Double Mutant Analysis with the Large Flower Mutant, ohbana1, to Explore the Regulatory Network Controlling the Flower and Seed Sizes in Arabidopsis thaliana
Abstract
1. Introduction
2. Results
2.1. Detection of Mutations Induced by Ar Ion Beam Irradiation
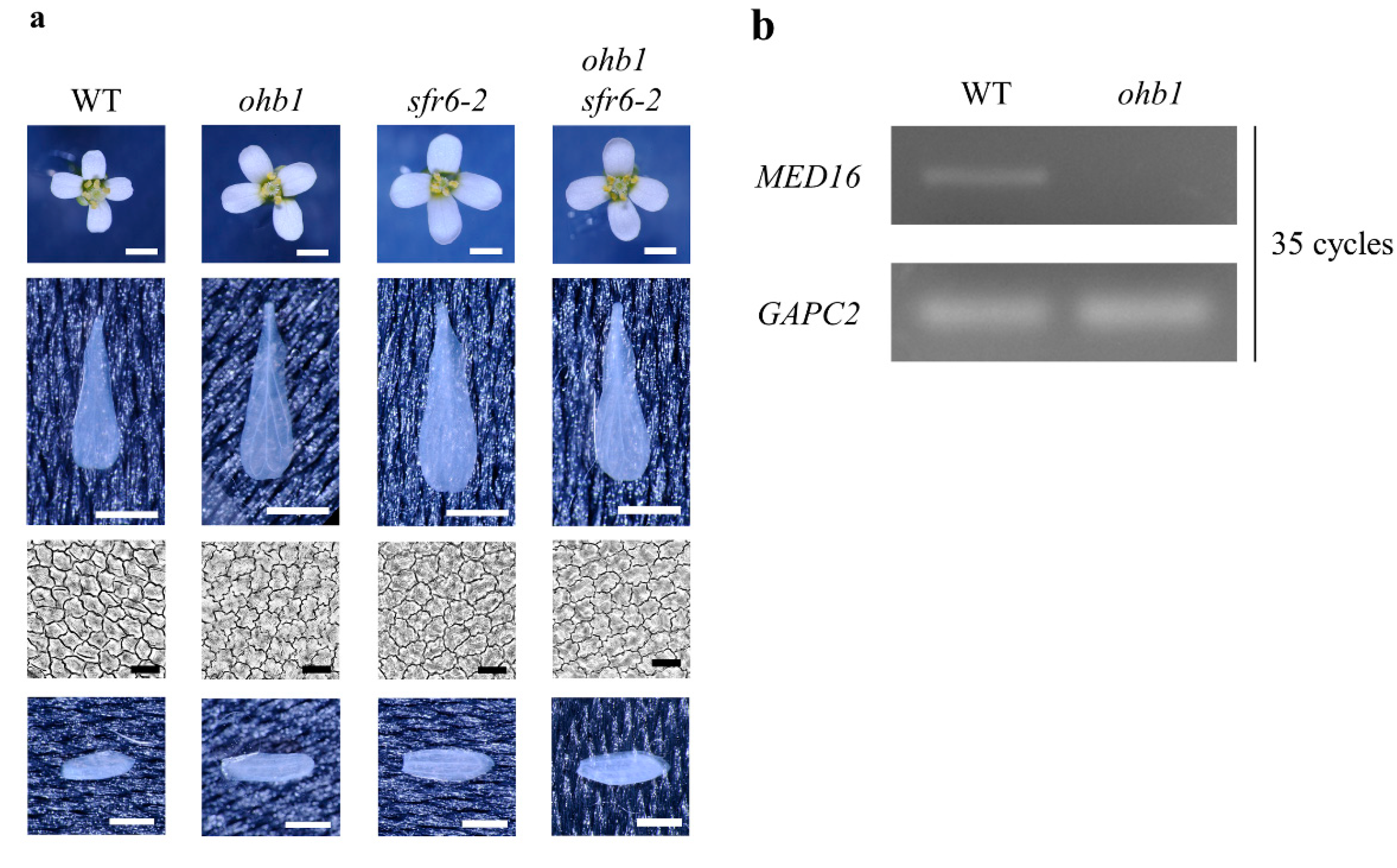
2.2. Characteristics of the Floral Organs in ohb1
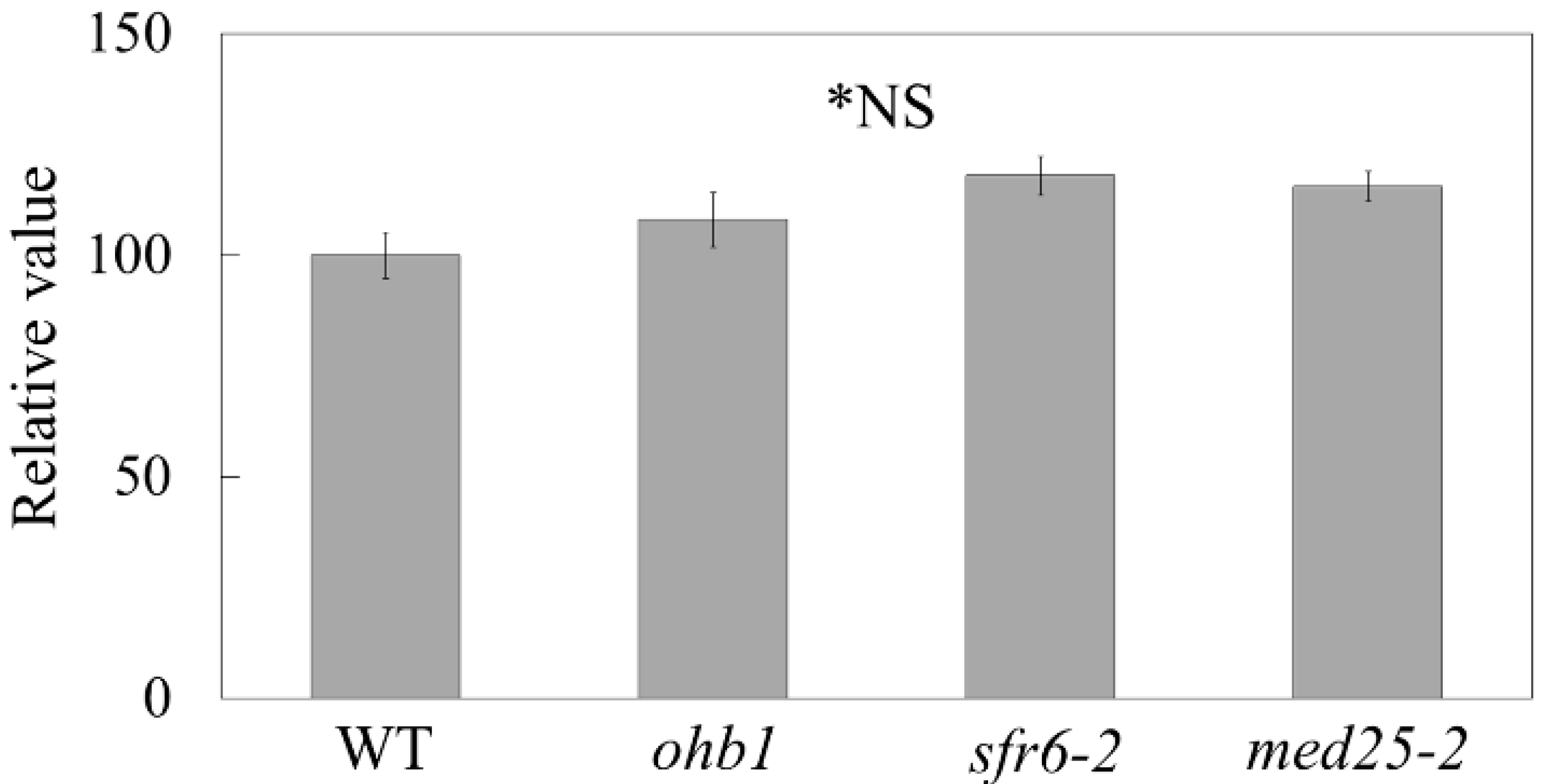
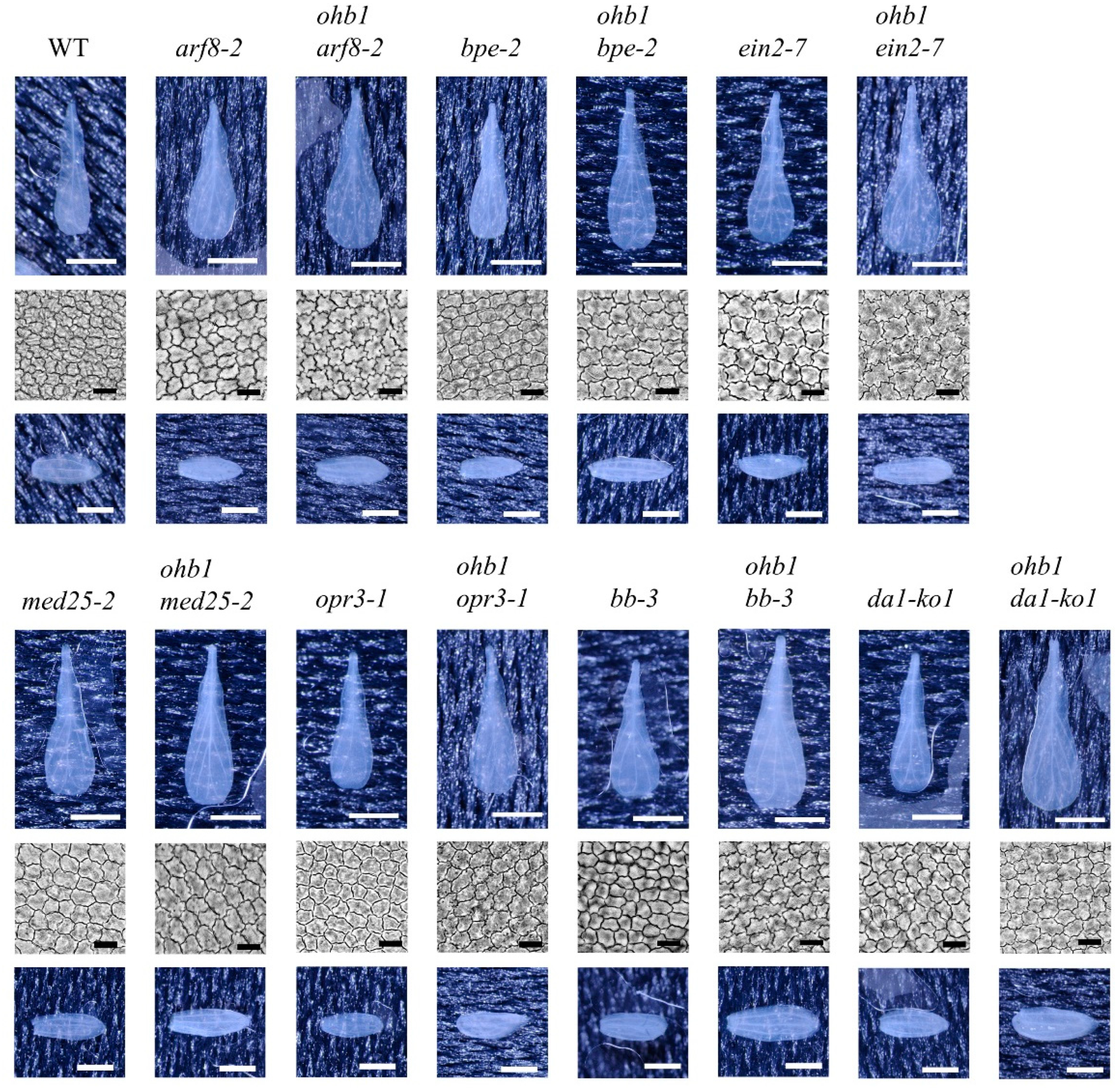
2.3. Double-Mutant Crosses between ohb1 and Large Flower Mutants Associated with Post-Mitotic Cell Expansion
2.4. Double-Mutant Crosses between ohb1 and Large Flower Mutants Associated with Cell Proliferation
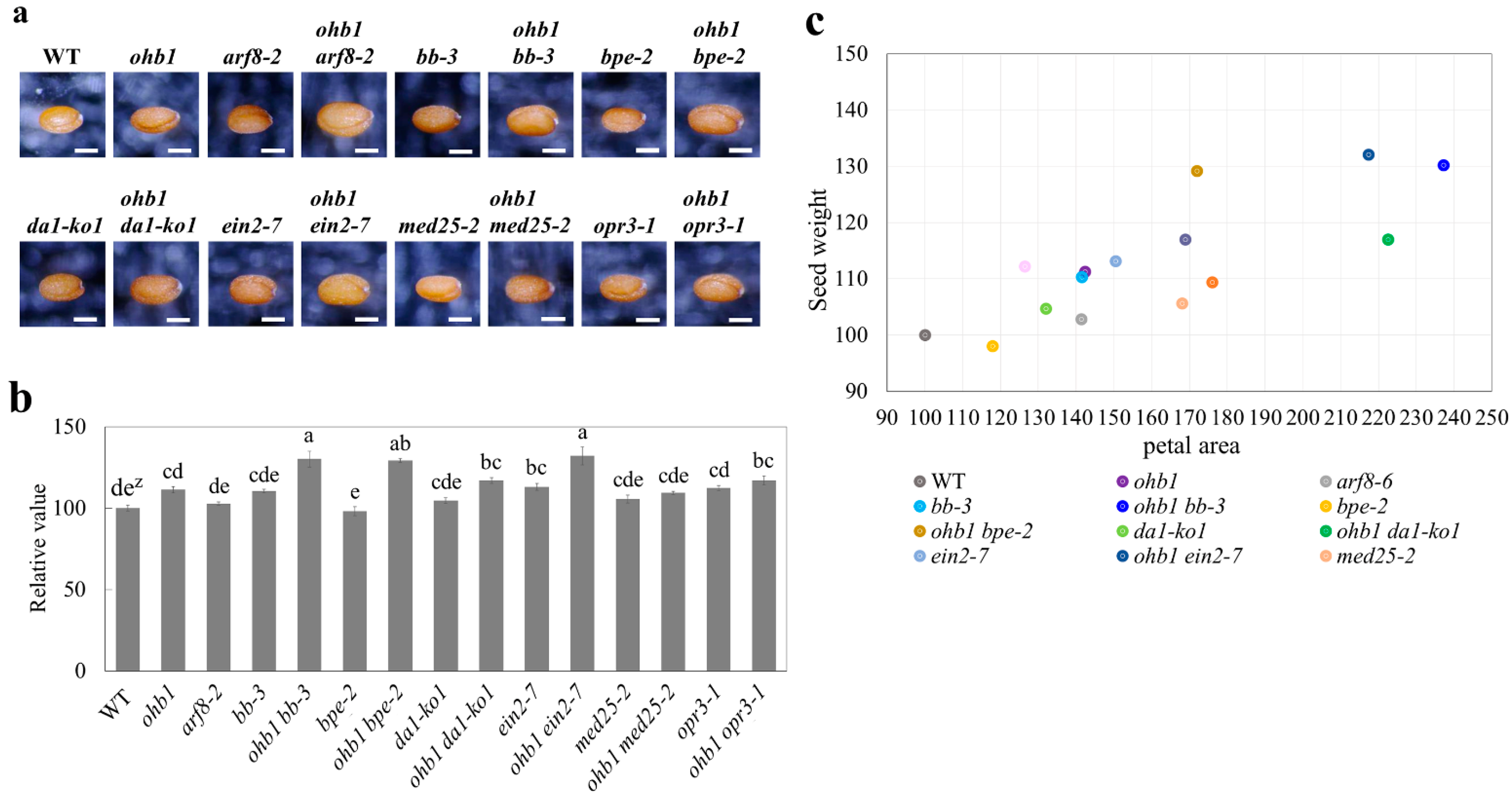
2.5. Seed Weights Produced by the Large Flower Mutants
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Whole-Genome Mutation Analysis
4.3. RT-PCR
4.4. Morphological Analysis of the Floral Organs
4.5. Flow Cytometric Analysis
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hepworth, J.; Lenhard, M. Regulation of plant lateral-organ growth by modulating cell number and size. Curr. Opin. Plant. Biol. 2014, 17, 36–42. [Google Scholar] [CrossRef]
- Mizukami, Y. A matter of size: Developmental control of organ size in plant. Curr. Opin. Plant. Biol. 2001, 4, 533–539. [Google Scholar] [CrossRef]
- Krizek, B.A. Ectopic expression of AINTEGUMENTA in Arabidopsis plants results in increased growth of floral organs. Dev. Genet. 1999, 25, 224–236. [Google Scholar] [CrossRef]
- Mizukami, Y.; Fischer, R.L. Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. USA 2000, 97, 942–947. [Google Scholar] [CrossRef] [PubMed]
- Ohno, C.K.; Reddy, G.V.; Heisler, M.G.B.; Meyerowitz, E.M. The Arabidopsis JAGGED gene encodes a zinc finger protein that promotes leaf tissue development. Development 2004, 131, 1111–1122. [Google Scholar] [CrossRef]
- Disch, S.; Anastasiou, E.; Sharma, V.K.; Laux, T.; Fletcher, J.C. The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Curr. Biol. 2006, 16, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zheng, L.; Corke, F.; Smith, C.; Bevan, M.W. Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Gene. Dev. 2008, 22, 1331–1336. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.T.; Tsukaya, H.; Uchimiya, H. The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Gene. Dev. 1998, 12, 2381–2911. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.T.; Tsukaya, H.; Saito, Y.; Uchimiya, H. Changes in the shapes of leaves and flowers upon overexpression of cytochrome P450 in Arabidopsis. Proc. Natl. Acad. Sci. USA 1999, 96, 9433–9437. [Google Scholar] [CrossRef]
- Hu, Y.; Poh, H.M.; Chua, N.H. The Arabidopsis ARGOS-LIKE gene regulates cell expansion during organ growth. Plant. J. 2006, 47, 1–9. [Google Scholar] [CrossRef]
- Varaud, E.; Brioudes, F.; Szécsi, J.; Leroux, J.; Brown, S.; Perrot-Rechenmann, C.; Bendahmanea, M. AUXIN RESPONSE FACTOR8 Regulates Arabidopsis Petal Growth by Interacting with the bHLH Transcription Factor BIGPETALp. Plant. Cell 2011, 23, 973–983. [Google Scholar] [CrossRef]
- Szécsi, J.; Joly, C.; Bordji, K.; Varaud, E.; Cock, J.M.; Dumas, C.; Bendahmane, M. BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. EMBO J. 2006, 25, 3912–3920. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; Li, N.; Dumenil, J.; Li, J.; Kamenski, A.; Bevan, M.W.; Gao, F.; Li, Y. The Ubiquitin Receptor DA1 Interacts with the E3 Ubiquitin Ligase DA2 to Regulate Seed and Organ Size in Arabidopsis. Plant. Cell 2013, 25, 3347–3359. [Google Scholar] [CrossRef]
- Brioudes, F.; Joly, C.; Szécsi, J.; Varaud, E.; Leroux, J.; Bellvert, F.; Bertrand, C.; Bendahmane, M. Jasmonate controls late development stages of petal growth in Arabidopsis thaliana. Plant. J. 2009, 60, 1070–1080. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Xie, Z.; Hu, C.; Zhang, J. A Review of Auxin Response Factors (ARFs) in Plants. Front. Plant. Sci. 2016, 7, 47. [Google Scholar] [CrossRef]
- Pei, H.; Ma, N.; Tian, J.; Luo, J.; Chen, J.; Li, J.; Zheng, Y.; Chen, X.; Fei, Z.; Gao, J. An NAC transcription factor controls ethylene-regulated cell expansion in flower petals. Plant. Physiol. 2013, 163, 775–791. [Google Scholar] [CrossRef]
- Feng, G.; Liu, G.; Xiao, J. The Arabidopsis EIN2 restricts organ growth by retarding cell expansion. Plant. Signal. Behav. 2015, 10, e1017169. [Google Scholar] [CrossRef][Green Version]
- He, X.J.; Mu, R.L.; Cao, W.H.; Zhang, Z.G.; Zhang, J.S.; Chen, S.Y. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant. J. 2005, 44, 903–916. [Google Scholar] [CrossRef] [PubMed]
- Bäckström, S.; Elfving, N.; Nilsson, R.; Wingsle, G.; Björklund, S. Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol. Cell 2007, 26, 717–729. [Google Scholar] [CrossRef]
- Mathur, S.; Vyas, S.; Kapoor, S.; Tyagi, A.K. The Mediator complex in plants: Structure, phylogeny, and expression profiling of representative genes in a dicot (Arabidopsis) and a monocot (Rice) during reproduction and abiotic stress. Plant. Physiol. 2011, 157, 1609–1627. [Google Scholar] [CrossRef]
- Wathugala, D.L.; Hemsley, P.A.; Moffat, C.S.; Cremelie, P.; Knight, M.R.; Knight, H. The Mediator subunit SFR6/MED16 controls defence gene expression mediated by salicylic acid and jasmonate responsive pathways. New Phytol. 2012, 195, 217–230. [Google Scholar] [CrossRef] [PubMed]
- Buendía-Monreal, M.; Gillmor, C.S. Mediator: A key regulator of plant development. Dev. Biol. 2016, 419, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Li, Y. Control of final organ size by Mediator complex subunit 25 in Arabidopsis thaliana. Development 2011, 138, 4545–4554. [Google Scholar] [CrossRef]
- Xu, R.; Li, Y. The Mediator complex subunit 8 regulates organ size in Arabidopsis thaliana. Plant. Signal. Behav. 2012, 7, 182–183. [Google Scholar] [CrossRef]
- Yang, Y.; Ou, B.; Zhang, J.; Si, W.; Gu, H.; Qin, G.; Qu, L.J. The Arabidopsis Mediator subunit MED16 regulates iron homeostasis by associating with EIN3/EIL1 through subunit MED25. Plant. J. 2014, 77, 838–851. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, G.; Gao, F.; Xu, R.; Li, N.; Zhang, Y.; Li, Y. Transcriptional repression of the APC/C activator genes CCS52A1/A2 by the mediator complex subunit MED16 controls endoreduplication and cell growth in Arabidopsis. Plant. Cell 2019, 31, 1899–1912. [Google Scholar] [CrossRef]
- Li, N.; Li, Y. Maternal control of seed size in plants. J. Exp. Bot. 2015, 66, 1087–1097. [Google Scholar] [CrossRef]
- Orozco-Arroyo, G.; Paolo, D.; Ezquer, I.; Colombo, L. Networks controlling seed size in Arabidopsis. Plant. Reprod. 2015, 28, 17–32. [Google Scholar] [CrossRef]
- Locascio, A.; Roig-Villanova, I.; Bernardi, J.; Varotto, S. Current perspectives on the hormonal control of seed development in Arabidopsis and maize: A focus on auxin. Front. Plant. Sci. 2014, 5, 412. [Google Scholar] [CrossRef]
- Tanaka, A.; Shikazono, N.; Hase, Y. Studies on biological effects of ion beams on lethality, molecular nature of mutation, mutation rate, and spectrum of mutation phenotype for mutation breeding in higher plants. J. Radiat Res. 2010, 51, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Abe, T.; Kazama, Y.; Hirano, T. Ion beam breeding and gene discovery for function analyses using mutants. Nucl. Phys. News 2015, 25, 30–35. [Google Scholar] [CrossRef]
- Sasaki, K.; Yamaguchi, H.; Aida, R.; Shikata, M.; Abe, T.; Ohtsubo, N. Mutation in Torenia fournieri Lind. UFO homolog confers loss of TfLFY interaction and results in a petal to sepal transformation. Plant. J. 2012, 71, 1002–1014. [Google Scholar] [CrossRef]
- Yasui, Y.; Mori, M.; Aii, J.; Abe, T.; Matsumoto, D.; Sato, S.; Hayashi, Y.; Ohnishi, O.; Ota, T. S-LOCUS EARLY FLOWERING 3 is exclusively present in the genomes of short-styled buckwheat plants that exhibit heteromorphic self-Incompatibility. PLoS ONE 2012, 7, e31264. [Google Scholar] [CrossRef]
- Kazama, Y.; Fujiwara, M.T.; Takehisa, H.; Ohbu, S.; Saito, H.; Ichida, H.; Hayashi, Y.; Abe, T. Characterization of a heavy-ion induced white flower mutant of allotetraploid Nicotiana tabacum. Plant. Cell Rep. 2013, 32, 11–19. [Google Scholar] [CrossRef]
- Maeda, S.; Gunji, S.; Hanai, K.; Hirano, T.; Kazama, Y.; Ohbayashi, I.; Abe, T.; Sawa, S.; Tsukaya, H.; Ferjani, A. The conflict between cell proliferation and expansion primarily affects stem organogenesis in Arabidopsis. Plant. Cell Physiol. 2014, 55, 1994–2007. [Google Scholar] [CrossRef]
- Katano, M.; Takahashi, K.; Hirano, T.; Kazama, Y.; Abe, T.; Tsukaya, H.; Ferjani, A. Suppressor screen and phenotype analyses revealed an emerging role of the monofunctional peroxisomal enoyl-CoA hydratase 2 in compensated cell enlargement. Front. Plant. Sci. 2016, 7, 132. [Google Scholar] [CrossRef]
- Kazama, Y.; Ishii, K.; Aonuma, W.; Ikeda, T.; Kawamoto, H.; Koizumi, A.; Filatov, D.A.; Chibalina, M.; Bergero, R.; Charlesworth, D.; et al. A new physical mapping approach refines the sex-determining gene positions on the Silene latifolia Y-chromosome. Sci. Rep. 2016, 6, 18917. [Google Scholar] [CrossRef]
- Shikazono, N.; Suzuki, C.; Kitamura, S.; Watanabe, H.; Tano, S.; Tanaka, A. Analysis of mutations induced by carbon ions in Arabidopsis thaliana. J. Exp. Bot. 2005, 56, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Kazama, Y.; Hirano, T.; Saito, H.; Liu, Y.; Ohbu, S.; Hayashi, Y.; Abe, T. Characterization of highly efficient heavy-ion mutagenesis in Arabidopsis thaliana. BMC Plant. Biol. 2011, 11, 161. [Google Scholar] [CrossRef] [PubMed]
- Hirano, T.; Kazama, Y.; Ohbu, S.; Shirakawa, Y.; Liu, Y.; Kambara, Y.; Fukunishi, N.; Abe, T. Molecular nature of mutation induced by high-LET irradiation with argon and carbon ions in Arabidopsis thaliana. Mutat. Res. 2012, 735, 19–31. [Google Scholar] [CrossRef]
- Kazama, Y.; Saito, H.; Yamamoto, Y.Y.; Hayashi, Y.; Ichida, H.; Ryuto, H.; Fukunishi, N.; Abe, T. LET-dependent effects of heavy-ion beam irradiation in Arabidopsis thaliana. Plant. Biotechnol. 2008, 25, 113–117. [Google Scholar] [CrossRef]
- Kazama, Y.; Hirano, T.; Nishihara, K.; Ohbu, S.; Shirakawa, Y.; Abe, T. Effect of high-LET Fe-ion beam irradiation on mutation induction in Arabidopsis thaliana. Genes Genet. Syst. 2013, 88, 189–197. [Google Scholar] [CrossRef]
- Knight, H.; Mugford, S.G.; Ülker1, B.; Gao, D.; Thorlby, G.; Knight, M.R. Identification of SFR6, a key component in cold acclimation acting post-translationally on CBF function. Plant. J. 2009, 58, 97–108. [Google Scholar] [CrossRef]
- Okushima, Y.; Overvoorde, P.J.; Arima, K.; Alonso, J.M.; Chan, A.; Chang, C.; Ecker, J.R.; Hughes, B.; Lui, A.; Nguyen, D.; et al. Functional Genomic Analysis of the AUXIN RESPONSE FACTOR Gene Family Members in Arabidopsis thaliana: Unique and Overlapping Functions of ARF7 and ARF19. Plant. Cell 2005, 17, 444–463. [Google Scholar] [CrossRef]
- Kazama, Y.; Ishii, K.; Hirano, T.; Wakana, T.; Yamada, M.; Ohbu, S.; Abe, T. Different mutational function of low- and high-linear energy transfer heavy-ion irradiation demonstrated by whole-genome resequencing of Arabidopsis mutants. Plant. J. 2017, 92, 1020–1030. [Google Scholar] [CrossRef] [PubMed]
- Knight, H.; Thomson, A.J.W.; McWatters, H.G. SENSITIVE TO FREEZING6 integrates cellular and environmental inputs to the plant circadian clock. Plant. Physiol. 2008, 148, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, C.; Zhang, Y.; Sun, Y.; Mou, Z. The Arabidopsis Mediator complex subunit16 positively regulates salicylate-mediated systemic acquired resistance and jasmonate/ethylene-induced defense pathways. Plant. Cell 2012, 24, 4294–4309. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yao, J.; Zhang, Y.; Sun, Y.; Mou, Z. The Arabidopsis Mediator complex subunits MED14/SWP and MED16/SFR6/IEN1 differentially regulate defense gene expression in plant immune responses. Plant. J. 2013, 75, 484–497. [Google Scholar] [CrossRef] [PubMed]
- Hemsley, P.A.; Hurst, C.H.; Kaliyadasa, E.; Lamb, R.; Knight, M.R.; Cothi, E.A.D.; Steele, J.F.; Knight, H. The Arabidopsis Mediator complex subunits MED16, MED14, and MED2 regulate Mediator and RNA polymerase II recruitment to CBF-responsive cold-regulated genes. Plant. Cell 2014, 26, 465–484. [Google Scholar] [CrossRef] [PubMed]
- Sorek, N.; Szemenyei, H.; Sorek, H.; Landers, A.; Knight, H.; Bauer, S.; Wemmer, D.E.; Somerville, C.R. Identification of MEDIATOR16 as the Arabidopsis COBRA suppressor MONGOOSE1. Proc. Natl. Acad. Sci. USA 2015, 112, 16048–16053. [Google Scholar] [CrossRef]
- Wang, C.; Yao, J.; Du, X.; Zhang, Y.; Sun, Y.; Rollins, J.; Mou, Z. The Arabidopsis mediator complex subunit16 is a key component of basal resistance against the necrotrophic fungal pathogen Sclerotinia sclerotiorum. Plant. Physiol. 2015, 169, 856–872. [Google Scholar] [CrossRef]
- Sugimoto-Shirasu, K.; Roberts, K. “Big it up”: Endoreduplication and cell-size control in plants. Curr. Opin. Plant. Biol. 2003, 6, 544–553. [Google Scholar] [CrossRef] [PubMed]
- Tsukaya, H. Has the impact of endoreduplication on cell size been overestimated? New Phytol. 2019, 223, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Bonawitz, N.D.; Kim, J.; Tobimatsu, Y.; Ciesielski, P.N.; Anderson, N.A.; Ximenes, E.; Maeda, J.; Ralph, J.; Donohoe, B.S.; Ladisch, M.; et al. Disruption of Mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature 2014, 509, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Hisanaga, T.; Kawade, K.; Tsukaya, H. Compensation: A key to clarifying the organ-level regulation of lateral organ size in plants. J. Exp. Bot. 2015, 66, 1055–1063. [Google Scholar] [CrossRef]
- Usami, T.; Horiguchi, G.; Yano, S.; Tsukaya, H. The more and smaller cells mutants of Arabidopsis thaliana identify novel roles for SQUAMOSA PROMOTER BINDING PROTEIN-LIKE genes in the control of heteroblasty. Development 2009, 136, 955–964. [Google Scholar] [CrossRef]
- Okushima, Y.; Mitina, I.; Quach, H.L.; Theologis, A. AUXIN RESPONSE FACTOR 2 (ARF2): A pleiotropic developmental regulator. Plant. J. 2005, 43, 29–46. [Google Scholar] [CrossRef]
- Schruff, M.C.; Spielman, M.; Tiwari, S.; Adams, S.; Fenby, N.; Scott, R.J. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 2006, 133, 251–261. [Google Scholar] [CrossRef]
- Nagpal, P.; Ellis, C.M.; Weber, H.; Ploense, S.E.; Barkawi, L.S.; Guilfoyle, T.J.; Hagen, G.; Alonso, J.M.; Cohen, J.D.; Farmer, E.E.; et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 2005, 132, 4107–4118. [Google Scholar] [CrossRef]
- Hirano, T.; Kazama, Y.; Ishii, K.; Ohbu, S.; Shirakawa, Y.; Abe, T. Comprehensive identification of mutations induced by heavy-ion beam irradiation in Arabidopsis thaliana. Plant. J. 2015, 82, 93–104. [Google Scholar] [CrossRef]
- Ishii, K.; Kazama, Y.; Hirano, T.; Hamada, M.; Ono, Y.; Yamada, M.; Abe, T. AMAP: A pipeline for whole-genome mutation detection in Arabidopsis thaliana. Genes Genet. Syst. 2016, 91, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Smyth, D.R.; Bowman, J.L.; Meyerowitz, E.M. Early flower development in Arabidopsis. Plant. Cell 1990, 2, 755–767. [Google Scholar] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]

| Chr. | Position | Locus | Reference Base | Mutated Base | Effect of Mutation |
|---|---|---|---|---|---|
| 2 | 19,044,495 | AT2G46400 | * | −42 bp | codon change codon deletion |
| 4 | 2,501,011 | AT4G04920 | * | +A | frame shift |
| 4 | 2,501,014 | AT4G04920 | T | A | non synonymous coding |
| 4 | 2,501,015 | AT4G04920 | * | −TTCGG | frame shift |
| 4 | 5,349,509 | AT4G08430 | G | A | non synonymous coding |
| Line | Petal | Sepal | ||||
|---|---|---|---|---|---|---|
| Length | Width | Area | Cell Area | Length | Width | |
| WT | 100.0 ± 2.3 e | 100.0 ± 2.9 e | 100.0 ± 4.5 g | 100.0 ± 2.5 h | 100.0 ± 3.1 g | 100.0 ± 3.5 f |
| ohb1 | 116.6 ± 2.4 d | 119.1 ± 3.6 d | 142.3 ± 7.9 de | 145.3 ± 2.5 def | 119.7 ± 3.7 cdef | 125.2 ± 4.4 cd |
| sfr6-2 | 127.4 ± 1.7 bc | 123.5 ± 2.3 cd | 159.3 ± 4.5 bcd | 157.7 ± 2.4 bcd | 122.8 ± 2.2 bcde | 121.9 ± 6.1 cde |
| ohb1 sfr6-2 | 119.7 ± 2.2 cd | 119.6 ± 2.4 d | 141.4 ± 4.4 def | 145.3 ± 3.5 def | 123.1 ± 2.8 bcde | 124.7 ± 2.8 cde |
| arf8-2 | 115.0 ± 1.3 d | 123.9 ± 2.3 cd | 141.4 ± 3.5 def | 117.1 ± 1.8 g | 109.9 ± 1.9 efg | 119.2 ± 5.2 cdef |
| ohb1 arf8-2 | 134.2 ± 1.6 b | 156.0 ± 2.3 a | 208.8 ± 5.9 a | 147.6 ± 1.5 cde | 128.1 ± 3.2 bc | 150.2 ± 4.8 ab |
| bpe-2 | 112.5 ± 1.7 d | 104.4 ± 1.6 e | 117.9 ± 3.1 fg | 118.7 ± 2.0 g | 106.7 ± 2.2 fg | 103.7 ± 4.1 ef |
| ohb1 bpe-2 | 133.8 ± 1.7 b | 128.2 ± 1.4 bcd | 171.9 ± 3.3 bc | 168.6 ± 2.6 b | 133.3 ± 1.9 ab | 124.5 ± 3.5 cde |
| ein2-7 | 126.4 ± 1.8 bc | 128.4 ± 2.0 bcd | 150.4 ± 4.2 cde | 156.8 ± 3.9 bcd | 113.3 ± 1.9 defg | 120.7 ± 4.7 cdef |
| ohb1 ein2-7 | 148.2 ± 1.8 a | 164.8 ± 2.7 a | 217.3 ± 4.7 a | 230.8 ± 5.0 a | 130.4 ± 1.8 abc | 134.1 ± 3.9 bcd |
| med25-2 | 128.3 ± 1.7 bc | 135.6 ± 2.3 bc | 168.0 ± 3.3 bc | 160.4 ± 2.0 bc | 124.4 ± 3.1 bcd | 133.0 ± 3.7 bcd |
| ohb1 med25-2 | 133.2 ± 1.9 b | 134.9 ± 1.9 bc | 175.9 ± 4.8 b | 166.7 ± 1.9 b | 126.9 ± 3.2 bc | 131.3 ± 3.6 bcd |
| opr3-1 | 113.1 ± 1.1 d | 118.5 ± 1.2 d | 126.5 ± 2.4 ef | 133.3 ± 2.1 f | 106.2 ± 2.0 g | 116.7 ± 3.3 def |
| ohb1 opr3-1 | 126.7 ± 1.9 bc | 137.5 ± 2.0 b | 168.9 ± 5.0 bc | 158.6 ± 2.1 bc | 124.7 ± 2.0 bcd | 136.0 ± 3.8 bcd |
| bb-3 | 113.9 ± 1.7 d | 134.3 ± 2.2 bc | 141.4 ± 3.7 def | 103.4 ± 2.6 h | 113.4 ± 2.6 defg | 139.7 ± 3.5 bc |
| ohb1 bb-3 | 144.4 ± 2.4 a | 170.4 ± 4.2 a | 237.2 ± 9.1 a | 139.8 ± 2.8 ef | 142.0 ± 4.4 a | 164.5 ± 5.5 a |
| da1-ko1ohb1 da1-ko1 | 115.9 ± 1.6 d | 117.5 ± 2.2 d | 132.0 ± 3.5 ef | 103.6 ± 1.8 h | 112.9 ± 2.2 defg | 127.1 ± 4.5 cd |
| 144.7 ± 1.5 a | 158.9 ± 3.1 a | 222.5 ± 5.2 a | 144.8 ± 2.5 def | 134.3 ± 2.4 ab | 152.7 ± 4.5 ab | |
| Line | 2C | 4C | 8C |
|---|---|---|---|
| WT | 72.5 ± 0.9 | 24.9 ± 0.7 | 2.6 ± 0.3 |
| ohb1 | 58.8 ± 2.7 * | 34.9 ± 2.1 * | 6.2 ± 0.7 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nhat, V.Q.; Kazama, Y.; Ishii, K.; Ohbu, S.; Kunitake, H.; Abe, T.; Hirano, T. Double Mutant Analysis with the Large Flower Mutant, ohbana1, to Explore the Regulatory Network Controlling the Flower and Seed Sizes in Arabidopsis thaliana. Plants 2021, 10, 1881. https://doi.org/10.3390/plants10091881
Nhat VQ, Kazama Y, Ishii K, Ohbu S, Kunitake H, Abe T, Hirano T. Double Mutant Analysis with the Large Flower Mutant, ohbana1, to Explore the Regulatory Network Controlling the Flower and Seed Sizes in Arabidopsis thaliana. Plants. 2021; 10(9):1881. https://doi.org/10.3390/plants10091881
Chicago/Turabian StyleNhat, Vuong Quoc, Yusuke Kazama, Kotaro Ishii, Sumie Ohbu, Hisato Kunitake, Tomoko Abe, and Tomonari Hirano. 2021. "Double Mutant Analysis with the Large Flower Mutant, ohbana1, to Explore the Regulatory Network Controlling the Flower and Seed Sizes in Arabidopsis thaliana" Plants 10, no. 9: 1881. https://doi.org/10.3390/plants10091881
APA StyleNhat, V. Q., Kazama, Y., Ishii, K., Ohbu, S., Kunitake, H., Abe, T., & Hirano, T. (2021). Double Mutant Analysis with the Large Flower Mutant, ohbana1, to Explore the Regulatory Network Controlling the Flower and Seed Sizes in Arabidopsis thaliana. Plants, 10(9), 1881. https://doi.org/10.3390/plants10091881







