Z. Molecular Insights into the genetic diversity of Garcinia cambogia Germplasm accessions. Braz. Arch Technol. 2015, 58, 765–772. [Google Scholar] [CrossRef]
- Li, P.; Wang, Y.; Sun, X.; Han, J. Using microsatellite (SSR) and morphological markers to assess the genetic diversity of 12 falcata (Medicago sativa spp. falcata) populations from Eurasia. Afr. J. Biotechnol. 2009, 8, 2102–2108. [Google Scholar]
- Olivier, L.; Chevalet, C.; Fqoulley, J.L. Utilisation des marqueurs pour la caractérisation des ressources génétiques. Prod. Anim. 2000, 247–252. Available online: https://hal.inrae.fr/hal-02695861 (accessed on 15 July 2021).
- Roldán-Ruiz, I.; De Riek, J.; Muylle, H.; Baert, J.; Ghesquiere, A.; Vandewalle, M. Les marqueurs moléculaires: Quelles utilisations possibles en cultures fourragères. Fourrages 2005, 183, 419–438. [Google Scholar]
- Zaccardelli, M.; Gnocchi, S.; Carelli, M.; Scotti, C. Variation among and within Italian alfalfa ecotypes by means of bio-agronomic characters and amplified fragment length polymorphism analyses. Plant Breed. 2003, 122, 61–65. [Google Scholar] [CrossRef]
- Keivani, M.; Ramezanpour, S.; Soltanloo, H.; Choukan, R.; Naghavi, M.; Ranjbar, M. Genetic diversity assessment of alfalfa (Medicago sativa L.) populations using AFLP markers. Aust. J. Crop. Sci. 2010, 4, 491. [Google Scholar]
- Litoriya, N.S.; Talati, J.G.; Kher, H.R. Identification of Lucerne (Medicago sativa L.) Varieties Using Randomly Amplified Polymorphic DNA Markers. Indian J. Agric Biochem. 2009, 22, 60–62. [Google Scholar]
- Touil, L.; Bao, A.; Wang, S.; Ferchichi, A. Genetic Diversity of Tunisian and Chinese Alfalfa (Medicago sativa L.) Revealed by RAPD and ISSR Markers. Am. J. Plant Sci. 2016, 7, 967–979. [Google Scholar] [CrossRef]
- Brouwer, D.J.; Osborn, T.C. Identification of RFLP markers linked to the unifoliolate leaf, cauliflower head mutation of alfalfa. J. Hered. 1997, 88, 150–152. [Google Scholar] [CrossRef][Green Version]
- Kidwell, K.K.; Hartweck, L.M.; Yandell, B.S.; Crump, P.M.; Brummer, J.E.; Moutray, J.; Osborn, T.C. Forage yields of alfalfa populations derived from parents selected on the basis of molecular marker diversity. CellBiol. Mol. Genet. 1999, 39, 223–227. [Google Scholar] [CrossRef]
- Falahati-Anbaran, M.; Habashi, A.A.; Esfahany, M.; Mohammadi, S.A.; Ghareyazie, B. Population genetic structure based on SSR markers in alfalfa (Medicago sativa L.) from various regions contiguous to the centers of origin of the species. J. Genet. 2007, 86, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Cholastova, T.; Knotova, D. Using Morphological and Microsatellite (SSR) Markers to Assess the Genetic Diversity in Alfalfa (Medicago sativa L.). Int. J. Agric. Biosyst. Eng. 2012, 6, 781–787. [Google Scholar] [CrossRef]
- Wang, Z.; Hongwei, H.; Fu, X.; Gao, H. Development of simple sequence repeat markers and diversity analysis in alfalfa (Medicago sativa L.). Mol. Biol. Rep. 2013, 40, 3291–3298. [Google Scholar] [CrossRef] [PubMed]
- Shivakumar, M.S.; Ramesh, S.A.; Mohan, R.; Udaykumar, H.R.; Keerthi, C.M. Cross Legume Species/Genera Transferability of SSR Markers and their Utility in Assessing Polymorphism among Advanced Breeding Lines in Dolichos Bean (Lablab purpureus L.). Int. J. Curr. Microbiol. App. Sci. 2017, 6, 656–668. [Google Scholar] [CrossRef]
- Wang, H.; Chen, P.; Liu, D. Assessment of genetic diversity of Yunnan, Tibetan and Xinjiang wheat using SSR markers. J. Genet. Genom. 2007, 34, 623–633. [Google Scholar] [CrossRef]
- Amouri, A.; Aoul, S.H. Molecular analysis of two genotypes of Medicago truncatula Gaertn. by the expressed sequence tag EST6SSR (MTIC124) in response to salinity. Int. J. Innov. Appl. Stud. 2016, 17, 627–631. [Google Scholar]
- Touil, L.; Guesmi, F.; Fares, K.; Zagrouba, C.; Ferchichi, A. Genetic diversity of some Mediterranean populations of the cultivated alfalfa (Medicago sativa L.) using SSR markers. Pak. J. Biol. Sci. 2008, 11, 1923–1928. [Google Scholar] [CrossRef] [PubMed]
- Diwan, N.; Bhagwat, A.A.; Bauchan, G.R.; Cregan, P.B. Simple sequence repeat DNA markers in alfalfa and perennial and annual Medicago species. Genomes 1997, 40, 887–895. [Google Scholar] [CrossRef]
- Julier, B.; Flajoulot, S.; Barre, P.; Cardinet, G.; Santoni, S.; Huguet, T.; Huyghe, C. Construction of two genetic linkage maps in cultivated tetraploid alfalfa (Medicago sativa) using microsatellite and AFLP markers. BMC Plant Biol. 2003, 3, 1–19. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Emberger, L. Une classification biogéographique des climats. Recherche et travaux de laboratoire géographique, botanique et zoologie. Fac. Sci. MonpellierFr. 1966, 7, 1–43. [Google Scholar]
- Kimura, M.; Crow, J.F. The number of alleles that can be maintained in a finite population. Genetics 1964, 49, 725–738. [Google Scholar] [CrossRef]
- Nei, M. analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef] [PubMed]
- Lewontin, R.C. The Apportionment of Human Diversity; Committee on Evolutionary Biology, University of Chicago: Chicago, IL, USA, 1972; pp. 381–398. [Google Scholar] [CrossRef]
- Nei, M. Genetic distance between populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Ferchichi, Y.; Rouz, S. Effet de l’amendement humique sur l’amélioration des paramètres agronomiques de quelques populations de Medicago tunetana cultivées sous stress abiotique. In Oral Communication in the International Scientific Day of Mograne ‘Sustainable Management of Natural Resources’; Higher School of Agriculture of Mograne: Mograne, Tunisia, 2017; p. 10. [Google Scholar]
- Julier, B.; Herrmann, D.; Flajoulot, S.; Barre, P.; Huyghe, C.; Ronfort, J. Structuration de la diversité génétique chez la luzerne cultivée, conséquence pour l’identification de gènes liés à des caractères agronomiques. Innov. Agron. 2014, 35, 13–18. [Google Scholar]
- Andru, J. Les Populations Invasives de Rongeurs en Milieu Agricole: Une Etude Menée Dans des Cultures de Grande Echelle, les Plantations de Palmiers à Huile en Indonésie—Approche Paysagère, Génétique et Ecotoxicologique, Thèse de Doctorat en Sciences-Santé; Université Claude Bernard Lyon: Lyon, France, 2012. [Google Scholar]
- Heyn, C.C. The Annual Species of Medicago. Scripta Hierosolymitana; University of Jerusalem: Jerusalem, Israel, 1963. [Google Scholar]
- Abdelkefi, A.; Boussaid, M.; Djemal, F.; Cherifi, K.; Marrakchi, M. Approche analytique de la variabilité chez deux espèces spontanées du genre Medicago. In Oral Communication, Soc. Tun. Chim. Biol.; Monastir, Tunisie, 1990. [Google Scholar]
- Julier, B.; Bare, P. Les cartes génétiques chez les espèces fourragères pérennes des régions tempérées. Fourrages 2005, 183, 389–403. [Google Scholar]
- Havananda, T.; Charles Brummer, E.; Doyle, J. Complex patterns of autopolyploid evolution in alfalfa and allies (Medicago sativa; Leguminosae). Am. J. Bot. 2011, 98, 1–14. [Google Scholar] [CrossRef]
- Gardon, N.G.; Alarcon, Y.; Moreno, M.V.; Arolfo, V.; Orodizzi, A.; Basigalup, D.H.; Gieco, J.O.; Bruno, C.I. Genetic diversity among alfalfa genotypes (Medicago sativa L.) of non-dormant cultivars using SSR markers and agronomic traits. Rev. FCA UNCUYO 2013, 45, 181–195. [Google Scholar]
- Rouz, S.; Ben Jeddi, F.; Zouaghi, M.; Ghrabi-Gammar, Z. Diversité Génétique des Accessions de Bersim (Trifolium alexandrinum L., Fabaceae) par la Technique SDS PAGE. Revue de l’I.N.R.A.T. 2012, 27, 145–161. [Google Scholar]
- Saghai-Maroof, M.A.; Soliman, K.M.; Jorgensen, R.A.; Allard, R.W. Ribosomal DNA sepacer-length polymorphism in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA 1984, 81, 8014–8019. [Google Scholar] [CrossRef] [PubMed]
- Hammer, O.; Harper, D.A.T.; Rayan, P.D. Past: Palaeontogical statistics software package for education and data analysis. Palaeontol. Electron. 2011, 4, 9. [Google Scholar]
| N° | Eco-Geographical Factors | Accessions | |||||||
|---|---|---|---|---|---|---|---|---|---|
| MT1 | MT2 | MT3 | MT4 | MT5 | MT6 | MT7 | MS1 | ||
| 1 | Origin | Siliana, Bargou | Siliana, Makthar | Siliana, Kesra | Kef, Sekiet Sidi Youssef | Kasserine, Thala | Kasserine, Thala | Kasserine, El Ayoun | Zaghouan, Mograne |
| 2 | Latitude (dms) | 36°2′47.15” | 35°49′43.77” | 35°49′43.774” | 36°14′32.532” | 35°30′39.39” | 35°29′5.55” | 35°20′50.42” | 36°42′82.72” |
| 3 | Longitude (dms) | 9°40′34.32” | 9°21′28.38” | 9°21′28.205” | 8°22′8.291” | 8°42′58.45” | 8°44′30.34” | 8°34′31.08” | 10°09′20.49” |
| 4 | Altitude (m) | 523 | 868 | 1010 | 899 | 1003 | 1041 | 903 | 156 |
| 5 | Bioclimatic zone | SSACW | SSACW | SSACW | SHCW | MSACW | MSACW | MSACW | SSACW |
| 6 | Rainy days (In no) | 63.44 | 58.50 | 61.84 | 59.52 | 46.08 | 84.46 | 54.83 | 85.68 |
| 7 | Annual precipitation (mm) | 522.50 | 468.73 | 494.89 | 464.80 | 340.83 | 327 | 299 | 415.7 |
| 8 | Texture | Clay | Silty clay loam | Silty clay loam | Silty clay | Clay | Clay | Silty clay loam | nd |
| 9 | Fine silt (%) | 19.85 b | 23.49 c | 23.32 c | 26.28 d | 20.05 b | 17.95 a | 26.06 d | nd |
| 10 | Coarse silt (%) | 2.55 a | 6.81 d | 6.15 c | 15.20 f | 5.30 b | 9.91 e | 9.96 e | nd |
| 11 | Clay (%) | 28.68 c | 37.2 de | 36.35 d | 25.16 b | 28.62 c | 21.87 a | 37.88 e | nd |
| 12 | Sand (%) | 42.21 e | 31.5 b | 33.29 c | 38.43 d | 42.74 f | 46.2 g | 27.12 a | nd |
| 13 | K (ppm) | 38 d | 102 ab | 297 c | 052 a | 137 b | 292 c | 247 c | nd |
| 14 | Organic Matter (%) | 4.91 c | 2 a | 8.6 e | 6.72 d | 2.41 a | 3.55 b | 4.32 bc | nd |
| 15 | pH | 7.47 ab | 7.2 a | 8.36 e | 7.92 cd | 8.1 de | 8.01 cd | 7.71 bc | nd |
| 16 | Total limestone (%) | 35.21 a | 50 cd | 34.4 a | 44.00 bc | 53.19 d | 39.42 ab | 49.68 cd | nd |
| 17 | Active limestone (%) | 8.46 a | 30.7 d | 11.74 ab | 18.51 bc | 21.96 c | 14.51abc | 20.62 c | nd |
| N° | Traits | MT1 | MT2 | MT3 | MT4 | MT5 | MT6 | MT7 |
|---|---|---|---|---|---|---|---|---|
| 1 | LL ** | 12.00 a | nd | 17.06 b | 14.94 ab | 11.62 a | nd | nd |
| 2 | LW *** | 4.50 a | nd | 6.75 b | 6.31 b | 3.31 a | nd | nd |
| 3 | PL | 8.00 | nd | 7.06 | 7.87 | 5.00 | nd | nd |
| 4 | PeL | 1.69 | nd | 1.87 | 1.69 | 1.25 | nd | nd |
| 5 | StL | 7.06 | nd | 7.31 | 7.31 | 5.69 | nd | nd |
| 6 | PoL ** | 3.64 a | nd | 4.98 b | 3.51 a | 4.40 ab | nd | nd |
| 7 | PoW *** | 6.04 a | nd | 8.91 c | 6.81 b | 6.04 a | nd | nd |
| 8 | NPT *** | 4.24 b | nd | 3.33 k | 2.60 a | 2.70 a | nd | nd |
| 9 | SL *** | 2.73 b | nd | 3.01 c | 2.63 ab | 2.55 a | nd | nd |
| 10 | SW | 1.91 | nd | 2.07 | 2.08 | 1.99 | nd | nd |
| 11 | ST *** | 1.08 b | nd | 1.06 b | 0.84 a | 0.86 a | nd | nd |
| No. | Markers | No. of Alleles | No. of Polymorphic Bands | Mean NoBands/Locus | PIC |
|---|---|---|---|---|---|
| 1 | FMT13 | 7 | 5 | 1.4 | 0.21 |
| 2 | MTIC338 | 12 | 8 | 1.5 | 0.38 |
| 3 | MTIC343 | 19 | 6 | 3.16 | 0.49 |
| 4 | MTIC82 | 6 | 4 | 1.5 | 0.26 |
| 5 | B14B03 | 10 | 5 | 2 | 0.31 |
| Total | 54 | 28 | - | - | |
| Mean | 10.8 | 5.6 | 1.19 | 0.33 |
| Accessions | na * | ne * | I * | h * |
|---|---|---|---|---|
| MT1 | 2.0000 | 1.6756 | 0.4032 | 0.5930 |
| MT2 | 2.0000 | 1.6756 | 0.4032 | 0.5930 |
| MT3 | 2.0000 | 1.2677 | 0.2112 | 0.3669 |
| MT4 | 2.0000 | 1.9231 | 0.4800 | 0.6730 |
| MT5 | 2.0000 | 1.7705 | 0.4352 | 0.6269 |
| MT6 | 2.0000 | 1.9231 | 0.4800 | 0.6730 |
| MT7 | 2.0000 | 1.5743 | 0.3648 | 0.5511 |
| MS1 | 2.0000 | 1.2677 | 0.2112 | 0.3669 |
| Mean | 2.0000 | 1.6347 | 0.3736 | 0.5555 |
| St. Dev | 0.0000 | 0.2566 | 0.1076 | 0.1234 |
| Pop ID | MT1 | MT2 | MT3 | MT4 | MT5 | MT6 | MT7 | MS1 |
|---|---|---|---|---|---|---|---|---|
| MT1 | ---- | 0.7600 | 0.6000 | 0.5600 | 0.6400 | 0.6400 | 0.4800 | 0.6800 |
| MT2 | 0.2744 | ---- | 0.6800 | 0.6400 | 0.8000 | 0.6400 | 0.7200 | 0.6800 |
| MT3 | 0.5108 | 0.3857 | ---- | 0.4800 | 0.7200 | 0.6400 | 0.8800 | 0.7600 |
| MT4 | 0.5798 | 0.4463 | 0.7340 | ---- | 0.6800 | 0.5200 | 0.5200 | 0.5600 |
| MT5 | 0.4463 | 0.2231 | 0.3285 | 0.3857 | ---- | 0.6000 | 0.7600 | 0.6400 |
| MT6 | 0.4463 | 0.4463 | 0.4463 | 0.6539 | 0.5108 | ---- | 0.6800 | 0.4800 |
| MT7 | 0.7340 | 0.3285 | 0.1278 | 0.6539 | 0.2744 | 0.3857 | ---- | 0.6400 |
| MS1 | 0.3857 | 0.3857 | 0.2744 | 0.5798 | 0.4463 | 0.7340 | 0.4463 | ---- |
| No. | Primers | Sequences 5′-3′ | Repeated Motif | Ta (°C) |
|---|---|---|---|---|
| 1 | MTIC451 | F: CGATCGGAACGAGGACTTTA | (AAG)6 | 52 |
| R: CCCCGTTTTTCTTCTCTCCT | ||||
| 2 | FMT13 | F: GATGAGAAAATGAAAAGAAC | (GA)2GG(GA)9 | 52 |
| R: CAAAAACTCACTCTAACACAC | ||||
| 3 | MTIC338 | F: TCCCCTTAAGCTTCACTCTTTTC | (CTT)5 | 56 |
| R: CATTGGTGGACGAGGTCTCT | ||||
| 4 | MTIC343 | F: TCCGATCTTGCGTCCTAACT | (GAA)8 | 56 |
| R: CCATTGCGGTGGCTACTCT | ||||
| 5 | MTIC82 | F: CACTTTCCACACTCAAACCA | (TC)11 | 55 |
| R: GAGAGGATTTCGGTGATGT | ||||
| 6 | MTIC432 | F: TGGAATTTGGGATATAGGAA | (AG)6 | 55 |
| R: GGCCATAAGAACTTCCACTT | ||||
| 7 | B14B03 | F: GCTTGTTCTTCTTCAAGCTC | (CA)9 | 55 |
| R: ACCTGACTTGTGTTTTATGC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferchichi, Y.; Sakhraoui, A.; Ltaeif, H.B.; Ben Mhara, Y.; Elimem, M.; Ben Naceur, M.; Ghrabi-Gammar, Z.; Rouz, S. Eco-Geographical, Morphological and Molecular Characterization of a Collection of the Perennial Endemic Species Medicago tunetana (Murb.) A.W. Hill (Fabaceae) from Tunisia. Plants 2021, 10, 1923. https://doi.org/10.3390/plants10091923
Ferchichi Y, Sakhraoui A, Ltaeif HB, Ben Mhara Y, Elimem M, Ben Naceur M, Ghrabi-Gammar Z, Rouz S. Eco-Geographical, Morphological and Molecular Characterization of a Collection of the Perennial Endemic Species Medicago tunetana (Murb.) A.W. Hill (Fabaceae) from Tunisia. Plants. 2021; 10(9):1923. https://doi.org/10.3390/plants10091923
Chicago/Turabian StyleFerchichi, Yosr, Anis Sakhraoui, Hela Belhaj Ltaeif, Yosr Ben Mhara, Mohamed Elimem, M’barek Ben Naceur, Zeineb Ghrabi-Gammar, and Slim Rouz. 2021. "Eco-Geographical, Morphological and Molecular Characterization of a Collection of the Perennial Endemic Species Medicago tunetana (Murb.) A.W. Hill (Fabaceae) from Tunisia" Plants 10, no. 9: 1923. https://doi.org/10.3390/plants10091923
APA StyleFerchichi, Y., Sakhraoui, A., Ltaeif, H. B., Ben Mhara, Y., Elimem, M., Ben Naceur, M., Ghrabi-Gammar, Z., & Rouz, S. (2021). Eco-Geographical, Morphological and Molecular Characterization of a Collection of the Perennial Endemic Species Medicago tunetana (Murb.) A.W. Hill (Fabaceae) from Tunisia. Plants, 10(9), 1923. https://doi.org/10.3390/plants10091923








