Morphological Diversity, Genetic Characterization, and Phytochemical Assessment of the Cypriot Tomato Germplasm
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Cultivation Scheme
2.3. Analysis of Phenotypic Data
2.4. DNA Extraction
2.5. PCR and SSR Genotyping
2.6. Molecular Data Analysis
2.7. Tomato Fruits Sample Preparation
2.8. Quantification of Total Phenols (TP)
2.9. Vitamin C Assessment
2.10. Lycopene and β-Carotene Quantification
2.11. Total Soluble Solids (TSS), pH and Titratable Acidity
2.12. Macro and Micro Nutrient Content in Tomato Fruits
2.13. Generalized Procrustes Analysis (GPA)
3. Results
3.1. Morpho-Agronomical Variation across Cypriot Tomato Landraces
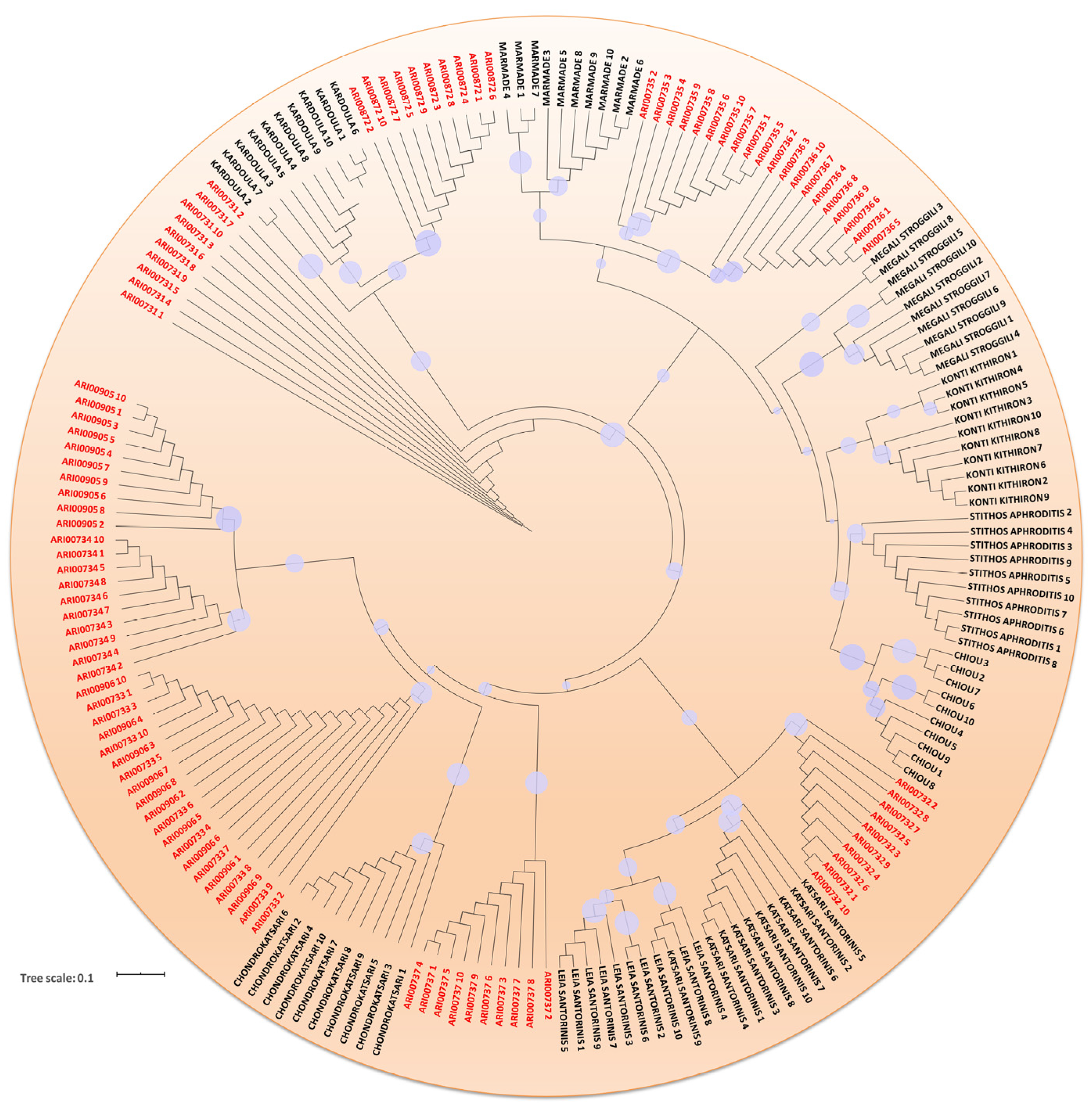
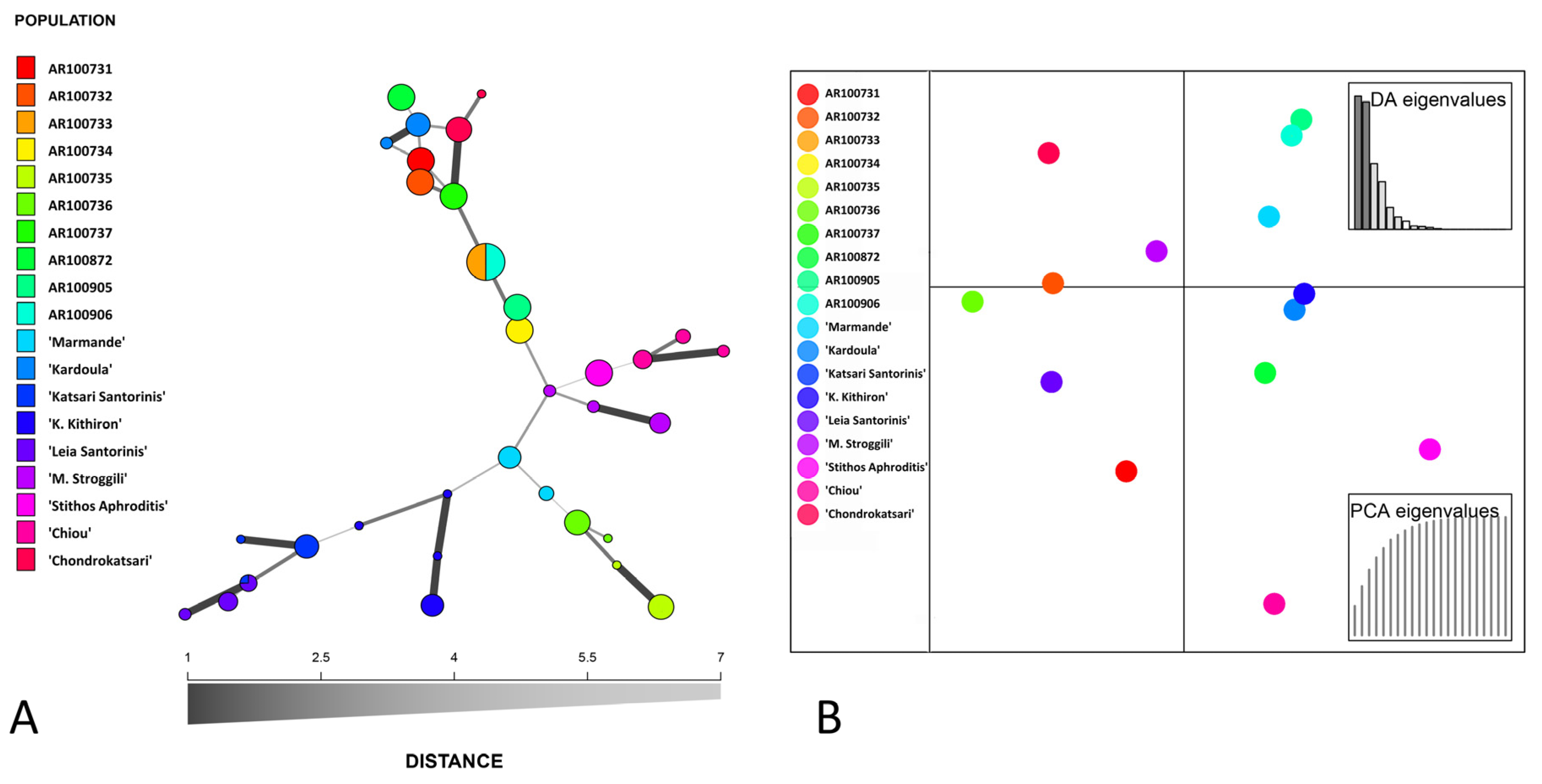
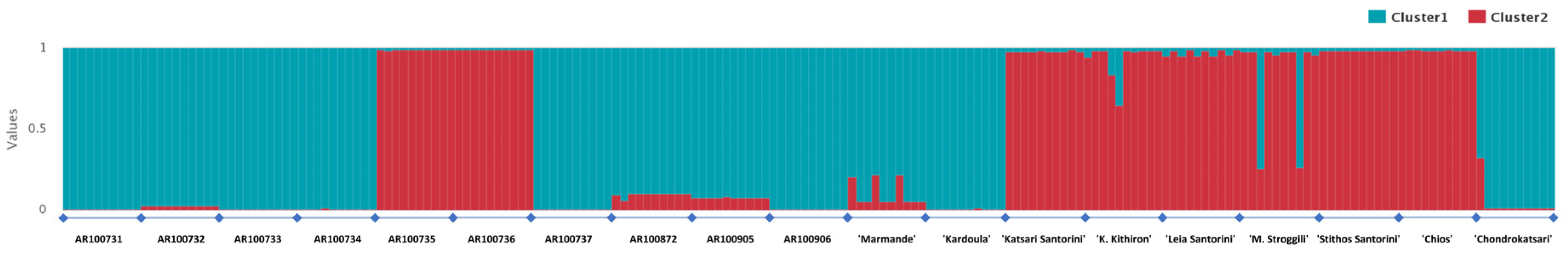
3.2. DNA Fingerprinting, Diversity Indexes and Genetic Relationship across the Tomato Germplasm
3.3. Physicochemical Characterization of Cypriot Tomato Landraces
3.3.1. Mineral Composition
3.3.2. Tomato Fruit Qualitative Characteristics
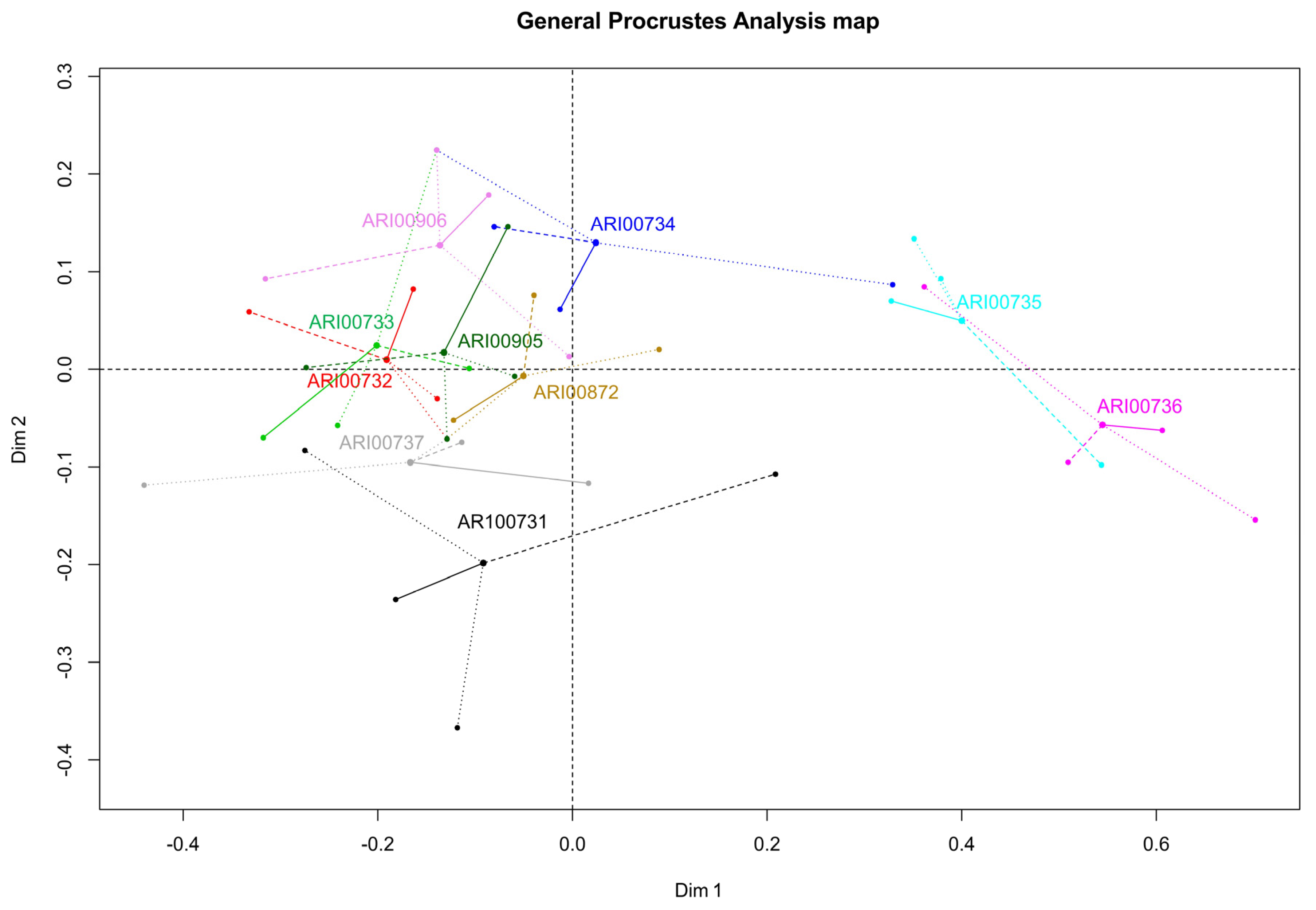
3.3.3. Generalized Procrustes Analysis (GPA)
4. Discussion
4.1. Morphological Parameters
4.2. Genetic Variation
4.3. Phytochemical Characterization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Bergougnoux, V. The history of tomato: From domestication to biopharming. Biotechnol. Adv. 2014, 32, 170–189. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations. Available online: http://www.fao.org/home/en (accessed on 24 April 2021).
- Ohyama, A.; Shirasawa, K.; Matsunaga, H.; Negoro, S.; Miyatake, K.; Yamaguchi, H.; Nunome, T.; Iwata, H.; Fukuoka, H.; Hayashi, T. Bayesian QTL mapping using genome-wide SSR markers and segregating population derived from a cross of two commercial F1 hybrids of tomato. Theor. Appl. Genet. 2017, 130, 1601–1616. [Google Scholar] [CrossRef] [PubMed]
- Tieman, D.; Zhu, G.; Resende, M.F.R., Jr.; Lin, T.; Nguyen, C.; Bies, D.; Rambla, J.L.; Beltran, K.S.O.; Taylor, M.; Zhang, B.; et al. A chemical genetic roadmap to improved tomato flavor. Science 2017, 355, 391–394. [Google Scholar] [CrossRef] [PubMed]
- Klee, H.J.; Giovannoni, J.J. Genetics and control of tomato fruit ripening and quality attributes. Annu. Rev. Genet. 2011, 45, 41–59. [Google Scholar] [CrossRef] [PubMed]
- Schilmiller, A.; Shi, F.; Kim, J.; Charbonneau, A.; Holmes, D.; Jones, A.D.; Last, R.L. Mass spectrometry screening reveals widespread diversity in trichome specialized metabolites of tomato chromosomal substitution lines. Plant J. 2010, 62, 391–403. [Google Scholar] [CrossRef] [Green Version]
- Rosli, H.G.; Martin, G.B. Functional genomics of tomato for the study of plant immunity. Brief. Funct. Genom. 2015, 14, 291–301. [Google Scholar] [CrossRef] [Green Version]
- Nikoloudakis, N.; Pappi, P.; Markakis, E.A.; Charova, S.N.; Fanourakis, D.; Paschalidis, K.; Delis, C.; Tzortzakakis, E.A.; Tsaniklidis, G. Structural diversity and highly specific host-pathogen transcriptional regulation of defensin genes is revealed in tomato. Int. J. Mol. Sci. 2020, 21, 9380. [Google Scholar] [CrossRef]
- Blanca, J.; Cañizares, J.; Cordero, L.; Pascual, L.; Diez, M.J.; Nuez, F. Variation revealed by SNP genotyping and morphology provides insight into the origin of the tomato. PLoS ONE 2012, 7, e48198. [Google Scholar] [CrossRef] [Green Version]
- Bauchet, G.; Causse, M. Genetic Diversity in Tomato (Solanum lycopersicum) and Its Wild Relatives. Guillaume Bauchet and Mathilde Causse; IntechOpen: London, UK, 2012. [Google Scholar]
- Costa, J.M.; Heuvelink, E. Introduction: The tomato crop and industry. In Tomatoes; CABI Publishing: Oxfordshire, UK, 2005. [Google Scholar]
- Alpert, K.B.; Grandillo, S.; Tanksley, S.D. fw 2.2: A major QTL controlling fruit weight is common to both red- and green-fruited tomato species. Theor. Appl. Genet. 1995, 91, 994–1000. [Google Scholar] [CrossRef]
- Liu, X.; Geng, X.; Zhang, H.; Shen, H.; Yang, W. Association and genetic identification of loci for four fruit traits in tomato using InDel markers. Front. Plant Sci. 2017, 8, 1269. [Google Scholar] [CrossRef] [Green Version]
- Grandillo, S.; Ku, H.M.; Tanksley, S.D. Identifying the loci responsible for natural variation in fruit size and shape in tomato. Theor. Appl. Genet. 1999, 99, 978–987. [Google Scholar] [CrossRef]
- Bai, Y.; Lindhout, P. Domestication and breeding of tomatoes: What have we gained and what can we gain in the future? Ann. Bot. 2007, 100, 1085–1094. [Google Scholar] [CrossRef]
- Hyman, J.R.; Gaus, J.; Foolad, M.R. A rapid and accurate method for estimating tomato lycopene content by measuring chromaticity values of fruit purée. J. Am. Soc. Hortic. Sci. 2004, 129, 717–723. [Google Scholar] [CrossRef] [Green Version]
- Miller, J.C.; Tanksley, S.D. RFLP analysis of phylogenetic relationships and genetic variation in the genus Lycopersicon. Theor. Appl. Genet. 1990, 80, 437–448. [Google Scholar] [CrossRef]
- Tamburino, R.; Sannino, L.; Cafasso, D.; Cantarella, C.; Orrù, L.; Cardi, T.; Cozzolino, S.; D’Agostino, N.; Scotti, N. Cultivated tomato (Solanum lycopersicum L.) suffered a severe cytoplasmic bottleneck during domestication: Implications from chloroplast genomes. Plants 2020, 9, 1443. [Google Scholar] [CrossRef]
- Berni, R.; Romi, M.; Parrotta, L.; Cai, G.; Cantini, C. Ancient tomato (Solanum lycopersicum L.) varieties of tuscany have high contents of bioactive compounds. Horticulturae 2018, 4, 51. [Google Scholar] [CrossRef] [Green Version]
- Periago, M.J.; García-Alonso, J.; Jacob, K.; Olivares, A.B.; Bernal, M.J.; Iniesta, M.D.; Martínez, C.; Ros, G. Bioactive compounds, folates and antioxidant properties of tomatoes (Lycopersicum esculentum) during vine ripening. Int. J. Food Sci. Nutr. 2009, 60, 694–708. [Google Scholar] [CrossRef] [PubMed]
- Firuzi, O.; Miri, R.; Tavakkoli, M.; Saso, L. Antioxidant therapy: Current status and future prospects. Curr. Med. Chem. 2011, 18, 3871–3888. [Google Scholar] [CrossRef] [PubMed]
- Mavromatis, A.G.; Athanasouli, V.; Vellios, E.; Khah, E.; Georgiadou, E.C.; Pavli, O.I.; Arvanitoyannis, I.S. Characterization of tomato landraces grown under organic conditions based on molecular marker analysis and determination of fruit quality parameters. J. Agric. Sci. 2013, 5, 239–252. [Google Scholar] [CrossRef]
- Fernie, A.R.; Tadmor, Y.; Zamir, D. Natural genetic variation for improving crop quality. Curr. Opin. Plant Biol. 2006, 9, 196–202. [Google Scholar] [CrossRef] [PubMed]
- McCouch, S.; Baute, G.J.; Bradeen, J.; Bramel, P.; Bretting, P.K.; Buckler, E.; Burke, J.M.; Charest, D.; Cloutier, S.; Cole, G.; et al. Feeding the future. Nat. Cell Biol. 2013, 499, 23–24. [Google Scholar] [CrossRef]
- Fiorani, F.; Schurr, U. Future scenarios for plant phenotyping. Annu. Rev. Plant Biol. 2013, 64, 267–291. [Google Scholar] [CrossRef] [Green Version]
- García-Martínez, S.; Corrado, G.; Ruiz, J.J.; Rao, R. Diversity and structure of a sample of traditional Italian and Spanish tomato accessions. Genet. Resour. Crop. Evol. 2013, 60, 789–798. [Google Scholar] [CrossRef]
- Chavent, M.; Kuentz-Simonet, V.; Labenne, A.; Saracco, J. Multivariate analysis of mixed data: The PCAmixdata R package. arXiv 2014, arXiv:1411.4911. [Google Scholar]
- Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000, 18, 233–234. [Google Scholar] [CrossRef]
- Matschiner, M.; Salzburger, W. TANDEM: Integrating automated allele binning into genetics and genomics workflows. Bioinformatics 2009, 25, 1982–1983. [Google Scholar] [CrossRef] [Green Version]
- Peakall, R.O.D.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Kamvar, Z.N.; Brooks, J.C.; Grünwald, N.J. Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Front. Genet. 2015, 6, 208. [Google Scholar] [CrossRef] [Green Version]
- Clark, L.V.; Jasieniuk, M. polysat: An R package for polyploid microsatellite analysis. Mol. Ecol. Resour. 2011, 11, 562–566. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [Green Version]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Ochoa-Velasco, C.E.; Valadez-Blanco, R.; Salas-Coronado, R.; Sustaita-Rivera, F.; Hernández-Carlos, B.; García-Ortega, S.; Santos-Sánchez, N.F. Effect of nitrogen fertilization and Bacillus licheniformis biofertilizer addition on the antioxidants compounds and antioxidant activity of greenhouse cultivated tomato fruits (Solanum lycopersicum L. var. Sheva). Sci. Hortic. 2016, 201, 338–345. [Google Scholar] [CrossRef]
- Nagata, M.; Yamashita, I. Simple method for simultaneous determination of chlorophyll and carotenoids in tomato fruit. Nippon Shokuhin Kogyo Gakkaishi 1992, 39, 925–928. [Google Scholar] [CrossRef] [Green Version]
- Lagroix, F.; Borradaile, G.J. Tectonics of the circum-Troodos sedimentary cover of Cyprus, from rock magnetic and structural observations. J. Struct. Geol. 2000, 22, 453–469. [Google Scholar] [CrossRef]
- Terzopoulos, P.J.; Bebeli, P.J. DNA and morphological diversity of selected Greek tomato (Solanum lycopersicum L.) landraces. Sci. Hortic. 2008, 116, 354–361. [Google Scholar] [CrossRef]
- Mazzucato, A.; Ficcadenti, N.; Caioni, M.; Mosconi, P.; Piccinini, E.; Reddy Sanampudi, V.R.; Sestili, S.; Ferrari, V. Genetic diversity and distinctiveness in tomato (Solanum lycopersicum L.) landraces: The Italian case study of “A pera Abruzzese”. Sci. Hortic. 2010, 125, 55–62. [Google Scholar] [CrossRef]
- Corrado, G.; Caramante, M.; Piffanelli, P.; Rao, R. Genetic diversity in Italian tomato landraces: Implications for the development of a core collection. Sci. Hortic. 2014, 168, 138–144. [Google Scholar] [CrossRef]
- Renna, M.; D’Imperio, M.; Gonnella, M.; Durante, M.; Parente, A.; Mita, G.; Santamaria, P.; Serio, F. Morphological and chemical profile of three tomato (Solanum lycopersicum L.) landraces of a semi-arid mediterranean environment. Plants 2019, 8, 273. [Google Scholar] [CrossRef] [Green Version]
- Cebolla-Cornejo, J.; Roselló, S.; Nuez, F. Phenotypic and genetic diversity of Spanish tomato landraces. Sci. Hortic. 2013, 162, 150–164. [Google Scholar] [CrossRef] [Green Version]
- Parisi, M.; Aversano, R.; Graziani, G.; Ruggieri, V.; Senape, V.; Sigillo, L.; Barone, A. Phenotypic and molecular diversity in a collection of ‘Pomodoro di Sorrento’’ ‘Italian tomato landrace’. Sci. Hortic. 2016, 203, 143–151. [Google Scholar] [CrossRef]
- Londoño-Giraldo, L.M.; Baena-Pedroza, A.M.; Martinez-Seidel, F.; Corpas-Iguarán, E.; Taborda-Ocampo, G. Gone wild: Integration of antioxidative, physicochemical, volatilomic and sensorial profiles ratify rustic relatives of cherry tomato as ideal mating partners. Sci. Hortic. 2021, 277, 109814. [Google Scholar] [CrossRef]
- Lázaro, A. Tomato landraces: An analysis of diversity and preferences. Plant Genet. Resour. 2018, 16, 315–324. [Google Scholar] [CrossRef]
- Terzopoulos, P.; Bebeli, P. Phenotypic diversity in Greek tomato (Solanum lycopersicum L.) landraces. Sci. Hortic. 2010, 126, 138–144. [Google Scholar] [CrossRef]
- Cortés-Olmos, C.; Valcárcel, J.V.; Roselló, J.; Díez, M.J.; Cebolla-Cornejo, J. Traditional eastern Spanish varieties of tomato. Sci. Agric. 2015, 72, 420–431. [Google Scholar] [CrossRef] [Green Version]
- Elkind, Y.; Galper, O.B.-O.; Scott, J.W.; Kedar, N. Genotype by environment interaction of tomato blossom-end scar size. Euphytica 1990, 50, 91–95. [Google Scholar] [CrossRef]
- Bauchet, G.; Grenier, S.; Samson, N.; Bonnet, J.; Grivet, L.; Causse, M. Use of modern tomato breeding germplasm for deciphering the genetic control of agronomical traits by Genome Wide Association study. Theor. Appl. Genet. 2017, 130, 875–889. [Google Scholar] [CrossRef]
- Cattáneo, R.A.; McCarthy, A.N.; Feingold, S.E. Evidence of genetic diversity within Solanum Lycopersicum, L. ‘Platense’ landrace and identification of various subpopulations. Genet. Resour. Crop. Evol. 2020, 67, 2057–2069. [Google Scholar] [CrossRef]
- Castellana, S.; Ranzino, L.; Beritognolo, I.; Cherubini, M.; Luneia, R.; Villani, F.; Mattioni, C. Genetic characterization and molecular fingerprint of traditional Umbrian tomato (Solanum lycopersicum L.) landraces through SSR markers and application for varietal identification. Genet. Resour. Crop. Evol. 2020, 67, 1807–1820. [Google Scholar] [CrossRef]
- Gonias, E.D.; Ganopoulos, I.; Mellidou, I.; Bibi, A.C.; Kalivas, A.; Mylona, P.V.; Osanthanunkul, M.; Tsaftaris, A.; Madesis, P.; Doulis, A.G. Exploring genetic diversity of tomato (Solanum lycopersicum L.) germplasm of genebank collection employing SSR and SCAR markers. Genet. Resour. Crop. Evol. 2019, 66, 1295–1309. [Google Scholar] [CrossRef]
- Todorovska, E.; Ivanova, A.; Ganeva, D.; Pevicharova, G.; Molle, E.; Bojinov, B.; Radkova, M.; Danailov, Z. Assessment of genetic variation in Bulgarian tomato (Solanum lycopersicum L.) genotypes, using fluorescent SSR genotyping platform. Biotechnol. Biotechnol. Equip. 2014, 28, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Constandinou, S.; Nikoloudakis, N.; Kyratzis, A.C.; Katsiotis, A. Genetic diversity of Avena ventricosa populations along an ecogeographical transect in Cyprus is correlated to environmental variables. PLoS ONE 2018, 13, e0193885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwal, S.; Rao, A.V. Tomato lycopene and its role in human health and chronic diseases. CMAJ 2000, 163, 739–744. [Google Scholar] [PubMed]
- Stevens, R.; Buret, M.; Duffé, P.; Garchery, C.; Baldet, P.; Rothan, C.; Causse, M. Candidate genes and quantitative trait loci affecting fruit ascorbic acid content in three tomato populations. Plant Physiol. 2007, 143, 1943–1953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carli, P.; Arima, S.; Fogliano, V.; Tardella, L.; Frusciante, L.; Ercolano, M.R. Use of network analysis to capture key traits affecting tomato organoleptic quality. J. Exp. Bot. 2009, 60, 3379–3386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carli, P.; Barone, A.; Fogliano, V.; Frusciante, L.; Ercolano, M.R. Dissection of genetic and environmental factors involved in tomato organoleptic quality. BMC Plant Biol. 2011, 11, 58. [Google Scholar] [CrossRef] [Green Version]
- Figàs, M.R.; Prohens, J.; Raigón, M.D.; Fita, A.; García-Martínez, M.D.; Casanova, C.; Borràs, D.; Plazas, M.; Andújar, I.; Soler, S. Characterization of composition traits related to organoleptic and functional quality for the differentiation, selection and enhancement of local varieties of tomato from different cultivar groups. Food Chem. 2015, 187, 517–524. [Google Scholar] [CrossRef] [Green Version]
- Baldina, S.; Picarella, M.E.; Troise, A.D.; Pucci, A.; Ruggieri, V.; Ferracane, R.; Barone, A.; Fogliano, V.; Mazzucato, A. Metabolite profiling of Italian tomato landraces with different fruit types. Front. Plant Sci. 2016, 7, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Toor, R.K.; Savage, G.P. Antioxidant activity in different fractions of tomatoes. Food Res. Int. 2005, 38, 487–494. [Google Scholar] [CrossRef]
- Tsaniklidis, G.; Delis, C.; Nikoloudakis, N.; Katinakis, P.; Aivalakis, G. Low temperature storage affects the ascorbic acid metabolism of cherry tomato fruits. Plant Physiol. Biochem. 2014, 84, 149–157. [Google Scholar] [CrossRef]
- Scarano, A.; Olivieri, F.; Gerardi, C.; Liso, M.; Chiesa, M.; Chieppa, M.; Frusciante, L.; Barone, A.; Santino, A.; Rigano, M.M. Selection of tomato landraces with high fruit yield and nutritional quality under elevated temperatures. J. Sci. Food Agric. 2020, 100, 2791–2799. [Google Scholar] [CrossRef] [PubMed]
- Kasim, M.U.; Kasim, R. Postharvest UV-B treatments increased fructose content of tomato (Solanum lycopersicon L. cv. Tayfun F1) harvested at different ripening stages. Food Sci. Technol. 2015, 35, 742–749. [Google Scholar] [CrossRef]
- Gautier, H.; Lopez-Lauri, F.; Massot, C.; Murshed, R.; Marty, I.; Grasselly, D.; Keller, C.; Sallanon, H.; Genard, M. Impact of ripening and salinity on tomato fruit ascorbate content and enzymatic activities related to ascorbate recycling. Funct. Plant Sci. Biotechnol. 2010, 4, 66–75. [Google Scholar]
- Luengwilai, K.; Fiehn, O.E.; Beckles, D.M. Comparison of leaf and fruit metabolism in two tomato (Solanum lycopersicum L.) genotypes varying in total soluble solids. J. Agric. Food Chem. 2010, 58, 11790–11800. [Google Scholar] [CrossRef] [PubMed]
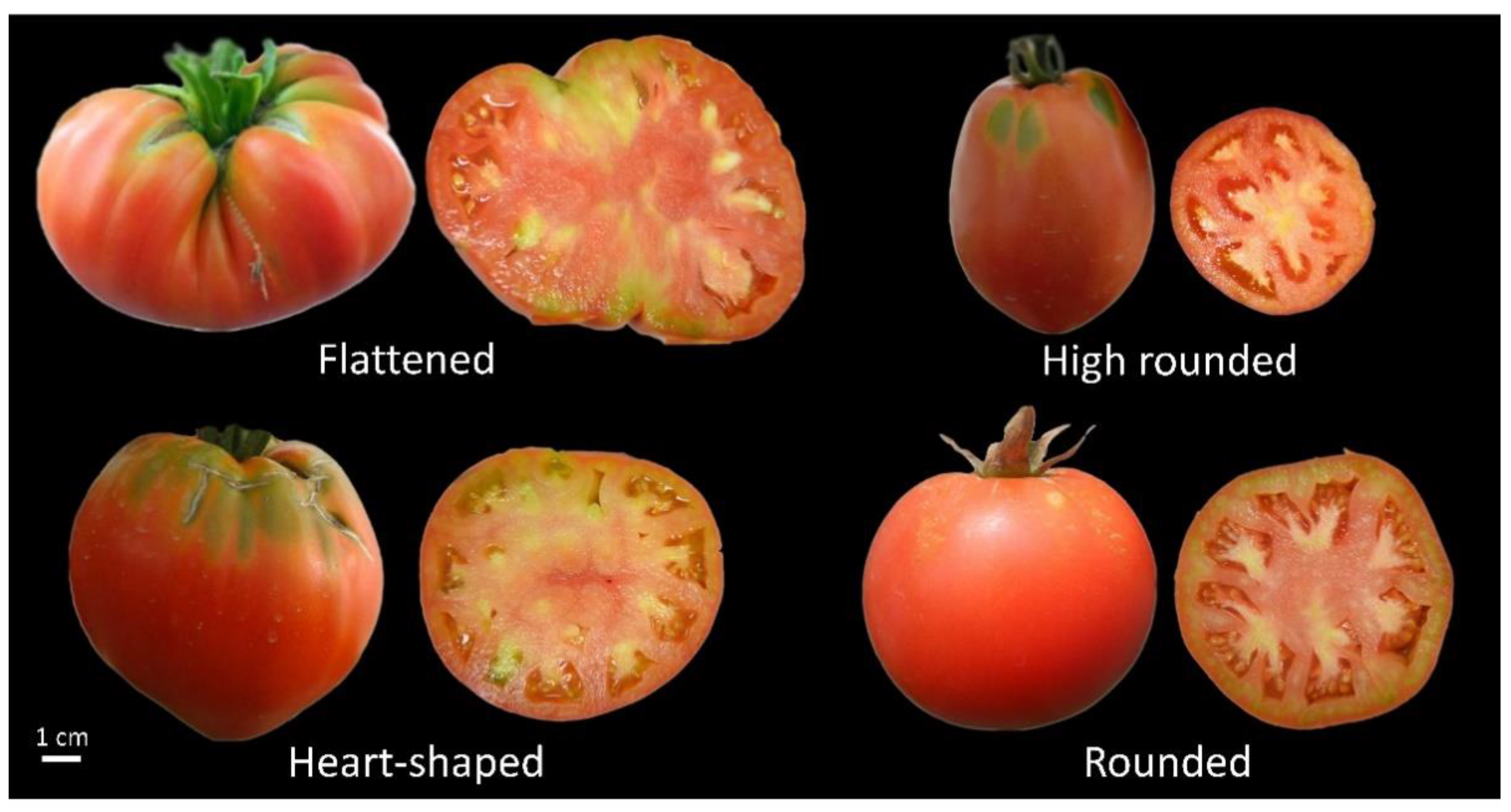


| No | Code | Origin | Fruit Shape | Fruit Size |
|---|---|---|---|---|
| 1. | AR100731 | Cypriot | Flattened | Very large |
| 2. | ARI00732 | Cypriot | Flattened | Very large |
| 3. | ARI00733 | Cypriot | Heart-shaped | Large |
| 4. | ARI00734 | Cypriot | High rounded | Small |
| 5. | ARI00735 | Cypriot | Rounded | Medium |
| 6. | ARI00736 | Cypriot | Rounded | Medium |
| 7. | ARI00737 | Cypriot | Flattened | Very large |
| 8. | ARI00872 | Cypriot | Flattened | Very large |
| 9. | ARI00905 | Cypriot | Heart-shaped | Large |
| 10. | ARI00906 | Cypriot | Heart-shaped | Large |
| 11. | ‘Marmande’ | French | Rounded | Medium |
| 12. | ‘Kardoula’ | Greek | Heart-shaped | Medium |
| 13. | ‘Katsari Santorinis’ | Greek | Rounded | Small |
| 14. | ‘Konti Kithiron’ | Greek | Elliptical shaped | Medium |
| 15. | ‘Leia Santorinis’ | Greek | Rounded | Small |
| 16. | ‘Megali Stroggili’ | Greek | Rounded | Large |
| 17. | ‘Stithos Aphroditis’ | Greek | Elongated | Medium |
| 18. | ‘Chiou’ | Greek | Rounded | Small |
| 19. | ‘Chondrokatsari’ | Greek | Flattened | Large |
| Genotypes | N | Na | Ne | I | Ho | He | uHe | F | MLGs |
|---|---|---|---|---|---|---|---|---|---|
| ARI00731 | 10 | 1.400 | 1.400 | 0.277 | 0.400 | 0.200 | 0.211 | −1.000 | 1 |
| ARI00732 | 10 | 1.300 | 1.300 | 0.208 | 0.300 | 0.150 | 0.158 | −1.000 | 1 |
| ARI00733 | 10 | 1.200 | 1.200 | 0.139 | 0.200 | 0.100 | 0.105 | −1.000 | 1 |
| ARI00734 | 10 | 1.200 | 1.200 | 0.139 | 0.200 | 0.100 | 0.105 | −1.000 | 1 |
| ARI00735 | 10 | 1.300 | 1.210 | 0.158 | 0.210 | 0.110 | 0.115 | −0.684 | 2 |
| ARI00736 | 10 | 1.400 | 1.320 | 0.240 | 0.290 | 0.168 | 0.176 | −0.455 | 2 |
| ARI00737 | 10 | 1.300 | 1.300 | 0.208 | 0.300 | 0.150 | 0.158 | −1.000 | 1 |
| ARI00872 | 10 | 1.300 | 1.300 | 0.208 | 0.300 | 0.150 | 0.158 | −1.000 | 1 |
| ARI00905 | 10 | 1.300 | 1.300 | 0.208 | 0.300 | 0.150 | 0.158 | −1.000 | 1 |
| ARI00906 | 10 | 1.200 | 1.200 | 0.139 | 0.200 | 0.100 | 0.105 | −1.000 | 1 |
| ‘Marmande’ | 10 | 1.400 | 1.345 | 0.261 | 0.200 | 0.184 | 0.194 | 0.000 | 2 |
| ‘Kardoula’ | 10 | 1.200 | 1.192 | 0.137 | 0.180 | 0.098 | 0.103 | −0.833 | 2 |
| ‘Katsari Santorinis’ | 10 | 1.400 | 1.242 | 0.187 | 0.200 | 0.123 | 0.129 | −0.278 | 3 |
| ‘Konti Kithiron’ | 10 | 1.500 | 1.281 | 0.235 | 0.230 | 0.157 | 0.165 | −0.274 | 4 |
| ‘Leia Santorinis’ | 10 | 1.500 | 1.422 | 0.310 | 0.220 | 0.218 | 0.229 | −0.022 | 3 |
| ‘Megali Stroggili’ | 10 | 1.500 | 1.332 | 0.269 | 0.240 | 0.178 | 0.187 | −0.217 | 3 |
| ‘Stithos Aphroditis’ | 10 | 1.100 | 1.100 | 0.069 | 0.100 | 0.050 | 0.053 | −1.000 | 1 |
| ‘Chiou’ | 10 | 1.300 | 1.265 | 0.198 | 0.180 | 0.140 | 0.147 | −0.222 | 3 |
| ‘Chondrokatsari’ | 10 | 1.300 | 1.222 | 0.171 | 0.200 | 0.118 | 0.124 | −0.333 | 2 |
| Average | 10 | 1.321 | 1.270 | 0.198 | 0.234 | 0.139 | 0.146 | −0.648 | 33 (Total) |
| Locus | Number of Alleles | PIC | Dp | Hs | Ht | Gst | Gprime_st | D |
|---|---|---|---|---|---|---|---|---|
| EST253712 | 3 | 0.346 | 0.889 | 0.023 | 0.302 | 0.923 | 0.949 | 0.301 |
| EST258529 | 4 | 0.372 | 0.803 | 0.414 | 0.593 | 0.301 | 0.534 | 0.322 |
| LE20592 | 3 | 0.352 | 0.877 | 0.055 | 0.193 | 0.712 | 0.766 | 0.153 |
| LELEUZIP | 4 | 0.375 | 0.753 | 0.528 | 0.598 | 0.117 | 0.260 | 0.156 |
| LEMDDNA | 3 | 0.374 | 0.749 | 0.290 | 0.590 | 0.509 | 0.735 | 0.446 |
| LESSRPSPGb | 3 | 0.347 | 0.888 | 0.048 | 0.640 | 0.924 | 0.975 | 0.657 |
| TMS42 | 3 | 0.346 | 0.856 | 0.005 | 0.409 | 0.987 | 0.993 | 0.429 |
| TMS52 | 4 | 0.310 | 0.937 | 0.005 | 0.732 | 0.993 | 0.998 | 0.771 |
| TMS58 | 2 | 0.294 | 0.724 | 0.018 | 0.081 | 0.780 | 0.804 | 0.068 |
| TMS59 | 3 | 0.351 | 0.889 | 0.076 | 0.322 | 0.763 | 0.836 | 0.281 |
| Average | 3.2 | 0.347 | 0.837 | 0.146 | 0.446 | 0.672 | 0.801 | 0.370 |
| Landrace | N (g/kg) | P (g/kg) | Ca (g/kg) | Mg (g/kg) | K (g/kg) | Na (g/kg) | Zn (mg/kg) | Mn (mg/kg) | Cu (mg/kg) |
|---|---|---|---|---|---|---|---|---|---|
| AR100731 | 21.0 ± 0.27 bc | 7.12 ± 0.11 b | 2.64 ± 0.09 b | 1.66 ± 0.02 b | 36.32 ± 3.54 b | 1.34 ± 0.01 de | 23.14 ± 1.60 gh | 17.22 ± 0.25 b | 8.61 ± 0.18 de |
| ARI00732 | 20.2 ± 0.48 c | 4.90 ± 0.04 f | 1.38 ± 0.16 e | 1.60 ± 0.01 bc | 31.93 ± 0.98 cd | 1.39 ± 0.00 c | 29.62 ± 0.94 de | 13.50 ± 0.51 bc | 6.55 ± 0.45 ef |
| ARI00733 | 18.6 ± 0.32 d | 3.61 ± 0.05 g | 1.64 ± 0.05 de | 1.43 ± 0.02 d | 31.86 ± 0.23 cd | 1.12 ± 0.00 g | 22.08 ± 1.81 h | 12.53 ± 0.55 c | 9.63 ± 1.32 d |
| ARI00734 | 21.6 ± 0.22 b | 5.49 ± 0.09 d | 2.19 ± 0.14 c | 1.46 ± 0.03 d | 32.50 ± 0.45 bd | 1.20 ± 0.01 f | 30.53 ± 0.54 cd | 14.08 ± 1.48 bc | 12.40 ± 0.95 c |
| ARI00735 | 21.3 ± 0.17 b | 7.22 ± 0.03 b | 4.24 ± 0.02 a | 1.65 ± 0.05 bc | 32.17 ± 0.03 bd | 1.37 ± 0.01 cd | 36.54 ± 0.45 b | 22.56 ± 1.53 a | 16.76 ± 1.25 b |
| ARI00736 | 20.2 ± 0.28 c | 8.00 ± 0.08 a | 3.96 ± 0.13 a | 2.08 ± 0.03 a | 43.23 ± 0.26 a | 1.33 ± 0.03 e | 41.65 ± 0.35 a | 17.21 ± 1.06 b | 19.83 ± 0.30 a |
| ARI00737 | 22.0 ± 0.069 b | 6.49 ± 0.13 c | 1.85 ± 0.03 d | 1.65 ± 0.01 bc | 33.21 ± 1.69 bd | 1.33 ± 0.01 e | 25.85 ± 1.53 fg | 17.28 ± 1.15 b | 18.12 ± 0.13 ab |
| ARI00872 | 21.0 ± 0.14 bc | 6.61 ± 0.08 c | 2.24 ± 0.01 c | 1.56 ± 0.01 c | 34.82 ± 0.08 bc | 1.36 ± 0.01 ce | 27.25 ± 0.35 ef | 12.18 ± 2.06 c | 10.20 ± 0.12 cd |
| ARI00905 | 21.0 ± 0.15 bc | 4.89 ± 0.03 f | 1.72 ± 0.06 d | 1.56 ± 0.06 c | 32.18 ± 0.21 bd | 1.68 ± 0.03 a | 33.25 ± 1.20 c | 14.52 ± 0.53 bc | 5.91 ± 0.78 f |
| ARI00906 | 23.6 ± 0.55 a | 5.25 ± 0.07 e | 1.34 ± 0.07 e | 1.63 ± 0.02 bc | 29.73 ± 0.82 d | 1.57 ± 0.01 b | 31.87 ± 1.36 cd | 14.42 ± 1.13 bc | 8.65 ± 0.25 de |
| Average | 21.5 ± 1.38 | 5.96 ± 1.35 | 2.32 ± 1.00 | 1.63 ± 0.18 | 33.79 ± 3.90 | 1.37 ± 0.16 | 30.17 ± 5.92 | 15.53 ± 3.20 | 11.66 ± 4.81 |
| CV% | 6.55 | 22.59 | 43.13 | 10.76 | 11.57 | 11.48 | 19.75 | 20.79 | 41.40 |
| ANOVA p value | p = 8 × 10−8 | p = 5.8 × 10−10 | p = 5.2 × 10−9 | p = 1.8 × 10−6 | p = 0.0015 | p = 1.2 × 10−9 | p = 4 × 10−7 | p = 0.0011 | p = 4.5 × 10−7 |
| Landrace | pH | TA (g/L) | TSS (°Brix) | TSS/TA | Vitamin C (mg/100 g FW) | TP (mg GAE/100 g FW) | Lycopene (mg/100 g FW) | β-Carotene (mg/100 g FW) |
|---|---|---|---|---|---|---|---|---|
| AR100731 | 4.56 ± 0.02 d | 3.04 ± 0.05 d | 3.80 ± 0.10 d | 12.48 ± 0.25 bc | 39.60 ± 1.55 b | 7.95 ± 0.64 ab | 4.37 ± 0.19 d | 0.68 ± 0.04 bcd |
| ARI00732 | 4.55 ± 0.02 d | 3.90 ± 0.03 a | 5.07 ± 0.06 ab | 12.83 ± 0.12 b | 24.40 ± 2.98 d | 5.67 ± 0.22 c | 5.47 ± 0.03 b | 0.65 ± 0.03 bcd |
| ARI00733 | 4.59 ± 0.01 cd | 3.63 ± 0.07 b | 4.80 ± 0.06 ab | 13.22 ± 0.37 b | 30.80 ± 0.19 c | 5.36 ± 0.31 cd | 4.52 ± 0.03 cd | 0.42 ± 0.07 e |
| ARI00734 | 4.66 ± 0.02 ab | 3.06 ± 0.05 d | 3.33 ± 0.07 ef | 10.89 ± 0.13 d | 30.00 ± 3.22 c | 4.50 ± 0.16 d | 1.42 ± 0.05 g | 0.54 ± 0.04 ce |
| ARI00735 | 4.68 ± 0.01 a | 3.05 ± 0.06 d | 3.20 ± 0.15 f | 10.49 ± 0.35 d | 48.02 ± 0.94 a | 8.87 ± 0.29 a | 1.49 ± 0.07 g | 0.92 ± 0.02 a |
| ARI00736 | 4.63 ± 0.03 ac | 3.12 ± 0.03 d | 3.53 ± 0.07 de | 11.33 ± 0.28 cd | 47.30 ± 1.65 a | 7.21 ± 0.21 b | 1.55 ± 0.05 g | 0.79 ± 0.10 ab |
| ARI00737 | 4.60 ± 0.01 bcd | 3.31 ± 0.04 c | 5.07 ± 0.18 a | 15.29 ± 0.35 a | 31.12 ± 0.41 c | 7.78 ± 0.57 ab | 4.84 ± 0.10 c | 0.69 ± 0.04 bc |
| ARI00872 | 4.67 ± 0.01 a | 2.85 ± 0.05 e | 3.23 ± 0.06 f | 11.32 ± 0.16 cd | 31.45 ± 0.23 c | 7.33 ± 0.34 b | 2.22 ± 0.24 f | 0.63 ± 0.03 bcd |
| ARI00905 | 4.59 ± 0.01 cd | 3.32 ± 0.03 c | 4.37 ± 0.03 c | 13.16 ± 0.02 b | 26.17 ± 0.76 cd | 7.91 ± 0.20 ab | 5.85 ± 0.04 a | 0.73 ± 0.02 b |
| ARI00906 | 4.56 ± 0.03 d | 3.70 ± 0.02 b | 4.73 ± 0.04 b | 12.81 ± 0.17 b | 24.37 ± 1.40 d | 6.93 ± 0.13 b | 3.83 ± 0.11 e | 0.49 ± 0.11 de |
| Average | 4.61 ± 0.05 | 3.30 ± 0.33 | 4.11 ± 0.75 | 12.40 ± 1.43 | 33.28 ± 8.78 | 6.95 ± 1.40 | 3.55 ± 1.67 | 0.65 ± 0.16 |
| CV% | 0.99 | 9.83 | 17.89 | 10.89 | 24.88 | 18.58 | 46.11 | 21.21 |
| ANOVA p value | p = 0.00018 | p = 2.2 × 10−12 | p = 5.3 × 10−13 | p = 4.5 × 10−10 | p = 2.5 × 10−09 | p = 3.7 × 10−7 | p = 2.2 × 10−16 | p = 0.00037 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Athinodorou, F.; Foukas, P.; Tsaniklidis, G.; Kotsiras, A.; Chrysargyris, A.; Delis, C.; Kyratzis, A.C.; Tzortzakis, N.; Nikoloudakis, N. Morphological Diversity, Genetic Characterization, and Phytochemical Assessment of the Cypriot Tomato Germplasm. Plants 2021, 10, 1698. https://doi.org/10.3390/plants10081698
Athinodorou F, Foukas P, Tsaniklidis G, Kotsiras A, Chrysargyris A, Delis C, Kyratzis AC, Tzortzakis N, Nikoloudakis N. Morphological Diversity, Genetic Characterization, and Phytochemical Assessment of the Cypriot Tomato Germplasm. Plants. 2021; 10(8):1698. https://doi.org/10.3390/plants10081698
Chicago/Turabian StyleAthinodorou, Filio, Petros Foukas, Georgios Tsaniklidis, Anastasios Kotsiras, Antonios Chrysargyris, Costas Delis, Angelos C. Kyratzis, Nikolaos Tzortzakis, and Nikolaos Nikoloudakis. 2021. "Morphological Diversity, Genetic Characterization, and Phytochemical Assessment of the Cypriot Tomato Germplasm" Plants 10, no. 8: 1698. https://doi.org/10.3390/plants10081698
APA StyleAthinodorou, F., Foukas, P., Tsaniklidis, G., Kotsiras, A., Chrysargyris, A., Delis, C., Kyratzis, A. C., Tzortzakis, N., & Nikoloudakis, N. (2021). Morphological Diversity, Genetic Characterization, and Phytochemical Assessment of the Cypriot Tomato Germplasm. Plants, 10(8), 1698. https://doi.org/10.3390/plants10081698










