Genome-Wide Analysis of Glycoside Hydrolase Family 35 Genes and Their Potential Roles in Cell Wall Development in Medicago truncatula
Abstract
:1. Introduction
2. Results
2.1. Identification and Analyses of BGAL Gene Family Members in M. truncatula
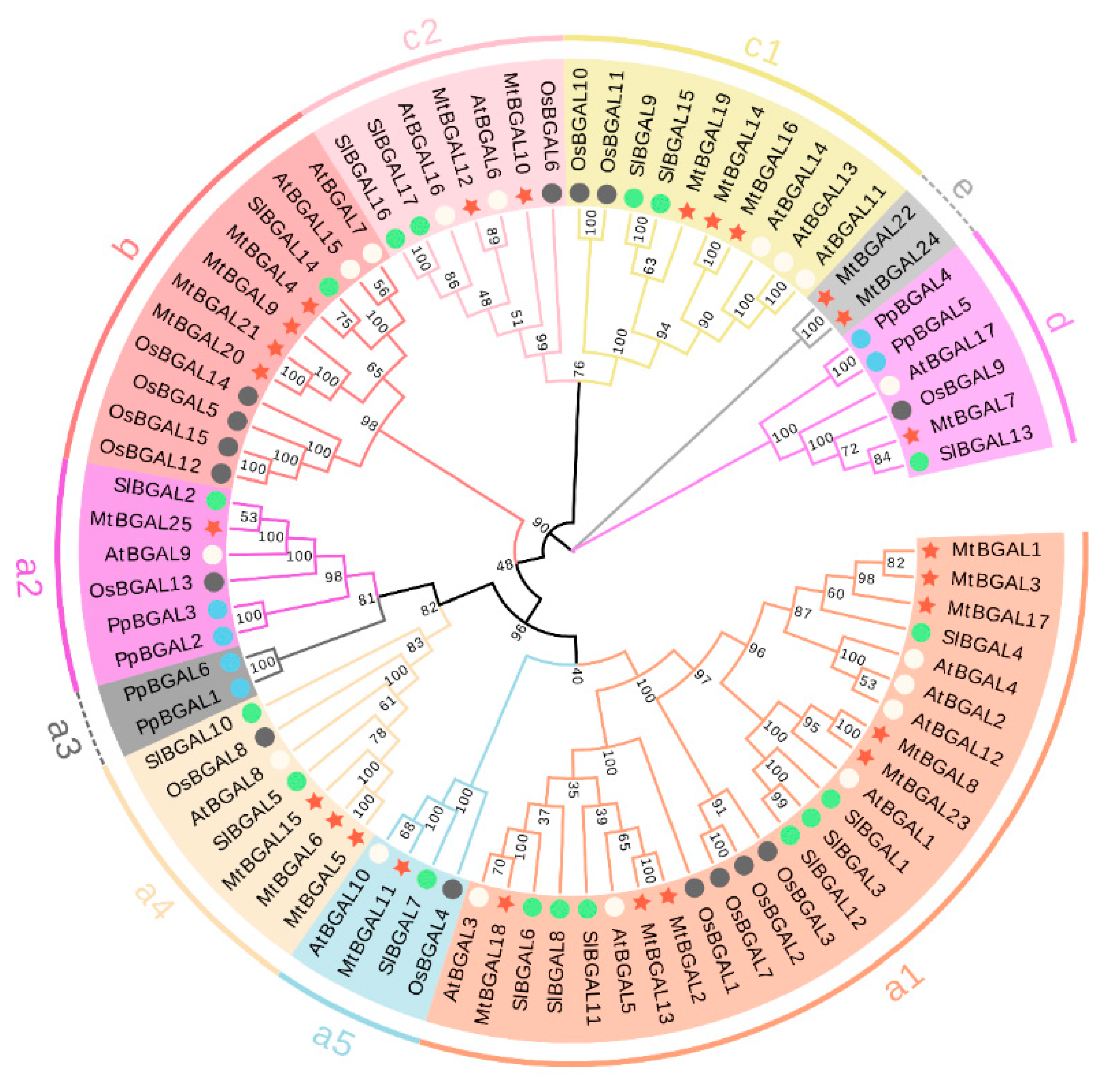
2.2. Multiple Sequence Alignment, Phylogenetic Analysis and Classification of the MtBGAL Genes
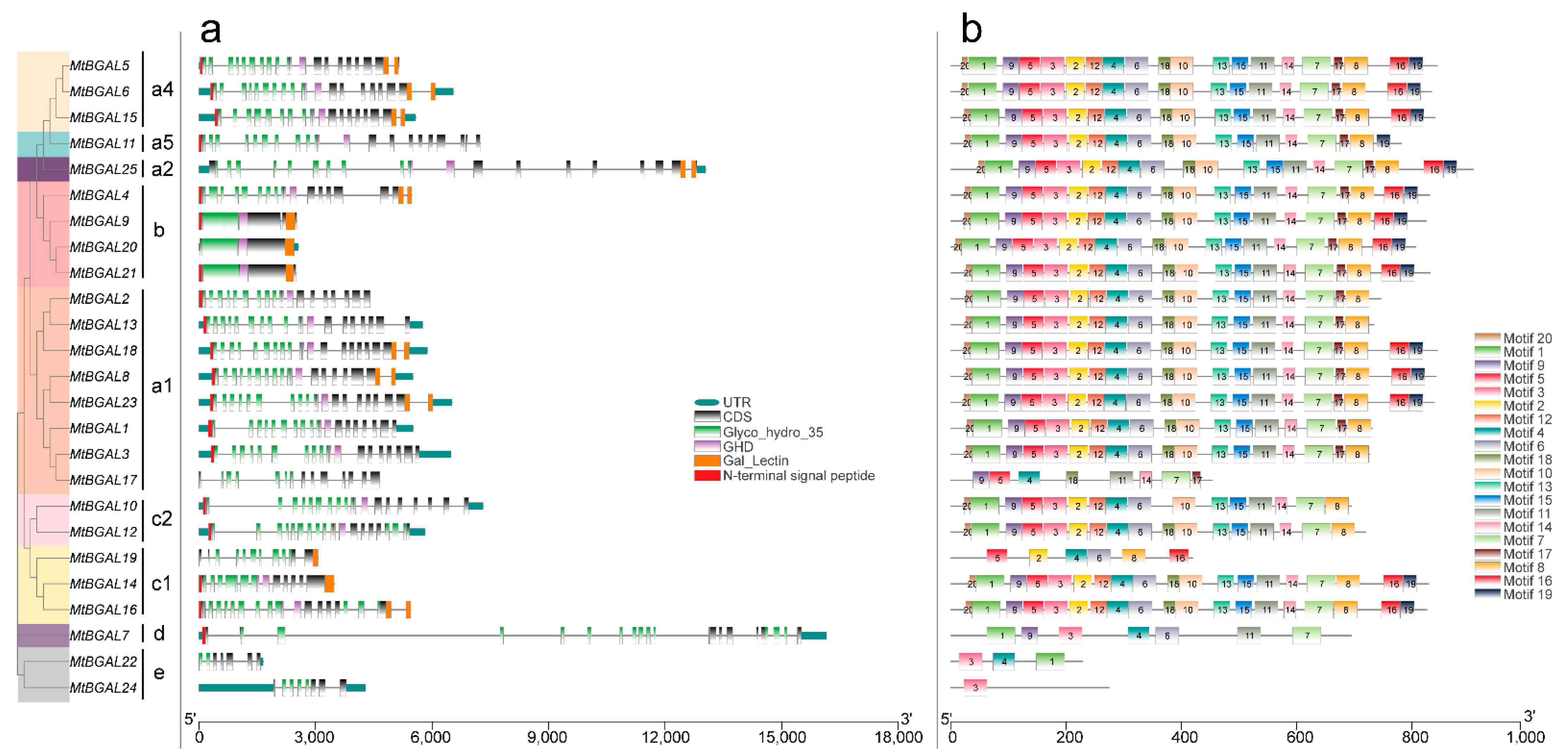
2.3. Analyses of Gene Structure, Conserved Domain and Motif Pattern of MtBGAL Genes
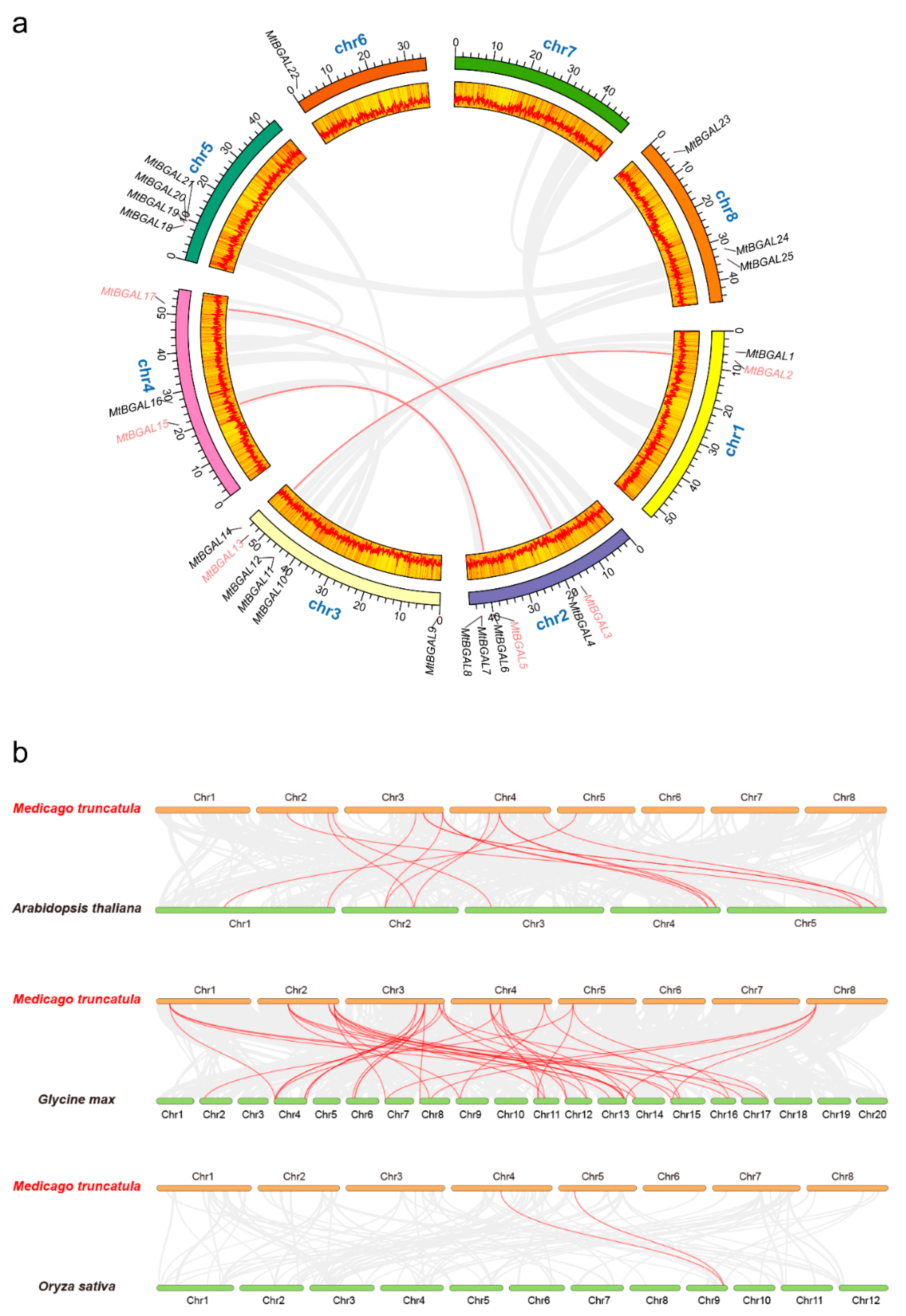
2.4. Chromosomal Distribution and Synteny Analysis of the MtBGAL Genes
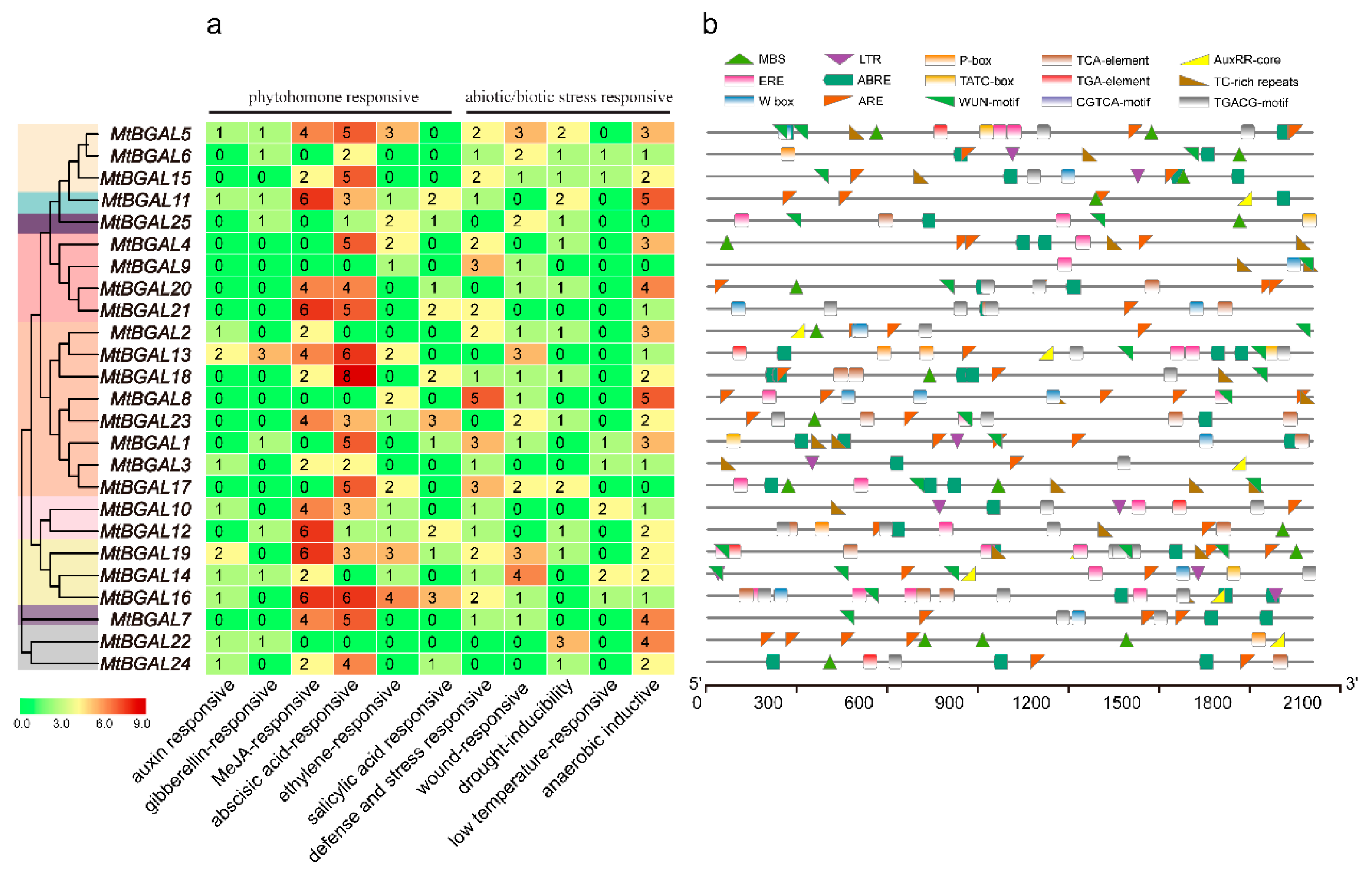
2.5. Analysis of the Cis-Acting Elements in the Promoter Regions of MtBGAL Genes
2.6. Analysis of Co-Expression Network of Cell Wall Genes with WGCNA
2.7. Expression Profiles of the MtBGAL Genes and Identification of Stem-Specific MtBGAL Genes
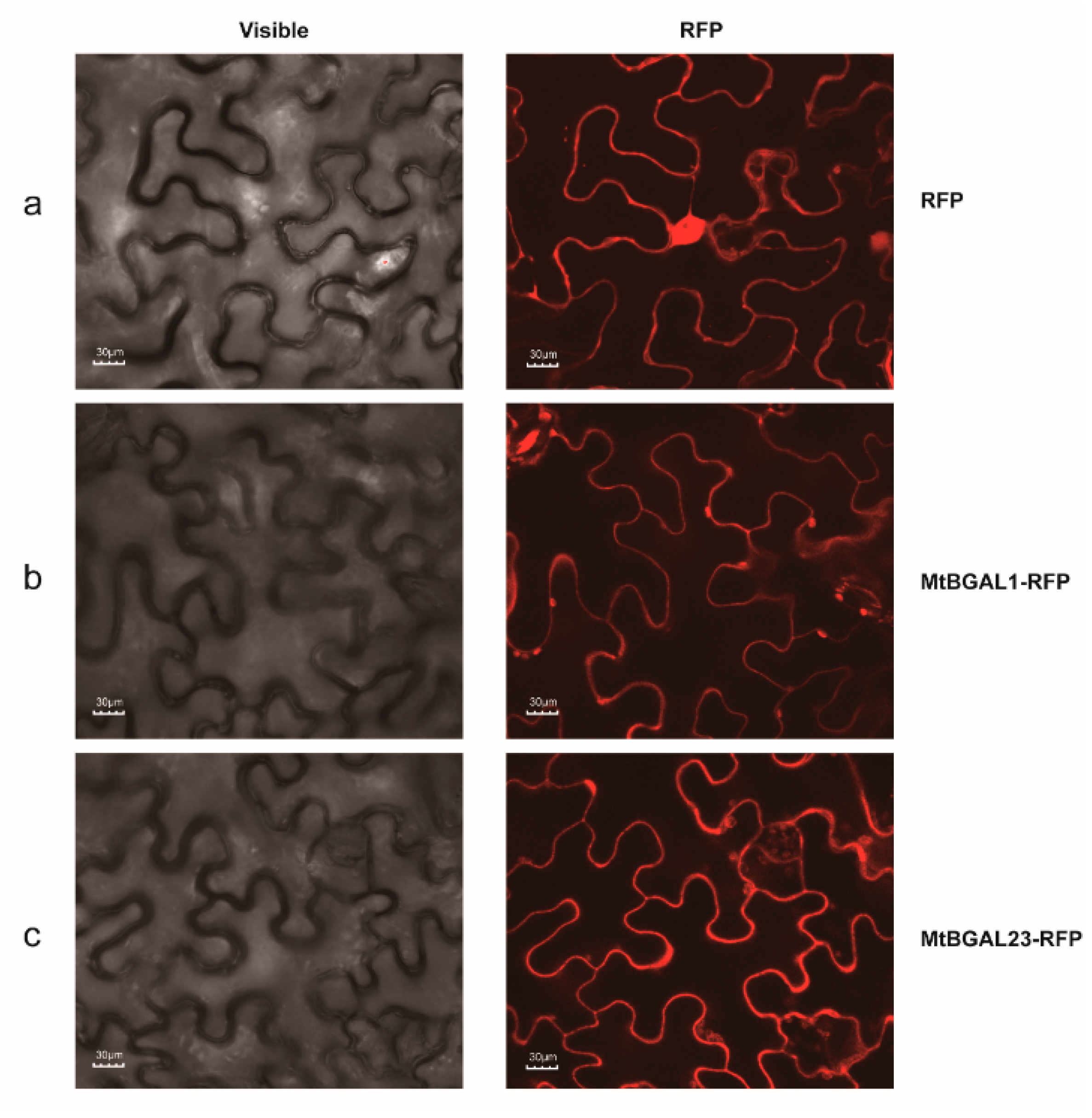
2.8. Subcellular Localization of MtBGAL1 and MtBGAL23
3. Discussion
4. Materials and Methods
4.1. Identification of MtBGAL Genes of the GH35 Family in the M. truncatula Genome
4.2. Sequence Analyses and Structural Characterization of the MtBGAL Genes
4.3. Phylogenetic Analysis and Classification of the MtBGAL Genes
4.4. Analyses of the Chromosomal Distribution and Gene Duplication of the MtBGAL Genes
4.5. Analysis of Cis-Acting Elements in the Promoter Region of the MtBGAL Genes
4.6. Analysis on Gene Co-Expression Network and GO/KEGG Enrichment
4.7. Plasmid Construction and Agrobacterium-Infection Assays in Nicotiana Benthamiana
4.8. Plant Materials
4.9. RNA Extraction and Gene Expression Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lombard, V.; Golaconda, R.H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, 490–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; McCarter, J.D.; Okamura-Oho, Y.; Yaghi, F.; Hinek, A.; Withers, S.G.; Callahan, J.W. Kinetic mechanism and characterization of human β-galactosidase precursor secreted by permanently transfected Chinese hamster ovary cells. Biochem. J. 1994, 15, 281–288. [Google Scholar] [CrossRef]
- Rojas, A.L.; Nagem, R.A.; Neustroev, K.N.; Arand, M.; Adamska, M.; Eneyskaya, E.V.; Kulminskaya, A.A.; Garratt, R.C.; Golubev, A.M.; Polikarpov, I. Crystal structures of β-galactosidase from Penicillium sp. and its complex with galactose. J. Mol. Cell Biol. 2004, 343, 1281–1292. [Google Scholar] [CrossRef]
- Paniagua, C.; Blanco-Portales, R.; Barcelo-Munoz, M.; Garcia-Gago, J.A.; Waldron, K.W.; Quesada, M.A.; Munoz-Blanco, J.; Mercado, J.A. Antisense down-regulation of the strawberry beta-galactosidase gene FabetaGal4 increases cell wall galactose levels and reduces fruit softening. J. Exp. Bot. 2016, 67, 619–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eda, M.; Matsumoto, T.; Ishimaru, M.; Tada, T. Structural and functional analysis of tomato β-galactosidase 4: Insight into the substrate specificity of the fruit softening-related enzyme. Plant J. 2016, 86, 300–307. [Google Scholar] [CrossRef] [Green Version]
- Javier, S.; Cristina, G.; Natalia, L.; Esteban, G.; Gloria, R.; Ignacio, Z. AtBGAL10 is the main xyloglucan β-galactosidase in Arabidopsis, and its absence results in unusual xyloglucan subunits and growth defects. Plant Physiol. 2012, 158, 1146–1157. [Google Scholar]
- Moneo-Sánchez, M.; Alonso-Chico, A.; Knox, J.P.; Dopico, B.; Labrador, E.; Martín, I. β-(1,4)-Galactan remodelling in Arabidopsis cell walls affects the xyloglucan structure during elongation. Planta 2019, 249, 351–362. [Google Scholar] [CrossRef] [Green Version]
- Roach, M.J.; Mokshina, N.Y.; Badhan, A.; Snegireva, A.V.; Hobson, N.; Deyholos, M.K.; Gorshkova, T.A. Development of cellulosic secondary walls in flax fibers requires β-galactosidase. Plant Physiol. 2011, 156, 1351–1363. [Google Scholar] [CrossRef] [Green Version]
- Dean, G.H.; Zheng, H.; Tewari, J.; Huang, J.; Young, D.S.; Hwang, Y.T.; Western, T.L.; Carpita, N.C.; McCann, M.C.; Mansfield, S.D.; et al. The Arabidopsis MUM2 gene encodes a beta-galactosidase required for the production of seed coat mucilage with correct hydration properties. Plant Cell 2007, 19, 4007–4021. [Google Scholar] [CrossRef] [Green Version]
- Kotake, T.; Dina, S.; Konishi, T.; Kaneko, S.; Igarashi, K.; Samejima, M.; Watanabe, Y.; Kimura, K.; Tsumuraya, Y. Molecular cloning of a β-galactosidase from radish that specifically hydrolyzes β-(1→)- and β-(1→6)-galactosyl residues of Arabinogalactan protein. Plant Physiol. 2005, 138, 1563–1576. [Google Scholar] [CrossRef] [Green Version]
- Macquet, A.; Ralet, M.C.; Loudet, O.; Kronenberger, J.; Mouille, G.; Marion-Poll, A.; North, H.M. A naturally occurring mutation in an Arabidopsis accession affects a beta-D-galactosidase that increases the hydrophilic potential of rhamnogalacturonan I in seed mucilage. Plant Cell 2007, 19, 3990–4006. [Google Scholar] [CrossRef] [Green Version]
- Megumi, I.; Smith, D.L.; Mort, A.J.; Gross, K.C. Enzymatic activity and substrate specificity of recombinant tomato β-galactosidases 4 and 5. J. Plant Physiol. 2009, 229, 447–456. [Google Scholar]
- Aung, B.; Gruber, M.Y.; Amyot, L.; Omari, K.; Bertrand, A.; Hannoufa, A. MicroRNA156 as a promising tool for alfalfa improvement. Plant Biotechnol. J. 2015, 13, 779–790. [Google Scholar] [CrossRef]
- Annicchiarico, P.; Nazzicari, N.; Li, X.; Wei, Y.; Pecetti, L.; Brummer, E.C. Accuracy of genomic selection for alfalfa biomass yield in different reference populations. BMC Genom. 2015, 16, 1020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Weng, J.K.; Chapple, C. Improvement of biomass through lignin modification. Plant J. 2008, 54, 569–581. [Google Scholar] [CrossRef]
- Chen, F.; Dixon, R.A. Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 2007, 25, 759–761. [Google Scholar] [CrossRef] [PubMed]
- Reddy, M.S.; Chen, F.; Shadle, G.; Jackson, L.; Aljoe, H.; Dixon, R.A. Targeted down-regulation of cytochrome P450 enzymes for forage quality improvement in alfalfa (Medicago sativa L.). Proc. Natl. Acad. Sci. USA 2005, 102, 16573–16578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lima, D.U.D.; Buckeridge, M.S. Interaction between cellulose and storage xyloglucans: The influence of the degree of galactosylation. Carbohyd. Polym. 2001, 46, 157–163. [Google Scholar] [CrossRef]
- Zhou, C.; Han, L.; Pislariu, C.; Nakashima, J.; Fu, C. From model to crop: Functional analysis of a stay-green gene in the model legume Medicago truncatula and effective use of the gene for alfalfa improvement. Plant Physiol. 2011, 157, 1483–1496. [Google Scholar] [CrossRef] [Green Version]
- Gou, J.; Debnath, S.; Sun, L.; Flanagan, A.; Tang, Y.; Jiang, Q.; Wen, J.; Wang, Z.Y. From model to crop: Functional characterization of SPL8 in M. truncatula led to genetic improvement of biomass yield and abiotic stress tolerance in alfalfa. Plant Biotechnol. J. 2018, 16, 951–962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Chandrasekar, B.; Van der Hoorn, R.A. Beta galactosidases in Arabidopsis and tomato—A mini review. Biochem. Soc. Trans. 2016, 44, 150–158. [Google Scholar] [CrossRef] [Green Version]
- Hruba, P.; Honys, D.; Twell, D.; Capkova, V.; Tupy, J. Expression of β-galactosidase and β-xylosidase genes during microspore and pollen development. Planta 2005, 220, 931–940. [Google Scholar] [CrossRef]
- Ahn, Y.O.; Zheng, M.Y.; Bevan, D.R.; Esen, A.; Shiu, S.H.; Benson, J.; Peng, H.P.; Miller, J.T.; Cheng, C.L.; Poulton, J.E. Functional genomic analysis of Arabidopsis thaliana glycoside hydrolase family 35. Phytochemistry 2007, 68, 1510–1520. [Google Scholar] [CrossRef]
- Tanthanuch, W.; Chantarangsee, M.; Maneesan, J.; Ketudat-Cairns, J. Genomic and expression analysis of glycosyl hydrolase family 35 genes from rice (Oryza sativa L.). BMC Plant Biol. 2008, 8, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dwevedi, A.; Kayastha, A.M. Plant β-galactosidases: Physiological significance and recent advances in technological applications. J. Plant Biochem. Biot. 2010, 19, 9–20. [Google Scholar] [CrossRef]
- Moneo-Sanchez, M.; Izquierdo, L.; Martin, I.; Labrador, E.; Dopico, B. Subcellular location of Arabidopsis thaliana subfamily a1 β-galactosidases and developmental regulation of transcript levels of their coding genes. Plant Physiol. Biochem. 2016, 109, 137–145. [Google Scholar] [CrossRef]
- Albornos, L.; Martin, I.; Perez, P.; Marcos, R.; Dopico, B.; Labrador, E. Promoter activities of genes encoding beta-galactosidases from Arabidopsis a1 subfamily. Plant Physiol. Bioch. 2012, 60, 223–232. [Google Scholar] [CrossRef]
- Martín, I.; Hernández-Nistal, J.; Albornos, L.; Labrador, E.; Dopico, B. βIII-Gal is involved in galactan reduction during phloem element differentiation in chickpea stems. Plant Cell Physiol. 2013, 54, 960–970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martín, I.; Jiménez, T.; Hernández-Nistal, J.; Dopico, B.; Labrador, E. The βI-galactosidase of Cicer arietinum is located in thickened cell walls such as those of collenchyma, sclerenchyma and vascular tissue. Plant Biol. 2011, 13, 777–783. [Google Scholar] [CrossRef] [PubMed]
- Martín, I.; Jiménez, T.; Esteban, R.; Dopico, B.; Labrador, E. Immunolocalization of a cell wall β-galactosidase reveals its developmentally regulated expression in Cicer arietinum and its relationship to vascular tissue. J. Plant Growth Regul. 2008, 27, 181–191. [Google Scholar] [CrossRef]
- Guo, D.; Chen, F.; Inoue, K.; Blount, J.W.; Dixon, R.A. Downregulation of caffeic acid 3-o-methyltransferase and caffeoyl CoA 3-o-methyltransferase in transgenic alfalfa: Impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 2001, 13, 73–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.Z.; Zhao, Q.; Chen, F.; Wang, M.Y.; Dixon, R.A. NAC domain function and transcriptional control of a secondary cell wall master switch. Plant J. 2011, 68, 1104–1114. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Jackson, L.; Shadle, G.; Nakashima, J.; Temple, S.; Chen, F.; Dixon, R.A. Distinct cinnamoyl CoA reductases involved in parallel routes to lignin in Medicago truncatula. Proc. Natl. Acad Sci. USA 2010, 107, 17803–17808. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Yang, J.H.; Chen, F.; Torres-Jerez, I.; Tang, Y.; Wang, M.; Du, Q.; Cheng, X.; Wen, J.; Dixon, R. Transcriptome analysis of secondary cell wall development in Medicago truncatula. BMC Genom. 2016, 17, 23. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Liu, J.; Dang, M.; Zhang, B.; Li, H.; Meng, R.; Qu, D.; Yang, Y.; Zhao, Z. Analysis of β-galactosidase during fruit development and ripening in two different texture types of apple cultivars. Front. Plant Sci. 2018, 9, 539. [Google Scholar] [CrossRef]
- Naoumkina, M.; Sumner, L.W.; Tang, Y.H.; Liu, C.J.; Dixon, R.A. Different mechanisms for phytoalexin induction by pathogen and wound signals in Medicago truncatula. Proc. Natl. Acad. Sci. USA 2007, 104, 17909–17915. [Google Scholar] [CrossRef] [Green Version]
- Bendtsen, J.D.; Henrik, N.; Heijne, G.V.; Brunak, S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004, 340, 783–795. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Smith, D.L.; Gross, K.C. A family of at least seven β-galactosidase genes is expressed during tomato fruit development. Plant Physiol. 2000, 123, 1173–1183. [Google Scholar] [CrossRef] [Green Version]
- Hobson, N.; Deyholos, M.K. Genomic and expression analysis of the flax (Linum usitatissimum) family of glycosyl hydrolase 35 genes. BMC Genom. 2013, 14, 344. [Google Scholar] [CrossRef] [Green Version]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, 1178–1186. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Peer, Y.V.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Ma, L.; Jiang, W.; Yao, Y.; Tang, Y.; Pang, Y. Comprehensive identification and characterization of abiotic stress and hormone responsive glycosyl hydrolase family 1 genes in Medicago truncatula. Plant Physiol. Biochem. 2021, 158, 21–33. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Gene ID | Pre-Protein | Mature Protein | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Aa | MW | Cleavage Site | Aa | MW | Theoretical pI | N-gly Site | Possible Destination | ||
| MtBGAL1 | Medtr1g018200 | 731 | 81.73 | 29–30 | 702 | 78.43 | 5.59 | 3 | Cell Wall |
| MtBGAL2 | Medtr1g023120 | 746 | 83.19 | 27–28 | 719 | 80.21 | 8.60 | 1 | Cell Wall |
| MtBGAL3 | Medtr2g039120 | 727 | 81.57 | 26–27 | 701 | 78.43 | 8.74 | 2 | Cell Wall |
| MtBGAL4 | Medtr2g042610 | 831 | 93.31 | 24–25 | 807 | 90.58 | 5.53 | 10 | Cell Wall |
| MtBGAL5 | Medtr2g094020 | 844 | 93.21 | 19–20 | 825 | 90.98 | 6.73 | 4 | Cell Wall |
| MtBGAL6 | Medtr2g094060 | 834 | 91.36 | 19–20 | 815 | 89.12 | 7.48 | 4 | Cell Wall |
| MtBGAL7 | Medtr2g100000 | 695 | 78.76 | 29–30 | 666 | 75.63 | 7.16 | 4 | Cell Wall |
| MtBGAL8 | Medtr2g100110 | 842 | 93.16 | 26–27 | 816 | 90.44 | 6.52 | 1 | Cell Wall |
| MtBGAL9 | Medtr3g005570 | 825 | 91.99 | 23–24 | 802 | 89.65 | 6.48 | 6 | Cell Wall |
| MtBGAL10 | Medtr3g088520 | 695 | 77.58 | 23–24 | 672 | 74.97 | 8.97 | 4 | Cell Wall |
| MtBGAL11 | Medtr3g096900 | 782 | 87.55 | 21–22 | 761 | 85.28 | 7.86 | 5 | Cell Wall |
| MtBGAL12 | Medtr3g096910 | 721 | 80.87 | 23–24 | 697 | 78.31 | 5.15 | 12 | Cell Wall |
| MtBGAL13 | Medtr3g112370 | 734 | 81.31 | 26–27 | 708 | 78.62 | 8.66 | 3 | Cell Wall |
| MtBGAL14 | Medtr3g117840 | 829 | 92.25 | 23–24 | 806 | 89.92 | 8.65 | 3 | Cell Wall |
| MtBGAL15 | Medtr4g059680 | 840 | 91.38 | 23–24 | 817 | 88.86 | 6.21 | 4 | Cell Wall |
| MtBGAL16 | Medtr4g073290 | 826 | 92.48 | 21–22 | 805 | 90.26 | 8.31 | 3 | Cell Wall |
| MtBGAL17 | Medtr4g126330 | 453 | 51.02 | - | - | - | 9.42 | 3 | Cell Wall |
| MtBGAL18 | Medtr5g021190 | 844 | 93.90 | 24–25 | 820 | 91.27 | 6.88 | 1 | Cell Wall |
| MtBGAL19 | Medtr5g022590 | 419 | 48.09 | - | - | - | 8.70 | 1 | Cell Wall |
| MtBGAL20 | Medtr5g024080 | 807 | 90.33 | - | - | - | 6.78 | 16 | Cell Wall |
| MtBGAL21 | Medtr5g025830 | 832 | 93.12 | 25–26 | 807 | 90.40 | 8.28 | 6 | Cell Wall |
| MtBGAL22 | Medtr6g007470 | 228 | 26.11 | - | - | - | 7.62 | 0 | Chloroplast, Nucleus |
| MtBGAL23 | Medtr8g016230 | 839 | 92.65 | 23–24 | 816 | 90.25 | 8.67 | 1 | Cell Wall |
| MtBGAL24 | Medtr8g076800 | 274 | 30.89 | - | - | - | 8.69 | 1 | Cell Membrane |
| MtBGAL25 | Medtr8g085210 | 907 | 101.17 | - | - | - | 7.02 | 6 | Cell Wall |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.; Li, Q.; Du, W.; Yao, Y.; Shen, G.; Jiang, W.; Pang, Y. Genome-Wide Analysis of Glycoside Hydrolase Family 35 Genes and Their Potential Roles in Cell Wall Development in Medicago truncatula. Plants 2021, 10, 1639. https://doi.org/10.3390/plants10081639
Yang J, Li Q, Du W, Yao Y, Shen G, Jiang W, Pang Y. Genome-Wide Analysis of Glycoside Hydrolase Family 35 Genes and Their Potential Roles in Cell Wall Development in Medicago truncatula. Plants. 2021; 10(8):1639. https://doi.org/10.3390/plants10081639
Chicago/Turabian StyleYang, Junfeng, Qian Li, Wenxuan Du, Yu Yao, Guoan Shen, Wenbo Jiang, and Yongzhen Pang. 2021. "Genome-Wide Analysis of Glycoside Hydrolase Family 35 Genes and Their Potential Roles in Cell Wall Development in Medicago truncatula" Plants 10, no. 8: 1639. https://doi.org/10.3390/plants10081639
APA StyleYang, J., Li, Q., Du, W., Yao, Y., Shen, G., Jiang, W., & Pang, Y. (2021). Genome-Wide Analysis of Glycoside Hydrolase Family 35 Genes and Their Potential Roles in Cell Wall Development in Medicago truncatula. Plants, 10(8), 1639. https://doi.org/10.3390/plants10081639




