The Breeding of Winter-Hardy Malting Barley
Abstract
1. Contextual Information for Understanding Malting Barley and Winter-Hardiness
1.1. The Life Cycle of Barley in Its Natural State
1.2. Malting Barley
1.3. Origin of Malting Barleys
1.4. Winter-Hardiness and the Winter Growth Habit
1.5. Regrowth in Spring Depends on Producing New Roots Following Extreme Cold
1.6. Winter Barley and Winter-Hardiness in North America
1.7. Breeding Winter Varieties of Cereals Traditionally Grown as Spring-Summer Annuals
1.8. Winter Malting Barley Development—Early Efforts
1.9. Genetic Analysis of Winter-Hardiness
1.10. Molecular-Genetic Analysis of Winter-Hardiness
1.11. Isolation of Genes Robustly Induced by Low Temperature in Plants
1.12. Mapping Loci Affecting COR Expression and Freezing Tolerance in Wheat and Barley
1.13. Identification of the CBFs in Arabidopsis
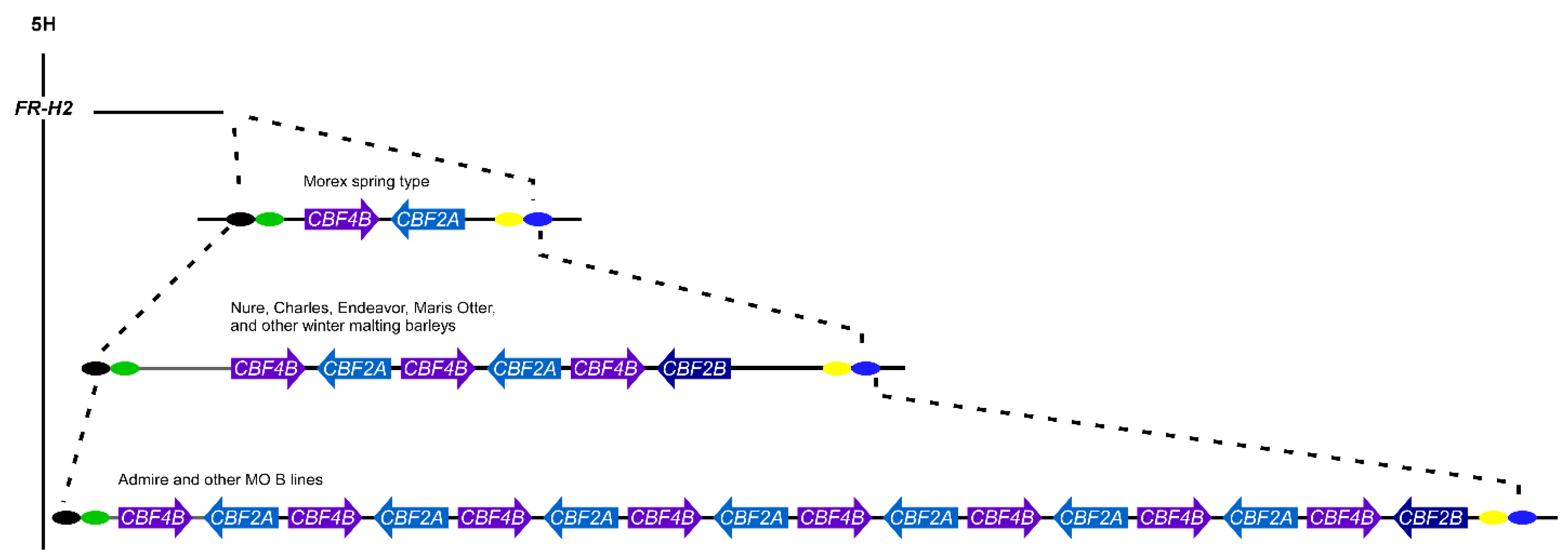
1.14. CBFs in the Cereals and Copy Number Variation
1.15. Winter-Hardiness and Its Connection to the Reproductive State of the Plant
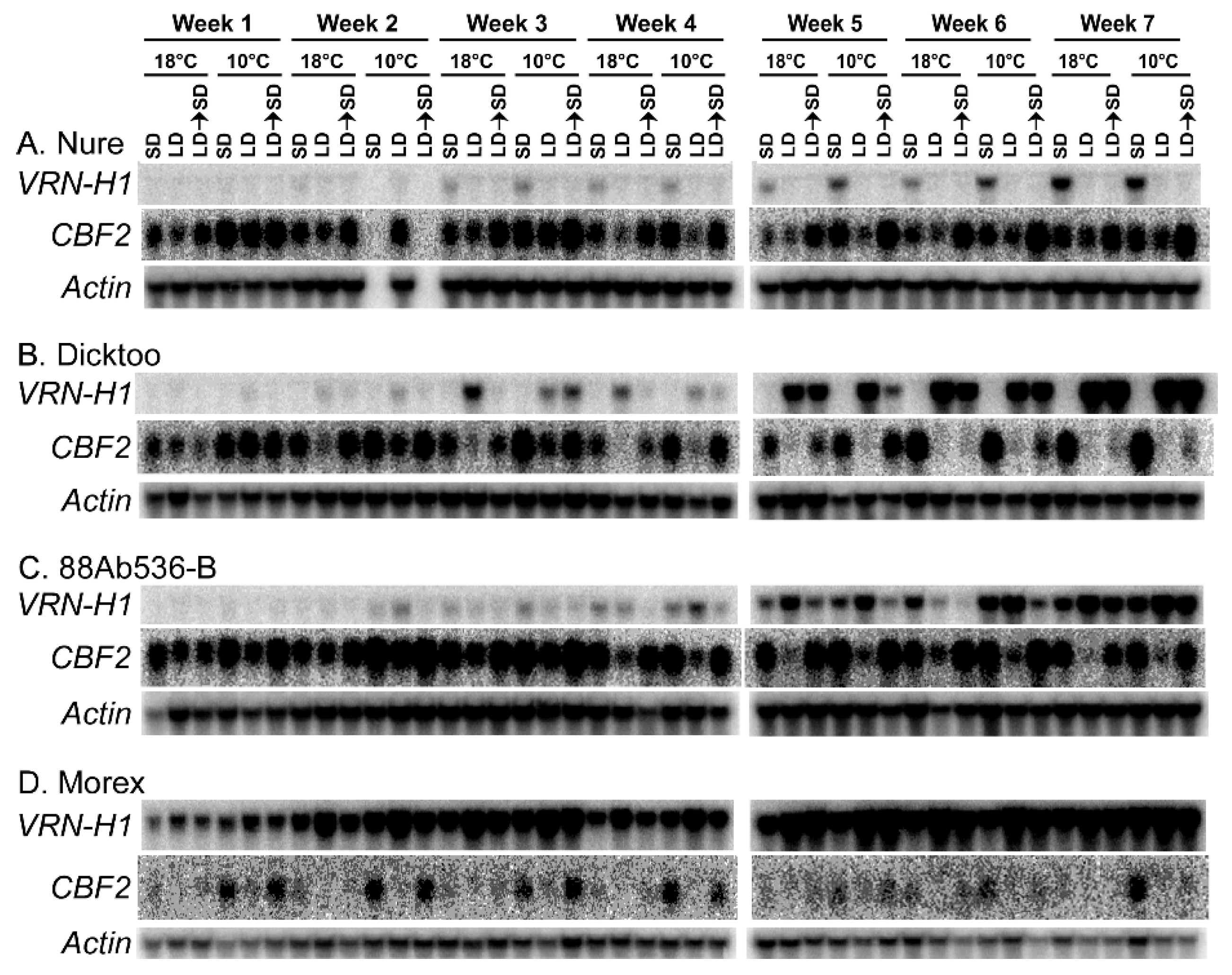
1.16. Relationship between Expression of VRN-H1 and CBFs
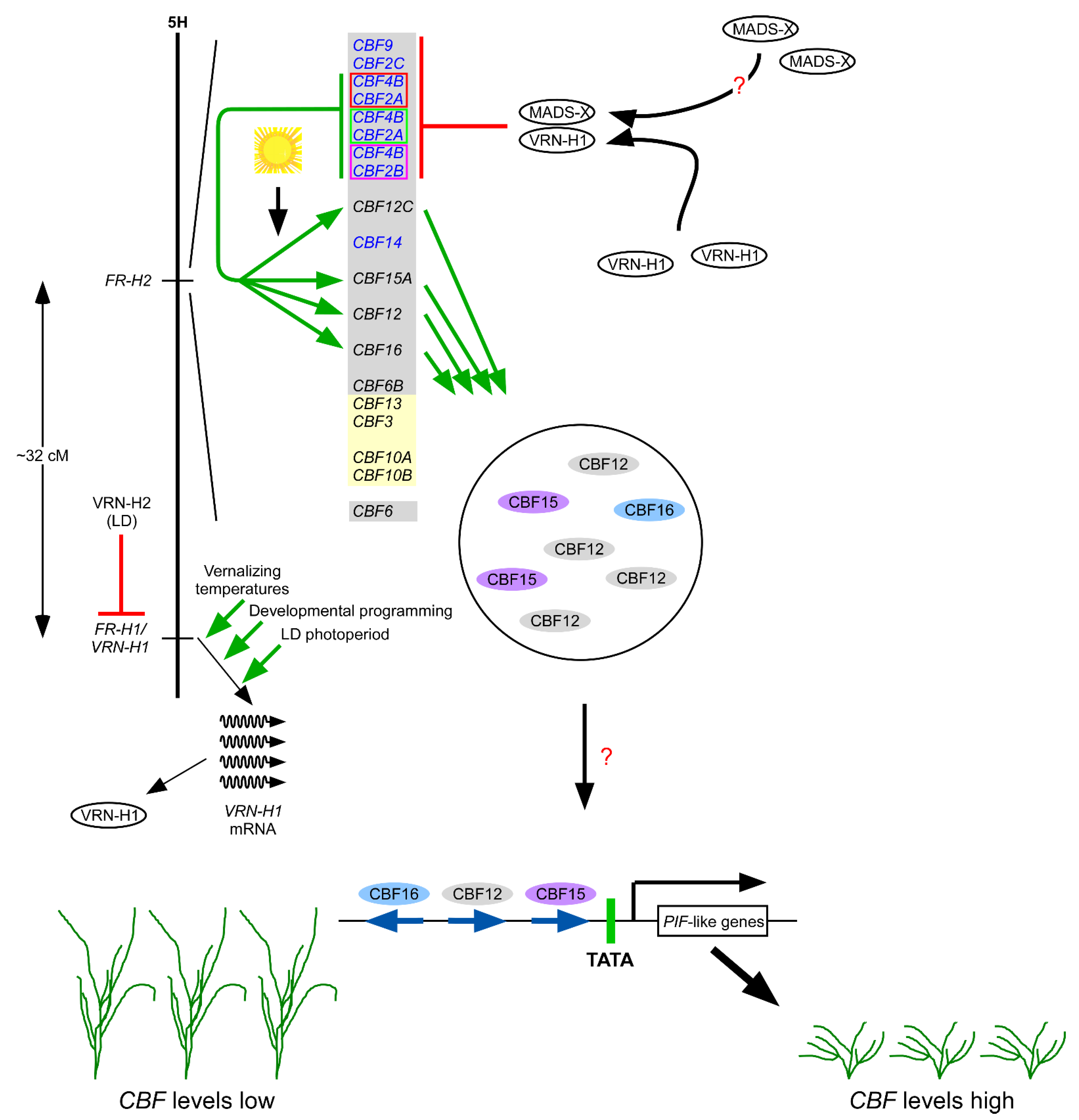
1.17. CBFs Activate Other CBFs
2. Winter-Hardiness and the Connection of the CBFs to the GA-GID1-DELLA Module
2.1. Morphological Traits Associated with Winter-Hardiness
2.2. CBFs Play a Role in Normal Growth and Development
2.3. The Semi-Dwarfing Wheat Rht-1 Genes, and Their Limitations in Stress Environments
2.4. Semi-Dwarfing Genes Used in Malting Barley and Their Limitations
2.5. The Connection between Winter-Hardiness and Gibberellic Acid in the Cereals
2.6. The Connection between the CBF Pathway and Gibberellic Acid in Arabidopsis
3. Future Directions
Understanding Winter-Hardiness and Predicting It in Future Breeding Populations
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zohary, D. The progenitors of wheat and barley in relation to domestication and agriculture dispersal in the Old World. In The Domestication and Exploitation of Plants and Animals; Ucko, P.J., Dimbleby, G.W., Eds.; Aldine Pub. Co.: Chicago, IL, USA, 1969; pp. 47–66. [Google Scholar]
- Salamini, F.; Ozkan, H.; Brandolini, A.; Schafer-Pregl, R.; Martin, W. Genetics and geography of wild cereal domestication in the near east. Nat. Rev. Genet. 2002, 3, 429–441. [Google Scholar] [CrossRef] [PubMed]
- von Bothmer, R.; Sato, K.; Komatsuda, T.; Yasuda, S.; Fischbeck, G. The domestication of cultivated barley. In Diversity in Barley (Hordeum vulgare), 1st ed.; von Bothmer, R., van Hintum, T., Knüpffer, H., Sato, K., Eds.; Elsevier: New York, NY, USA, 2003; pp. 9–27. [Google Scholar]
- Flood, R.G.; Halloran, G.M. Genetics and physiology of vernalization response in wheat. Adv. Agron. 1986, 39, 87–125. [Google Scholar]
- Matus, I.A.; Hayes, P.M. Genetic diversity in three groups of barley germplasm assessed by simple sequence repeats. Genome 2002, 45, 1095–1106. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Loukoianov, A.; Tranquilli, G.; Helguera, M.; Fahima, T.; Dubcovsky, J. Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. USA 2003, 100, 6263–6268. [Google Scholar] [CrossRef] [PubMed]
- Cockram, J.; Hones, H.; O’Sullivan, D.M. Genetic variation at flowering time loci in wild and cultivated barley. Plant Genet. Resour. 2011, 9, 264–267. [Google Scholar] [CrossRef][Green Version]
- Jones, H.; Civan, P.; Cockram, J.; Leigh, F.J.; Smith, L.M.; Jones, M.K.; Charles, M.P.; Molina-Cano, J.L.; Powell, W.; Jones, G.; et al. Evolutionary history of barley cultivation in Europe revealed by genetic analysis of extant landraces. BMC Evol. Biol. 2011, 11, 320. [Google Scholar] [CrossRef]
- Comadran, J.; Ramsay, L.; MacKenzie, K.; Hayes, P.; Close, T.J.; Muehlbauer, G.; Stein, N.; Waugh, R. Patterns of polymorphism and linkage disequilibrium in cultivated barley. Theor. Appl. Genet. 2011, 122, 523–531. [Google Scholar] [CrossRef][Green Version]
- Comadran, J.; Kilian, B.; Russell, J.; Ramsay, L.; Stein, N.; Ganal, M.; Shaw, P.; Bayer, M.; Thomas, W.; Marshall, D.; et al. Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nat. Genet. 2012, 44, 1388–1392. [Google Scholar] [CrossRef]
- Zohary, D. Monophyletic vs. polyphyletic origin of the crops on which agriculture was founded in the Near East. Genet. Resour. Crop Evol. 1999, 46, 133–142. [Google Scholar] [CrossRef]
- Badr, A.; Muller, K.; Schafer-Pregl, R.; El Rabey, H.; Effgen, S.; Ibrahim, H.H.; Pozzi, C.; Rohde, W.; Salamini, F. On the origin and domestication history of barley (Hordeum vulgare). Mol. Biol. Evol. 2000, 17, 499–510. [Google Scholar] [CrossRef]
- Molina-Cano, J.L.; Russell, J.R.; Moralejo, M.A.; Escacena, J.L.; Arias, G.; Powell, W. Chloroplast DNA microsatellite analysis supports a polyphyletic origin for barley. Theor. Appl. Genet. 2005, 110, 613–619. [Google Scholar] [CrossRef]
- Morrell, P.L.; Clegg, M.T. Genetic evidence for a second domestication of barley (Hordeum vulgare) east of the Fertile Crescent. Proc. Natl. Acad. Sci. USA 2007, 104, 3289–3294. [Google Scholar] [CrossRef] [PubMed]
- Azhaguvel, P.; Komatsuda, T. A phylogenetic analysis based on nucleotide sequence of a marker linked to the brittle rachis locus indicates a diphyletic origin of barley. Ann. Bot. 2007, 100, 1009–1015. [Google Scholar] [CrossRef] [PubMed]
- Saisho, D.; Purugganan, M.D. Molecular phylogeography of domesticated barley traces expansion of agriculture in the Old World. Genetics 2007, 177, 1765–1776. [Google Scholar] [CrossRef]
- Brown, T.A.; Jones, M.K.; Powell, W.; Allaby, R.G. The complex origins of domesticated crops in the Fertile Crescent. Trends Ecol. Evol. 2009, 24, 103–109. [Google Scholar] [CrossRef]
- Dai, F.; Nevo, E.; Wu, D.; Comadran, J.; Zhou, M.; Qiu, L.; Chen, Z.; Beiles, A.; Chen, G.; Zhang, G. Tibet is one of the centers of domestication of cultivated barley. Proc. Natl. Acad. Sci. USA 2012, 109, 16969–16973. [Google Scholar] [CrossRef] [PubMed]
- Igartua, E.; Moralejo, M.; Casas, A.M.; Torres, L.; Molina-Cano, J.L. Whole-genome analysis with SNPs from BOPA1 shows clearly defined groupings of Western Mediterranean, Ethiopian, and Fertile Crescent barleys. Genet. Resour. Crop Evol. 2013, 60, 251–264. [Google Scholar] [CrossRef]
- Poets, A.M.; Fang, Z.; Clegg, M.T.; Morrell, P.L. Barley landraces are characterized by geographically heterogeneous genomic origins. Genome Biol. 2015, 16, 173. [Google Scholar] [CrossRef]
- Fox, G.P.; Panozzo, J.F.; Li, C.D.; Lance, R.C.M.; Inkerman, P.A.; Henry, R.J. Molecular basis of barley quality. Aust. J. Agric. Res. 2003, 54, 1081–1101. [Google Scholar] [CrossRef]
- Hang, A.; Obert, D.; Gironella, A.I.N.; Burton, C.S. Barley amylose and β-glucan: Their relationships to protein, agronomic traits, and environmental factors. Crop Sci. 2007, 47, 1754–1760. [Google Scholar] [CrossRef]
- Burton, R.A.; Jobling, S.A.; Harvey, A.J.; Shirley, N.J.; Mather, D.E.; Bacic, A.; Fincher, G.B. The genetics and transcriptional profiles of the cellulose synthase-like HvCslF gene family in barley. Plant Physiol. 2008, 146, 1821–1833. [Google Scholar] [CrossRef] [PubMed]
- Burton, R.A.; Collins, H.M.; Fincher, G.B. The role of endosperm cell walls in barley malting quality. In Genetics and Improvement of Barley Malt Quality; Zhang, G., Li, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 190–237. [Google Scholar]
- Garcia-Gimenez, G.; Russell, J.; Aubert, M.K.; Fincher, G.B.; Burton, R.A.; Waugh, R.; Tucker, M.R.; Houston, K. Barley grain (1,3;1,4)-beta-glucan content: Effects of transcript and sequence variation in genes encoding the corresponding synthase and endohydrolase enzymes. Sci. Rep. 2019, 9, 11. [Google Scholar] [CrossRef] [PubMed]
- Chiapparino, E.; Donini, P.; Reeves, J.; Tuberosa, R.; O’Sullivan, D.M. Distribution of β-amylase I haplotypes among European cultivated barleys. Mol. Breed. 2006, 18, 341–354. [Google Scholar] [CrossRef]
- Von Wettstein, D.; Cochran, J.S.; Ullrich, S.E.; Kannangara, C.G.; Jitkov, V.A.; Burns, J.; Reisenauer, P.E.; Chen, X.; Jones, B.L. Registration of ‘Radiant’ barley. Crop Sci. 2004, 44, 1859–1860. [Google Scholar] [CrossRef]
- Hoki, T.; Saito, W.; Hirota, N.; Shirai, M.; Takoi, K.; Yoshida, S.; Shimase, M.; Saito, T.; Takaoka, T.; Kihara, M.; et al. Breeding of lipoxygenase-1-less malting barley variety CDC PolarStar and effect of lipoxygenase-1-less trait on beer quality at pilot and commercial scale brewing. Brew. Sci. 2013, 66, 37–45. [Google Scholar]
- Swanston, J.S.; Thomas, W.T.B.; Powell, W.; Young, G.R.; Lawrence, P.E.; Ramsay, L.; Waugh, R. Using molecular markers to determine barleys most suitable for malt whisky distilling. Mol. Breed. 1999, 5, 103–109. [Google Scholar] [CrossRef]
- Thomas, W.T.B. Prospects for molecular breeding of barley. Ann. Appl. Biol. 2003, 142, 1–12. [Google Scholar] [CrossRef]
- Rae, S.J.; Macaulay, M.; Ramsay, L.; Leigh, F.; Matthews, D.; O’Sullivan, D.M.; Donini, P.; Morris, P.C.; Powell, W.; Marshall, D.F.; et al. Molecular barley breeding. Euphytica 2007, 158, 295–303. [Google Scholar] [CrossRef]
- Mohammadi, M.; Blake, T.K.; Budde, A.D.; Chao, S.; Hayes, P.M.; Horsley, R.D.; Obert, D.E.; Ullrich, S.E.; Smith, K.P. A genome-wide association study of malting quality across eight U.S. barley breeding programs. Theor. Appl. Genet. 2015, 128, 705–721. [Google Scholar] [CrossRef]
- Poets, A.M.; Mohammadi, M.; Seth, K.; Wang, H.; Kono, T.J.; Fang, Z.; Muehlbauer, G.J.; Smith, K.P.; Morrell, P.L. The effects of both recent and long-term selection and genetic drift are readily evident in North American barley breeding populations. G3: Genes Genomes Genet. 2015, 6, 609–622. [Google Scholar] [CrossRef]
- Martin, J.M.; Blake, T.K.; Hockett, E.A. Diversity among North American spring barley cultivars based on coefficients of parentage. Crop Sci. 1991, 31, 1131–1137. [Google Scholar] [CrossRef]
- Eslick, R.F.; Hockett, E.A. Genetic engineering as a key to water-use efficieny. J. Agric. Meteorol. 1974, 14, 13–23. [Google Scholar] [CrossRef]
- Aufhammer, G. Barley Varieties, E.B.C, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 1958; p. 159. [Google Scholar]
- Lein, A. Breeding for malting quality In Barley Genetics I, Proceedings of the First International Barley Genetics Symposium, Wageningen, The Netherlands, 26–31 August 1963; Centre for Agricultural Publications and Documentation: Wageningen, The Netherlands, 1964; pp. 310–324. [Google Scholar]
- Fischbeck, G. Barley cultivar developmment in Europe–Success in the past and possible changes in the future. In Barley Genetics VI, Proceedings of the Sixth International Barley Genetics Symposium, Helsingborg, Sweden, 22–27 July 1991; Munksgaard International Publishers Inc.: Copenhagen, Denmark, 1992; pp. 885–901. [Google Scholar]
- Fischbeck, G. Diversification through breeding. In Diversity in Barley (Hordeum vulgare), 1st ed.; von Bothmer, R., van Hintum, T., Knüpffer, H., Sato, K., Eds.; Developments in Plant Genetics and Breeding; Elsevier: New York, NY, USA, 2003; pp. 29–52. [Google Scholar]
- Psota, V.; Hartmann, J.; Sejkorova, S.; Louckova, T.; Vejrazka, K. 50 years of progress in quality of malting barley grown in the Czech Republic. J. Inst. Brew. 2009, 115, 279–291. [Google Scholar] [CrossRef]
- Riggs, T.J.; Hanson, P.R.; Start, N.D.; Miles, D.M.; Morgan, C.L.; Ford, M.A. Comparison of spring barley varieties grown in England and Wales between 1880 and 1980. J. Agric. Sci. 1981, 97, 599–610. [Google Scholar] [CrossRef]
- Russell, J.R.; Ellis, R.P.; Thomas, W.T.B.; Waugh, R.; Provan, J.; Booth, A.; Fuller, J.; Lawrence, P.; Young, G.; Powell, W. A retrospective analysis of spring barley germplasm development from ‘foundation genotypes’ to currently successful cultivars. Mol. Breed. 2000, 6, 553–568. [Google Scholar] [CrossRef]
- Schittenhelm, S.; Okeno, J.A.; Friedt, W. Prospects of agronomic improvement in spring barley based on a comparison of old and new germplasm. J. Agron. Crop Sci.-Z. Acker Pflanzenbau 1996, 176, 295–303. [Google Scholar] [CrossRef]
- Wiebe, G.A.; Reid, D.A. Classification of Barley Varieties Grown in the United States and Canada in 1958; U.S. Department of Agriculture: Washington, DC, USA, 1961; Volume 1224, p. 234.
- Harlan, H.V.; Martini, M.L. Problems and Results in Barley Breeding. Yearb. Agric. U.S. Department of Agriculture, Washing-ton D.C. 1936, 303–347. Available online: http://naldr.nal.usda.gov/NALWeb/Agricola_Link.asp?Accession=IND43893522 (accessed on 2 February 2021).
- Åberg, E.; Wiebe, G.A. Classification of Barley Varieties Grown in the United States and Canada in 1945; U.S. Department of Agriculture: Washington, DC, USA, 1946; p. 190. [Google Scholar]
- Peterson, G.A.; Foster, A.E. Malting barley in the United States. Adv. Agron. 1973, 25, 327–378. [Google Scholar]
- Wych, R.D.; Rasmusson, D.C. Genetic improvement in malting barley cultivars since 1920. Crop Sci. 1983, 23, 1037–1040. [Google Scholar] [CrossRef]
- Jensen, N.F.; Edwards, L.H.; Smith, E.L.; Sorrells, M.E. Registration of Wintermalt barley. Crop Sci. 1982, 22, 157. [Google Scholar] [CrossRef]
- Gusta, L.V.; Wisniewski, M. Understanding plant cold hardiness: An opinion. Physiol. Plant. 2013, 147, 4–14. [Google Scholar] [CrossRef]
- Bell, G.D.H. The breeding of two-row winter-hardy barley. J. Agric. Sci. 1944, 34, 223–238. [Google Scholar] [CrossRef]
- Thomashow, M.F. Molecular basis of plant cold acclimation: Insights gained from studying the CBF cold response pathway. Plant Physiol. 2010, 154, 571–577. [Google Scholar] [CrossRef]
- Fowler, D.B.; Limin, A.E. Interactions among factors regulating phenological development and acclimation rate determine low-temperature tolerance in wheat. Ann. Bot. 2004, 94, 717–724. [Google Scholar] [CrossRef]
- von Zitzewitz, J.; Szucs, P.; Dubcovsky, J.; Yan, L.; Francia, E.; Pecchioni, N.; Casas, A.; Chen, T.H.; Hayes, P.M.; Skinner, J.S. Molecular and structural characterization of barley vernalization genes. Plant Mol. Biol. 2005, 59, 449–467. [Google Scholar] [CrossRef]
- Cockram, J.; Horsnell, R.; Soh, E.; Norris, C.; O’Sullivan, D.M. Molecular and phenotypic characterization of the alternative seasonal growth habit and flowering time in barley (Hordeum vulgare ssp. vulgare L.). Mol. Breed. 2015, 35, 165. [Google Scholar] [CrossRef]
- Gusta, L.V.; O’Connor, B.J.; Gao, Y.P.; Jana, S. A re-evaluation of controlled freeze-tests and controlled environment hardening conditions to estimate the winter survival potential of hardy winter wheats. Can. J. Plant Sci. 2001, 81, 241–246. [Google Scholar] [CrossRef]
- Olien, C.R. Freezing processes in the crown of ’Hudson’ barley, Hordeum vulgare (L., emend. Lam.) Hudson. Crop Sci. 1964, 4, 91–95. [Google Scholar] [CrossRef]
- Warnes, D.D.; Schmidt, J.W.; Johnson, V.A. Correlation of artificial crown freezing survival with natural survival in winter barley. In Barley genetics II. Proceedings of Second International Barley Genetics Symposium, Washington State University, Pullman Washington 1970; Washington State University Press: Pullman, WA, USA, 1971; pp. 364–377. [Google Scholar]
- Warnes, D.D.; Johnson, V.A. Crown-freezing and natural survival comparisons of F2–F4 bulk populations from 25 winter barley crosses. Crop Sci. 1972, 12, 403–405. [Google Scholar] [CrossRef]
- Chen, T.H.; Gusta, L.V.; Fowler, D.B. Freezing injury and root development in winter cereals. Plant Physiol. 1983, 73, 773–777. [Google Scholar] [CrossRef] [PubMed]
- Wiebe, G.A. Introduction of barley into the New World. Agric. Handb. USDA 1979, 338, 2–9. [Google Scholar]
- Poehlman, J.M. Breeding winter barley for hardiness and disease resistance. Econ. Bot. 1952, 6, 176–184. [Google Scholar] [CrossRef]
- Muñoz-Amatriaín, M.; Hernandez, F.; Herb, D.; Baenziger, P.S.; Bochard, A.; Capettini, F.; Casas, A.; Cuesta-Marcos, A.; Einfeldt, C.; Fisk, S.; et al. Perspectives on low temperature tolerance and vernalization sensitivity in barley: Prospects for facultative growth habit. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Wiebe, G.A.; Reid, D.A. Comparative Winter Hardiness of Barley Varieties; U.S. Department of Agriculture: Washington, DC, USA, 1958; Volume 1176, p. 20.
- Poehlman, J.M. Agronomic Characteristics and Disease Resistance of Winter Barleys Tested in Missouri, 1943 to 1948; University of Missouri, College of Agriculture, Agricultural Experiment Station Research Bulletin: Columbia, MO, USA, 1949; Volume 442, p. 28. [Google Scholar]
- Rohde, C.R.; Pulham, C.F. Heritability estimates of winter hardiness in winter barley determined by the standard unit method of regression analysis. Agron. J. 1960, 52, 584–586. [Google Scholar] [CrossRef]
- Lambert, J.W. Registration of barley varieties, XIV. Agron. J. 1958, 50, 708–711. [Google Scholar] [CrossRef]
- McGill, D.P.; Webster, O.J.; Warnes, D.D. Chase barley. Crop Sci. 1964, 4, 666. [Google Scholar] [CrossRef]
- Spillman, W.J. Quantitative studies on the transmisson of parental characters to hybrid offspring. In Proceedings of the Fifteenth Annual Convention of the Association of American Agricultureal Colleges and Experiment Stations, Washington, DC, USA, 12–14 November 1901; pp. 88–98. [Google Scholar]
- Tschermak-Seysenegg, E. A Noteworthy Two-Row Winter Barley; Deutsche Landwirtschaftliche Presse: Berlin, Germany, 1932; Volume 59, p. 423. [Google Scholar]
- National Institute of Agricultural Botany. Detailed Descriptions of Varieties of Wheat, Barley and Oats Recommended by the National Institute of Agricultural Botany; National Institute of Agricultural Botany: Cambridge, UK, 1965. [Google Scholar]
- Hornsey, I.S. Maris Otter. In The Oxford Companion to Beer; Oliver, G., Ed.; Oxford University Press: New York, NY, USA, 2011; p. 571. [Google Scholar]
- Duelos, L.A.; Poehlman, J.M.; Hoskins, P.H. Breeding 2-row winter-type malting barley. In Barley Genetics II, Proceedings of Second International Barley Genetics Symposium; Washington State University: Pullman, WA, USA, 1971; pp. 283–286. [Google Scholar]
- Poehlman, J.M.; Duclos, L.; Kruse, C. Progress in development of two-row winter malting barley. In Proceedings of the Barley Improvement Conference, Minneapolis, MN, USA, 18 January 1973; pp. 15–21. [Google Scholar]
- Suneson, C.A.; Stevens, H. Studies with Bulked Hybrid Populations of Barley; Tech. Bull; U.S. Department of Agriculture: Washington, DC, USA, 1953; pp. 1–15.
- Stockinger, E.J.; Skinner, J.S.; Gardner, K.G.; Francia, E.; Pecchioni, N. Expression levels of barley Cbf genes at the Frost resistance-H2 locus are dependent upon alleles at Fr-H1 and Fr-H2. Plant J. 2007, 51, 308–321. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N. Studies on the inheritance of the spring and winter growing habit in crosses between spring and winter barleys. [Chosen Govt. Gen.] Agr. Expt. Sta. Bul. 1925, 2, 1–7. [Google Scholar]
- Vavilov, N.I.; Kouznetsov, E.S. Translation of On the Genetic Nature of Winter and Spring Varieties of Plants; Imperial Bureau of Plant Genetics, School of Agriculture: Cambridge, UK, 1921; p. 38. [Google Scholar]
- Reid, D.A. Winter hardiness of progenies from winter x spring barley (Hordeum vulgare, L. emend. Lam.) crosses. Crop Sci. 1965, 5, 263–266. [Google Scholar] [CrossRef]
- Doll, H.; Haahr, V.; Søgaard, B. Relationship between vernalization requirement and winter hardiness in doubled haploids of barley. Euphytica 1989, 42, 209–213. [Google Scholar]
- Hayes, P.M.; Blake, T.; Chen, T.H.H.; Tragoonrung, S.; Chen, F.; Pan, A.; Liu, B. Quantitative trait loci on barley (Hordeum vulgare L.) chromosome 7 associated with components of winterhardiness. Genome 1993, 36, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Johnson, I.J. Registration of barley varieties. Agron. J. 1953, 45, 320–323. [Google Scholar] [CrossRef]
- Rasmusson, D.C.; Wilcoxson, R.W. Registration of Morex barley. Crop Sci. 1979, 19, 293. [Google Scholar] [CrossRef]
- Livingston, D.P., III.; Olien, C.R.; Freed, R.D. Sugar composition and freezing tolerance in barley crowns at varying carbohydrate levels. Crop Sci. 1989, 29, 1266–1270. [Google Scholar] [CrossRef]
- Linde-Laursen, I.; Heslop-Harrison, J.S.; Shepherd, K.W.; Taketa, S. The barley genome and its relationship with the wheat genomes. A survey with an internationally agreed recommendation for barley chromosome nomenclature. Hereditas (Landskrona) 1997, 126, 1–16. [Google Scholar] [CrossRef]
- Skinner, J.S.; Szucs, P.; von Zitzewitz, J.; Marquez-Cedillo, L.; Filichkin, T.; Stockinger, E.J.; Thomashow, M.F.; Chen, T.H.; Hayes, P.M. Mapping of barley homologs to genes that regulate low temperature tolerance in Arabidopsis. Theor. Appl. Genet. 2006, 112, 832–842. [Google Scholar] [CrossRef] [PubMed]
- Francia, E.; Rizza, F.; Cattivelli, L.; Stanca, A.M.; Galiba, G.; Toth, B.; Hayes, P.M.; Skinner, J.S.; Pecchioni, N. Two loci on chromosome 5H determine low-temperature tolerance in a ‘Nure’ (winter) × ‘Tremois’ (spring) barley map. Theor. Appl. Genet. 2004, 108, 670–680. [Google Scholar] [CrossRef] [PubMed]
- Fisk, S.P.; Cuesta-Marcos, A.; Cistue, L.; Russell, J.; Smith, K.P.; Baenziger, S.; Bedo, Z.; Corey, A.; Filichkin, T.; Karsai, I.; et al. FR-H3: A new QTL to assist in the development of fall-sown barley with superior low temperature tolerance. Theor. Appl. Genet. 2013, 126, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Visioni, A.; Tondelli, A.; Francia, E.; Pswarayi, A.; Malosetti, M.; Russell, J.; Thomas, W.; Waugh, R.; Pecchioni, N.; Romagosa, I.; et al. Genome-wide association mapping of frost tolerance in barley (Hordeum vulgare L.). BMC Genom. 2013, 14, 424. [Google Scholar] [CrossRef]
- Tondelli, A.; Pagani, D.; Ghafoori, I.N.; Rahimi, M.; Ataei, R.; Rizza, F.; Flavell, A.J.; Cattivelli, L. Allelic variation at Fr-H1/Vrn-H1 and Fr-H2 loci is the main determinant of frost tolerance in spring barley. Environ. Exp. Bot. 2014, 106, 148–155. [Google Scholar] [CrossRef]
- Brule-Babel, A.L.; Fowler, D.B. Genetic control of cold hardiness and vernalization requirement in winter wheat. Crop Sci. 1988, 28, 879–884. [Google Scholar] [CrossRef]
- Sutka, J.; Snape, J.W. Location of a gene for frost resistance on chromosome 5A of wheat. Euphytica 1989, 42, 41–44. [Google Scholar] [CrossRef]
- Roberts, D.W.A. Identification of loci on chromosome 5A of wheat involved in control of cold hardiness, vernalization, leaf length, rosette growth habit, and height of hardened plants. Genome 1990, 33, 247–259. [Google Scholar] [CrossRef]
- Galiba, G.; Quarrie, S.A.; Sutka, J.; Morounov, A.; Snape, J.W. RFLP mapping of the vernalization (Vrn1) and frost resistance (Fr1) genes on chromosome 5A of wheat. Theor. Appl. Genet. 1995, 90, 1174–1179. [Google Scholar] [CrossRef] [PubMed]
- Snape, J.W.; Semikhodskii, A.; Fish, L.; Sarma, R.N.; Quarrie, S.A.; Galiba, G.; Sutka, J. Mapping frost tolerance loci in wheat and comparative mapping with other cereals. Acta. Agric. Hung. 1997, 45, 265–270. [Google Scholar]
- Tóth, B.; Galiba, G.; Feher, E.; Sutka, J.; Snape, J.W. Mapping genes affecting flowering time and frost resistance on chromosome 5B of wheat. Theor. Appl. Genet. 2003, 107, 509–514. [Google Scholar] [CrossRef]
- Vágújfalvi, A.; Galiba, G.; Cattivelli, L.; Dubcovsky, J. The cold-regulated transcriptional activator Cbf3 is linked to the frost-tolerance locus Fr-A2 on wheat chromosome 5A. Mol. Genet. Genom. 2003, 269, 60–67. [Google Scholar] [CrossRef]
- Båga, M.; Chodaparambil, S.V.; Limin, A.E.; Pecar, M.; Fowler, D.B.; Chibbar, R.N. Identification of quantitative trait loci and associated candidate genes for low-temperature tolerance in cold-hardy winter wheat. Funct. Integr. Genom. 2007, 7, 53–68. [Google Scholar] [CrossRef]
- Francia, E.; Barabaschi, D.; Tondelli, A.; Laido, G.; Rizza, F.; Stanca, A.M.; Busconi, M.; Fogher, C.; Stockinger, E.J.; Pecchioni, N. Fine mapping of a HvCBF gene cluster at the frost resistance locus Fr-H2 in barley. Theor. Appl. Genet. 2007, 115, 1083–1091. [Google Scholar] [CrossRef]
- Knox, A.K.; Dhillon, T.; Cheng, H.; Tondelli, A.; Pecchioni, N.; Stockinger, E.J. CBF gene copy number variation at Frost Resistance-2 is associated with levels of freezing tolerance in temperate-climate cereals. Theor. Appl. Genet. 2010, 121, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Cattivelli, L.; Bartels, D. Molecular cloning and characterization of cold-regulated genes in barley. Plant Physiol. 1990, 93, 1504–1510. [Google Scholar] [CrossRef]
- Dunn, M.A.; Hughes, M.A.; Pearce, R.S.; Jack, P.L. Molecular characterization of a barley gene induced by cold treatment. J. Exp. Bot. 1990, 41, 1405–1413. [Google Scholar] [CrossRef]
- Hajela, R.K.; Horvath, D.P.; Gilmour, S.J.; Thomashow, M.F. Molecular cloning and expression of cor (Cold-Regulated) genes in Arabidopsis thaliana. Plant Physiol. 1990, 93, 1246–1252. [Google Scholar] [CrossRef] [PubMed]
- Houde, M.; Danyluk, J.; Laliberte, J.F.; Rassart, E.; Dhindsa, R.S.; Sarhan, F. Cloning, characterization, and expression of a cDNA encoding a 50-kilodalton protein specifically induced by cold acclimation in wheat. Plant Physiol. 1992, 99, 1381–1387. [Google Scholar] [CrossRef] [PubMed]
- Houde, M.; Dhindsa, R.S.; Sarhan, F. A molecular marker to select for freezing tolerance in Gramineae. Mol. Gen. Genet. 1992, 234, 43–48. [Google Scholar] [CrossRef]
- Artus, N.N.; Uemura, M.; Steponkus, P.L.; Gilmour, S.J.; Lin, C.; Thomashow, M.F. Constitutive expression of the cold-regulated Arabidopsis thaliana COR15a gene affects both chloroplast and protoplast freezing tolerance. Proc. Natl. Acad. Sci. USA 1996, 93, 13404–13409. [Google Scholar] [CrossRef] [PubMed]
- Danyluk, J.; Perron, A.; Houde, M.; Limin, A.; Fowler, B.; Benhamou, N.; Sarhan, F. Accumulation of an acidic dehydrin in the vicinity of the plasma membrane during cold acclimation of wheat. Plant Cell 1998, 10, 623–638. [Google Scholar] [CrossRef] [PubMed]
- Koag, M.C.; Fenton, R.D.; Wilkens, S.; Close, T.J. The binding of maize DHN1 to lipid vesicles. Gain of structure and lipid specificity. Plant Physiol. 2003, 131, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Thalhammer, A.; Hincha, D.K. A mechanistic model of COR15 protein function in plant freezing tolerance: Integration of structural and functional characteristics. Plant Signal. Behav. 2014, 9, e977722. [Google Scholar] [CrossRef] [PubMed]
- Crosatti, C.; Rizza, F.; Cattivelli, L. Accumulation and characterization of the 75 kDa protein induced by low temperature in barley. Plant Sci. 1994, 97, 39–46. [Google Scholar] [CrossRef]
- Crosatti, C.; Soncini, C.; Stanca, A.M.; Cattivelli, L. The accumulation of a cold-regulated chloroplastic protein is light-dependent. Planta 1995, 196, 458–463. [Google Scholar] [CrossRef]
- Thomashow, M.F. Genes induced during cold acclimation in higher plants. In Advances in Low-Temperature Biology, Volume 2; Steponkus, P.L., Ed.; JAI Press: London, UK, 1993; pp. 183–210. [Google Scholar]
- Thomashow, M.F. Characterization of genes induced during cold acclimation in Arabidopsis thaliana. In Plant Responses to Cellular Dehydration During Environmental Stress; Close, T.J., Bray, A.B., Eds.; American Society of Plant Physiologists: Rockville, MD, USA, 1993; pp. 137–143. [Google Scholar]
- Baker, S.S.; Wilhelm, K.S.; Thomashow, M.F. The 5’-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 1994, 24, 701–713. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 1994, 6, 251–264. [Google Scholar]
- Stockinger, E.J.; Cheng, H.; Skinner, J.S. Structural organization of barley CBF genes coincident with QTLs for cold hardiness. In Cold Hardiness in Plants: Molecular Genetics, Cell Biology and Physiology; Chen, T.H.H., Uemura, M., Fujikawa, S., Eds.; CABI Publishing Oxon: Wallingford, UK, 2006; pp. 53–63. [Google Scholar]
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar] [CrossRef]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef]
- Gilmour, S.J.; Zarka, D.G.; Stockinger, E.J.; Salazar, M.P.; Houghton, J.M.; Thomashow, M.F. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998, 16, 433–442. [Google Scholar] [CrossRef]
- Jaglo-Ottosen, K.R.; Gilmour, S.J.; Zarka, D.G.; Schabenberger, O.; Thomashow, M.F. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 1998, 280, 104–106. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, S.J.; Fowler, S.G.; Thomashow, M.F. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 2004, 54, 767–781. [Google Scholar] [CrossRef] [PubMed]
- Fowler, S.; Thomashow, M.F. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 2002, 14, 1675–1690. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.T.; Zarka, D.G.; Van Buskirk, H.A.; Fowler, S.G.; Thomashow, M.F. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 2005, 41, 195–211. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef] [PubMed]
- Jaglo, K.R.; Kleff, S.; Amundsen, K.L.; Zhang, X.; Haake, V.; Zhang, J.Z.; Deits, T.; Thomashow, M.F. Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol. 2001, 127, 910–917. [Google Scholar] [CrossRef] [PubMed]
- Canella, D.; Gilmour, S.J.; Kuhn, L.A.; Thomashow, M.F. DNA binding by the Arabidopsis CBF1 transcription factor requires the PKKP/RAGRxKFxETRHP signature sequence. Biochim. Biophys. Acta 2009, 1799, 454–462. [Google Scholar] [CrossRef]
- Magome, H.; Yamaguchi, S.; Hanada, A.; Kamiya, Y.; Oda, K. dwarf and delayed-flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J. 2004, 37, 720–729. [Google Scholar] [CrossRef] [PubMed]
- Lehti-Shiu, M.D.; Uygun, S.; Moghe, G.D.; Panchy, N.; Fang, L.; Hufnagel, D.E.; Jasicki, H.L.; Feig, M.; Shiu, S.H. Molecular evidence for functional divergence and decay of a transcription factor derived from whole-genome duplication in Arabidopsis thaliana. Plant Physiol. 2015, 168, 1717–1734. [Google Scholar] [CrossRef]
- Haake, V.; Cook, D.; Riechmann, J.L.; Pineda, O.; Thomashow, M.F.; Zhang, J.Z. Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol. 2002, 130, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Skinner, J.S.; von Zitzewitz, J.; Szucs, P.; Marquez-Cedillo, L.; Filichkin, T.; Amundsen, K.; Stockinger, E.J.; Thomashow, M.F.; Chen, T.H.; Hayes, P.M. Structural, functional, and phylogenetic characterization of a large CBF gene family in barley. Plant Mol. Biol. 2005, 59, 533–551. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.K.; Galiba, G.; Dubcovsky, J. A cluster of 11 CBF transcription factors is located at the frost tolerance locus Fr-Am2 in Triticum monococcum. Mol. Genet. Genom. 2006, 275, 193–203. [Google Scholar] [CrossRef]
- Badawi, M.; Danyluk, J.; Boucho, B.; Houde, M.; Sarhan, F. The CBF gene family in hexaploid wheat and its relationship to the phylogenetic complexity of cereal CBFs. Mol. Genet. Genom. 2007, 277, 533–554. [Google Scholar] [CrossRef]
- Knox, A.K.; Li, C.; Vagujfalvi, A.; Galiba, G.; Stockinger, E.J.; Dubcovsky, J. Identification of candidate CBF genes for the frost tolerance locus Fr-Am2 in Triticum monococcum. Plant Mol. Biol. 2008, 67, 257–270. [Google Scholar] [CrossRef]
- Pasquariello, M.; Barabaschi, D.; Himmelbach, A.; Steuernagel, B.; Ariyadasa, R.; Stein, N.; Gandolfi, F.; Tenedini, E.; Bernardis, I.; Tagliafico, E.; et al. The barley Frost resistance-H2 locus. Funct. Integr. Genom. 2014, 14, 85–100. [Google Scholar] [CrossRef]
- Dhillon, T.; Morohashi, K.; Stockinger, E.J. CBF2A-CBF4B genomic region copy numbers alongside the circadian clock play key regulatory mechanisms driving expression of FR-H2 CBFs. Plant Mol. Biol. 2017, 94, 333–347. [Google Scholar] [CrossRef] [PubMed]
- Francia, E.; Morcia, C.; Pasquariello, M.; Mazzamurro, V.; Milc, J.A.; Rizza, F.; Terzi, V.; Pecchioni, N. Copy number variation at the HvCBF4–HvCBF2 genomic segment is a major component of frost resistance in barley. Plant Mol. Biol. 2016, 92, 161–175. [Google Scholar] [CrossRef] [PubMed]
- Mareri, L.; Milc, J.; Laviano, L.; Buti, M.; Vautrin, S.; Cauet, S.; Mascagni, F.; Natali, L.; Cavallini, A.; Berges, H.; et al. Influence of CNV on transcript levels of HvCBF genes at Fr-H2 locus revealed by resequencing in resistant barley cv. ’Nure’ and expression analysis. Plant Sci. 2020, 290, 110305. [Google Scholar] [CrossRef]
- Wesenberg, D.M.; Baenziger, P.S.; Rasmusson, D.C.; Burrup, D.E.; Jones, B.L. Registration of 88Ab536-B barley germplasm. Crop Sci. 1998, 38, 559. [Google Scholar] [CrossRef]
- Munoz-Amatriain, M.; Cistue, L.; Xiong, Y.; Bilgic, H.; Budde, A.D.; Schmitt, M.R.; Smith, K.P.; Hayes, P.M.; Muehlbauer, G.J. Structural and functional characterization of a winter malting barley. Theor. Appl. Genet. 2010, 120, 971–984. [Google Scholar] [CrossRef]
- Fricano, A.; Rizza, F.; Faccioli, P.; Pagani, D.; Pavan, P.; Stella, A.; Rossini, L.; Piffanelli, P.; Cattivelli, L. Genetic variants of HvCbf14 are statistically associated with frost tolerance in a European germplasm collection of Hordeum vulgare. Theor. Appl. Genet. 2009, 119, 1335–1348. [Google Scholar] [CrossRef][Green Version]
- Distelfeld, A.; Li, C.; Dubcovsky, J. Regulation of flowering in temperate cereals. Curr. Opin. Plant Biol. 2009, 12, 178–184. [Google Scholar] [CrossRef]
- Greenup, A.; Peacock, W.J.; Dennis, E.S.; Trevaskis, B. The molecular biology of seasonal flowering-responses in Arabidopsis and the cereals. Ann. Bot. 2009, 103, 1165–1172. [Google Scholar] [CrossRef] [PubMed]
- Nitcher, R.; Distelfeld, A.; Tan, C.; Yan, L.; Dubcovsky, J. Increased copy number at the HvFT1 locus is associated with accelerated flowering time in barley. Mol. Genet. Genom. 2013, 288, 261–275. [Google Scholar] [CrossRef]
- Trevaskis, B.; Hemming, M.N.; Dennis, E.S.; Peacock, W.J. The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci. 2007, 12, 352–357. [Google Scholar] [CrossRef]
- Trevaskis, B. The central role of the VERNALIZATION1 gene in the vernalization response of cereals. Funct. Plant Biol. 2010, 37, 479–487. [Google Scholar] [CrossRef]
- Szűcs, P.; Skinner, J.S.; Karsai, I.; Cuesta-Marcos, A.; Haggard, K.G.; Corey, A.E.; Chen, T.H.; Hayes, P.M. Validation of the VRN-H2/VRN-H1 epistatic model in barley reveals that intron length variation in VRN-H1 may account for a continuum of vernalization sensitivity. Mol. Genet. Genom. 2007, 277, 249–261. [Google Scholar] [CrossRef]
- Karsai, I.; Mészáros, K.; Hayes, P.M.; Bedő, Z. Effects of loci on chromosomes 2 (2H) and 7 (5H) on developmental patterns in barley (Hordeum vulgare L.) under different photoperiod regimes. Theor. Appl. Genet. 1997, 94, 612–618. [Google Scholar] [CrossRef]
- Mahfoozi, S.; Limin, A.E.; Hayes, P.M.; Hucl, P.; Fowler, D.B. Influence of photoperiod response on the expression of cold hardiness in wheat and barley. Can. J. Plant Sci. 2000, 80, 721–724. [Google Scholar] [CrossRef]
- Fowler, D.B.; Breton, G.; Limin, A.E.; Mahfoozi, S.; Sarhan, F. Photoperiod and temperature interactions regulate low-temperature-induced gene expression in barley. Plant Physiol. 2001, 127, 1676–1681. [Google Scholar] [CrossRef] [PubMed]
- Danyluk, J.; Kane, N.A.; Breton, G.; Limin, A.E.; Fowler, D.B.; Sarhan, F. TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiol. 2003, 132, 1849–1860. [Google Scholar] [CrossRef] [PubMed]
- Fowler, D.B.; Chauvin, L.P.; Limin, A.E.; Sarhan, F. The regulatory role of vernalization in the expression of low-temperature-induced genes in wheat and rye. Theor. Appl. Genet. 1996, 93, 554–559. [Google Scholar] [CrossRef]
- Dhillon, T.; Pearce, S.; Stockinger, E.; Distelfeld, A.; Li, C.; Knox, A.K.; Vashegyi, I.; Vagujfalvi, A.; Galiba, G.; Dubcovsky, J. Regulation of freezing tolerance and flowering in temperate cereals: The VRN-1 connection. Plant Physiol. 2010, 153, 1846–1858. [Google Scholar] [CrossRef]
- Deng, W.; Casao, M.C.; Wang, P.; Sato, K.; Hayes, P.M.; Finnegan, E.J.; Trevaskis, B. Direct links between the vernalization response and other key traits of cereal crops. Nat. Commun. 2015, 6, 5882. [Google Scholar] [CrossRef] [PubMed]
- Dubcovsky, J.; Loukoianov, A.; Fu, D.; Valarik, M.; Sanchez, A.; Yan, L. Effect of photoperiod on the regulation of wheat vernalization genes VRN1 and VRN2. Plant Mol. Biol. 2006, 60, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Trevaskis, B.; Hemming, M.N.; Peacock, W.J.; Dennis, E.S. HvVRN2 responds to daylength, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiol. 2006, 140, 1397–1405. [Google Scholar] [CrossRef] [PubMed]
- Díaz, A.; Zikhali, M.; Turner, A.S.; Isaac, P.; Laurie, D.A. Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum). PLoS ONE 2012, 7, e33234. [Google Scholar] [CrossRef]
- Zhu, J.; Pearce, S.; Burke, A.; See, D.R.; Skinner, D.Z.; Dubcovsky, J.; Garland-Campbell, K. Copy number and haplotype variation at the VRN-A1 and central FR-A2 loci are associated with frost tolerance in hexaploid wheat. Theor. Appl. Genet. 2014. [Google Scholar] [CrossRef]
- Saisho, D.; Ishii, M.; Hori, K.; Sato, K. Natural variation of barley vernalization requirements: Implication of quantitative variation of winter growth habit as an adaptive trait in East Asia. Plant Cell. Physiol. 2011, 52, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Jeknić, Z.; Pillman, K.A.; Dhillon, T.; Skinner, J.S.; Veisz, O.; Cuesta-Marcos, A.; Hayes, P.M.; Jacobs, A.K.; Chen, T.H.H.; Stockinger, E.J. Hv-CBF2A overexpression in barley accelerates COR gene transcript accumulation and acquisition of freezing tolerance during cold acclimation. Plant Mol. Biol. 2014, 84, 67–82. [Google Scholar] [CrossRef] [PubMed]
- Soltész, A.; Smedley, M.; Vashegyi, I.; Galiba, G.; Harwood, W.; Vágújfalvi, A. Transgenic barley lines prove the involvement of TaCBF14 and TaCBF15 in the cold acclimation process and in frost tolerance. J. Exp. Bot. 2013, 64, 1849–1862. [Google Scholar] [CrossRef]
- Thomas, J.B.; Gaudet, D.A. The winter wheat breeding program at Lethbridge: Some ideas and approaches. In Proceedings of the New Frontiers in Winter Wheat Production, Saskatoon, SK, Canada, 20–22 June 1983; pp. 208–227. [Google Scholar]
- Thomas, J.B.; Schaalje, G.B.; Grant, M.N. Survival, height and genotype by environment interaction in winter wheat. Can. J. Plant Sci. 1993, 73, 417–427. [Google Scholar] [CrossRef]
- Thomas, J.B.; Schaalje, G.B. Winter survival and competition in a mixture of winter wheat cultivars. Crop Sci. 1997, 37, 732–738. [Google Scholar] [CrossRef]
- Klages, K.H. Metrical attributes and the physiology of hardy varieties of winter wheat. Agron. J. 1926, 18, 529–566. [Google Scholar] [CrossRef]
- Roberts, D.W.A. The effect of light on development of the rosette growth habit of winter wheat. Can. J. Bot. 1984, 62, 818–822. [Google Scholar] [CrossRef]
- Roberts, D.W.A. Induction of prostrate growth habit in winter wheat under artificial conditions. Res. Highlights Agric. Can. Res. Stn. Lethbridge 1982, 48–49. [Google Scholar]
- Roberts, D.W.A. Correlations between cold hardiness and plant growth characteristics in winter wheat. Res. Highlights Agric. Can. Res. Stn. Lethbridge 1980, 16–18. [Google Scholar]
- Gray, G.R.; Chauvin, L.P.; Sarhan, F.; Huner, N. Cold acclimation and freezing tolerance (a complex interaction of light and temperature). Plant Physiol. 1997, 114, 467–474. [Google Scholar] [CrossRef]
- Dhillon, T.; Stockinger, E.J. Cbf14 copy number variation in the A, B, and D genomes of diploid and polyploid wheat. Theor. Appl. Genet. 2013, 126, 2777–2789. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, Z.; Xie, S.; Si, T.; Li, Y.; Zhu, J.K. Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol. 2016, 171, 2744–2759. [Google Scholar] [CrossRef]
- Jia, Y.; Ding, Y.; Shi, Y.; Zhang, X.; Gong, Z.; Yang, S. The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 2016, 212, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Yan, Y.; Jiang, B.; Shi, Y.; Jia, Y.; Cheng, J.; Shi, Y.; Kang, J.; Li, H.; Zhang, D.; et al. The cold response regulator CBF1 promotes Arabidopsis hypocotyl growth at ambient temperatures. EMBO J. 2020, 39, e103630. [Google Scholar] [CrossRef]
- Leivar, P.; Quail, P.H. PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 2011, 16, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Mayba, O.; Pfeiffer, A.; Shi, H.; Tepperman, J.M.; Speed, T.P.; Quail, P.H. A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 2013, 9, e1003244. [Google Scholar] [CrossRef]
- Lee, C.M.; Thomashow, M.F. Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2012, 109, 15054–15059. [Google Scholar] [CrossRef]
- Jiang, B.; Shi, Y.; Zhang, X.; Xin, X.; Qi, L.; Guo, H.; Li, J.; Yang, S. PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E6695–E6702. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Shi, Y.; Peng, Y.; Jia, Y.; Yan, Y.; Dong, X.; Li, H.; Dong, J.; Li, J.; Gong, Z.; et al. Cold-induced CBF-PIF3 interaction enhances freezing tolerance by stabilizing the phyB thermosensor in Arabidopsis. Mol. Plant 2020, 13, 894–906. [Google Scholar] [CrossRef] [PubMed]
- Ahres, M.; Gierczik, K.; Boldizsar, A.; Vitamvas, P.; Galiba, G. Temperature and light-quality-dependent regulation of freezing tolerance in barley. Plants 2020, 9, 83. [Google Scholar] [CrossRef]
- Peng, J.; Richards, D.E.; Hartley, N.M.; Murphy, G.P.; Devos, K.M.; Flintham, J.E.; Beales, J.; Fish, L.J.; Worland, A.J.; Pelica, F.; et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 1999, 400, 256–261. [Google Scholar] [CrossRef]
- Pearce, S.; Saville, R.; Vaughan, S.P.; Chandler, P.M.; Wilhelm, E.P.; Sparks, C.A.; Al-Kaff, N.; Korolev, A.; Boulton, M.I.; Phillips, A.L.; et al. Molecular characterization of Rht-1 dwarfing genes in hexaploid wheat. Plant Physiol. 2011, 157, 1820–1831. [Google Scholar] [CrossRef]
- Chandler, P.M.; Marion-Poll, A.; Ellis, M.; Gubler, F. Mutants at the Slender1 locus of barley cv Himalaya. Molecular and physiological characterization. Plant Physiol. 2002, 129, 181–190. [Google Scholar] [CrossRef]
- Hedden, P.; Sponsel, V. A Century of Gibberellin Research. J. Plant Growth Regul. 2015, 34, 740–760. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Martinez, C.; Gusmaroli, G.; Wang, Y.; Zhou, J.; Wang, F.; Chen, L.; Yu, L.; Iglesias-Pedraz, J.M.; Kircher, S.; et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 2008, 451, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Nohales, M.A.; Kay, S.A. GIGANTEA gates gibberellin signaling through stabilization of the DELLA proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 21893–21899. [Google Scholar] [CrossRef] [PubMed]
- Blazquez, M.A.; Trenor, M.; Weigel, D. Independent control of gibberellin biosynthesis and flowering time by the circadian clock in Arabidopsis. Plant Physiol. 2002, 130, 1770–1775. [Google Scholar] [CrossRef]
- Zhao, X.; Yu, X.; Foo, E.; Symons, G.M.; Lopez, J.; Bendehakkalu, K.T.; Xiang, J.; Weller, J.L.; Liu, X.; Reid, J.B.; et al. A study of gibberellin homeostasis and cryptochrome-mediated blue light inhibition of hypocotyl elongation. Plant Physiol. 2007, 145, 106–118. [Google Scholar] [CrossRef]
- Arana, M.V.; Marin-de la Rosa, N.; Maloof, J.N.; Blazquez, M.A.; Alabadi, D. Circadian oscillation of gibberellin signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 9292–9297. [Google Scholar] [CrossRef]
- Achard, P.; Liao, L.; Jiang, C.; Desnos, T.; Bartlett, J.; Fu, X.; Harberd, N.P. DELLAs contribute to plant photomorphogenesis. Plant Physiol. 2007, 143, 1163–1172. [Google Scholar] [CrossRef]
- Fowler, D.B.; Gusta, L.V. Selection for winterhardiness in wheat. 1. Identification of genotypic variability. Crop Sci. 1979, 19, 769–772. [Google Scholar] [CrossRef]
- Gilliland, D.J.; Fowler, D.B. Effect of a Rht gene conditioning the semidwarf character on winterhardiness in winter wheat (Triticum aestivum L. em Thell). Can. J. Plant Sci. 1988, 68, 301–309. [Google Scholar] [CrossRef]
- Jatayev, S.; Sukhikh, I.; Vavilova, V.; Smolenskaya, S.E.; Goncharov, N.P.; Kurishbayev, A.; Zotova, L.; Absattarova, A.; Serikbay, D.; Hu, Y.G.; et al. Green revolution ‘stumbles’ in a dry environment: Dwarf wheat with Rht genes fails to produce higher grain yield than taller plants under drought. Plant Cell Environ. 2020, 43, 2355–2364. [Google Scholar] [CrossRef]
- Burleigh, J.R.; Allan, R.E.; Vogel, O.A. Varietal differences in seedling emergence of winter wheats as influenced by temperature and depth of plants. Agron. J. 1965, 57, 195–198. [Google Scholar] [CrossRef]
- Butler, J.D.; Byrne, P.F.; Mohammadi, V.; Chapman, P.L.; Haley, S.D. Agronomic performance of Rht alleles in a spring wheat population across a range of moisture levels. Crop Sci. 2005, 45, 939–947. [Google Scholar] [CrossRef]
- Guedira, M.; Brown-Guedira, G.; Van Sanford, D.; Sneller, C.; Souza, E.; Marshall, D. Distribution of Rht genes in modern and historic winter wheat cultivars from the eastern and central USA. Crop Sci. 2010, 50, 1811–1822. [Google Scholar] [CrossRef]
- Pereira, M.J.; Pfahler, P.T.; Barnett, R.D.; Blount, A.R.; Wofford, D.S.; Littell, R.C. Coleoptile length of dwarf wheat isolines: Gibberellic acid, temperature, and cultivar interactions. Crop Sci. 2002, 42, 1483–1487. [Google Scholar] [CrossRef]
- Thomas, W.T.B.; Forster, B.P.; Waugh, R. Natural and Induced Mutants of Barley: Single Nucleotide Polymorphisms in Genes Important for Breeding; Wiley-Blackwell: Chichester, UK, 2010; pp. 217–230. [Google Scholar]
- Ali, M.A.M.; Okiror, O.; Rasmusson, D.C. Performance of semidwarf barley. Crop Sci. 1978, 18, 418–422. [Google Scholar] [CrossRef]
- Mickelson, H.R.; Rasmusson, D.C. Genes for short stature in barley. Crop Sci. 1994, 34, 1180–1183. [Google Scholar] [CrossRef]
- Pakniyat, H.; Handley, L.L.; Thomas, W.T.B.; Connolly, T.; Macaulay, M.; Caligari, P.D.S.; Forster, B.P. Comparison of shoot dry weight, Na+ content and δ13C values of ari-e and other semi-dwarf barley mutants under salt-stress. Euphytica 1997, 94, 7–14. [Google Scholar] [CrossRef]
- Bouma, J. New Variety of Spring Barley ‘Diamant’ in Czechoslovakia; Deutsche Akademie der Wissenschaften zu Berlin: Berlin, Germany, 1967; Volume 2, pp. 177–182. [Google Scholar]
- Bouma, J.; Ohnoutka, Z. Importance and application of the mutant ‘Diamant’ in spring barley breeding. In Proceedings of the International Symposium on the Contribution of Plant Mutation Breeding to Crop Improvement, Vienna, Austria, 18–22 June 1990; pp. 127–133. [Google Scholar]
- Thomas, W.T.B.; Powell, W.; Swanston, J.S. The effects of major genes on quantitatively varying characters in barley. 4. The GPert and denso loci and quality characters. Heredity 1991, 66, 381–389. [Google Scholar] [CrossRef]
- Hellewell, K.B.; Rasmusson, D.C.; Gallo-Meagher, M. Enhancing yield of semidwarf barley. Crop Sci. 2000, 40, 352–358. [Google Scholar] [CrossRef]
- Jia, Q.; Zhang, J.; Westcott, S.; Zhang, X.Q.; Bellgard, M.; Lance, R.; Li, C. GA-20 oxidase as a candidate for the semidwarf gene sdw1/denso in barley. Funct. Integr. Genom. 2009, 9, 255–262. [Google Scholar] [CrossRef]
- Jia, Q.; Zhang, X.Q.; Westcott, S.; Broughton, S.; Cakir, M.; Yang, J.; Lance, R.; Li, C. Expression level of a gibberellin 20-oxidase gene is associated with multiple agronomic and quality traits in barley. Theor. Appl. Genet. 2011, 122, 1451–1460. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Jia, Q.; Zhou, G.; Zhang, X.Q.; Angessa, T.; Broughton, S.; Yan, G.; Zhang, W.; Li, C. Characterization of the sdw1 semi-dwarf gene in barley. BMC Plant. Biol. 2017, 17, 11. [Google Scholar] [CrossRef]
- Corns, W.G. Effects of seed treatments with gibberellin and dates of seeding on winter survival and vegetative yield of Kharkov wheat. Can. J. Plant Sci. 1959, 39, 293–296. [Google Scholar] [CrossRef]
- Jung, J. The effect of CCC on winter-hardiness and culm length of wheat. Z. Acker.-Pflanzenbau 1965, 122, 9–14. [Google Scholar]
- Wunsche, U. Influence of 2-chloroethyl trimethylammonium chloride and gibberellin A3 on frost hardiness of winter wheat. Naturwissenschaften 1966, 53, 386–387. [Google Scholar] [CrossRef]
- Roberts, D.W.A. The effect of CGC and gibberellins A3 and A7 on the cold hardiness of Kharkov 22 MC winter wheat. Can. J. Bot. 1971, 49, 705–711. [Google Scholar] [CrossRef]
- Gilmour, S.J.; Sebolt, A.M.; Salazar, M.P.; Everard, J.D.; Thomashow, M.F. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 2000, 124, 1854–1865. [Google Scholar] [CrossRef]
- Hsieh, T.H.; Lee, J.T.; Yang, P.T.; Chiu, L.H.; Charng, Y.Y.; Wang, Y.C.; Chan, M.T. Heterology expression of the Arabidopsis C-repeat/dehydration response element binding factor 1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiol. 2002, 129, 1086–1094. [Google Scholar] [CrossRef] [PubMed]
- Achard, P.; Gong, F.; Cheminant, S.; Alioua, M.; Hedden, P.; Genschik, P. The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 2008, 20, 2117–2129. [Google Scholar] [CrossRef]
- Magome, H.; Yamaguchi, S.; Hanada, A.; Kamiya, Y.; Oda, K. The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant J. 2008, 56, 613–626. [Google Scholar] [CrossRef]
- Zhou, M.; Chen, H.; Wei, D.; Ma, H.; Lin, J. Arabidopsis CBF3 and DELLAs positively regulate each other in response to low temperature. Sci. Rep. 2017, 7, 39819. [Google Scholar] [CrossRef]
- Lantzouni, O.; Alkofer, A.; Falter-Braun, P.; Schwechheimer, C. GROWTH-REGULATING FACTORS interact with DELLAs and regulate growth in cold stress. Plant Cell 2020, 32, 1018–1034. [Google Scholar] [CrossRef]
- Huang, W.; He, Y.; Yang, L.; Lu, C.; Zhu, Y.; Sun, C.; Ma, D.; Yin, J. Genome-wide analysis of growth-regulating factors (GRFs) in Triticum aestivum. Peer. J. 2021, 9, e10701. [Google Scholar] [CrossRef] [PubMed]
- Omidbakhshfard, M.A.; Proost, S.; Fujikura, U.; Mueller-Roeber, B. Growth-Regulating Factors (GRFs): A small transcription factor family with important functions in plant biology. Mol. Plant 2015, 8, 998–1010. [Google Scholar] [CrossRef]
- Bayer, M.M.; Rapazote-Flores, P.; Ganal, M.; Hedley, P.E.; Macaulay, M.; Plieske, J.; Ramsay, L.; Russell, J.; Shaw, P.D.; Thomas, W.; et al. Development and evaluation of a barley 50k iSelect SNP array. Front. Plant Sci. 2017, 8, 1792. [Google Scholar] [CrossRef]
- Pennycooke, J.C.; Cheng, H.; Roberts, S.M.; Yang, Q.; Rhee, S.Y.; Stockinger, E.J. The low temperature-responsive, Solanum CBF1 genes maintain high identity in their upstream regions in a genomic environment undergoing gene duplications, deletions, and rearrangements. Plant Mol. Biol. 2008, 67, 483–497. [Google Scholar] [CrossRef] [PubMed]
- Henrichsen, C.N.; Vinckenbosch, N.; Zollner, S.; Chaignat, E.; Pradervand, S.; Schutz, F.; Ruedi, M.; Kaessmann, H.; Reymond, A. Segmental copy number variation shapes tissue transcriptomes. Nat. Genet. 2009, 41, 424–429. [Google Scholar] [CrossRef] [PubMed]
- Gheldof, N.; Witwicki, R.M.; Migliavacca, E.; Leleu, M.; Didelot, G.; Harewood, L.; Rougemont, J.; Reymond, A. Structural variation-associated expression changes are paralleled by chromatin architecture modifications. PLoS ONE 2013, 8, e79973. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Kobayashi, M.; Itoh, H.; Tagiri, A.; Kayano, T.; Tanaka, H.; Iwahori, S.; Matsuoka, M. Expression of a gibberellin 2-oxidase gene around the shoot apex is related to phase transition in rice. Plant Physiol. 2001, 125, 1508–1516. [Google Scholar] [CrossRef] [PubMed]
- Pearce, S.; Vanzetti, L.S.; Dubcovsky, J. Exogenous gibberellins induce wheat spike development under short days only in the presence of VERNALIZATION1. Plant Physiol. 2013, 163, 1433–1445. [Google Scholar] [CrossRef] [PubMed]
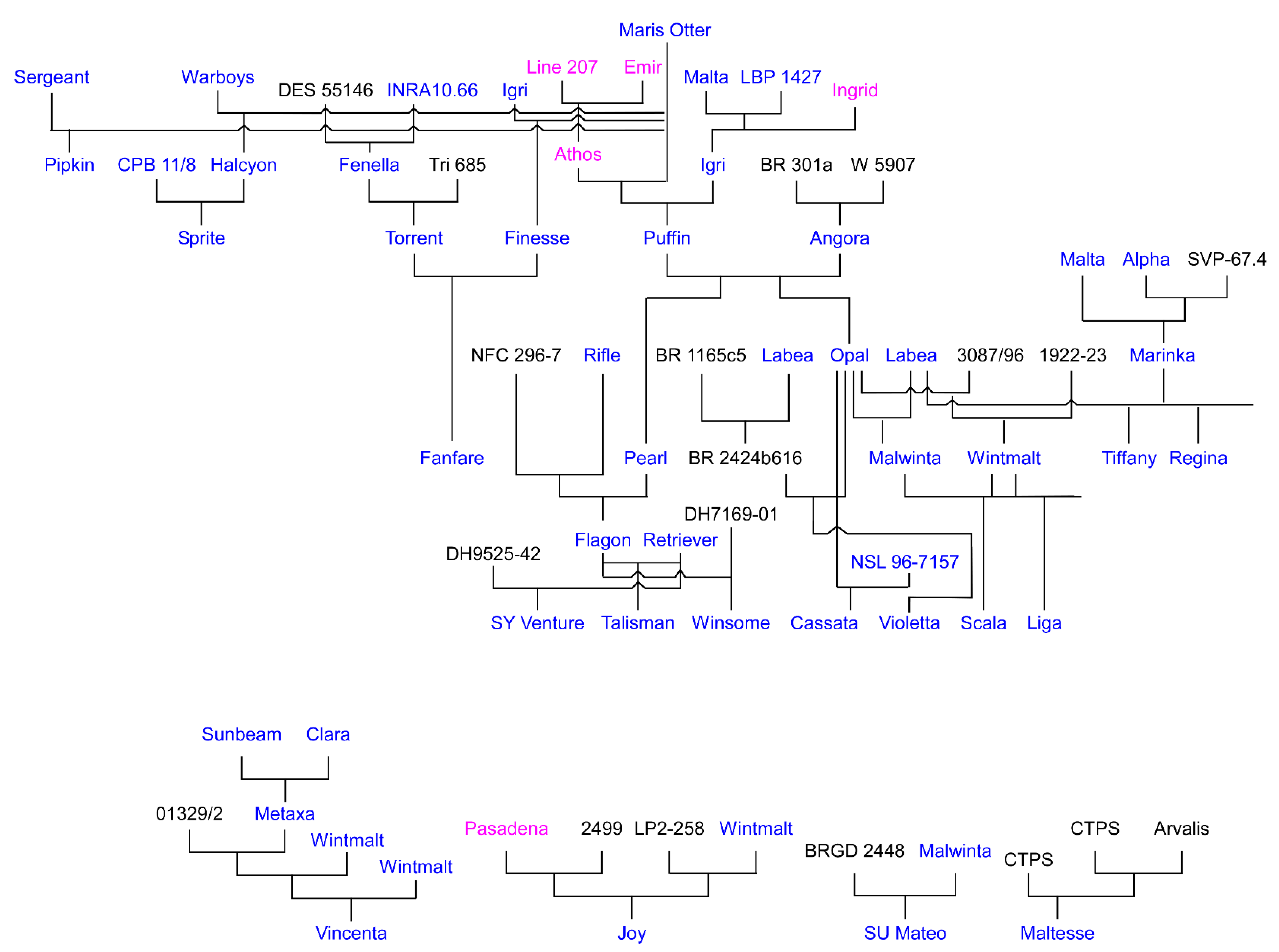
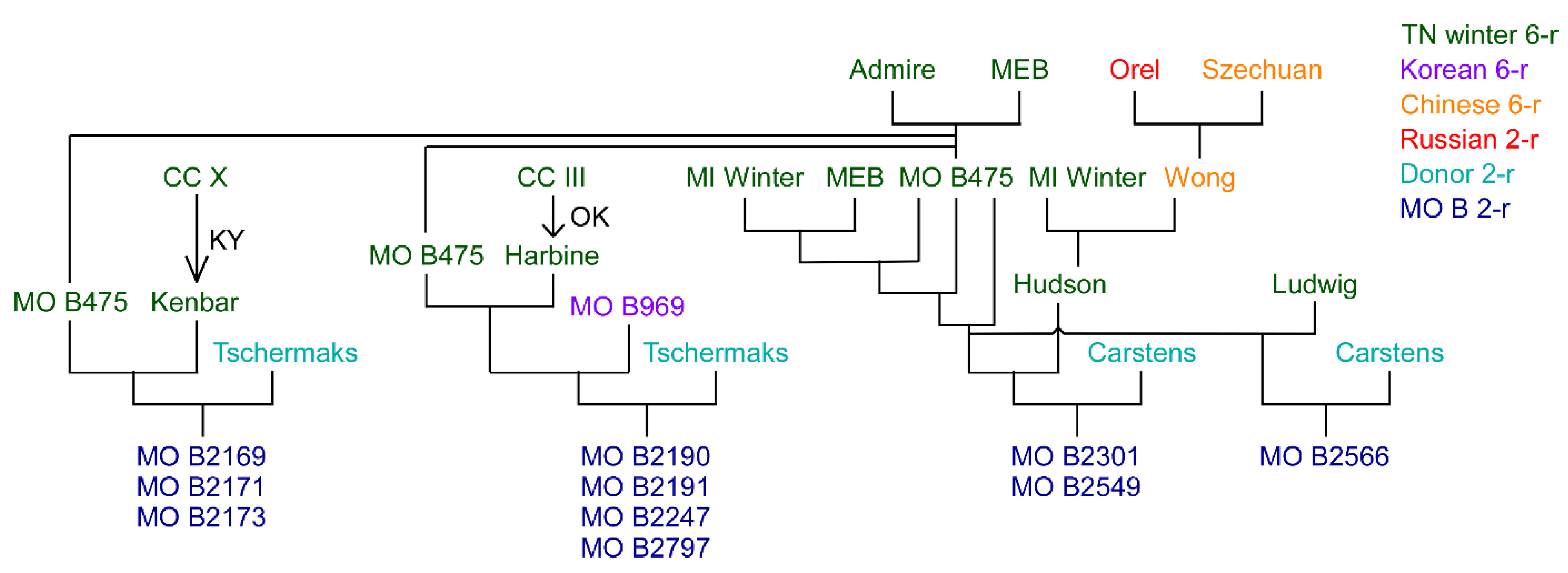
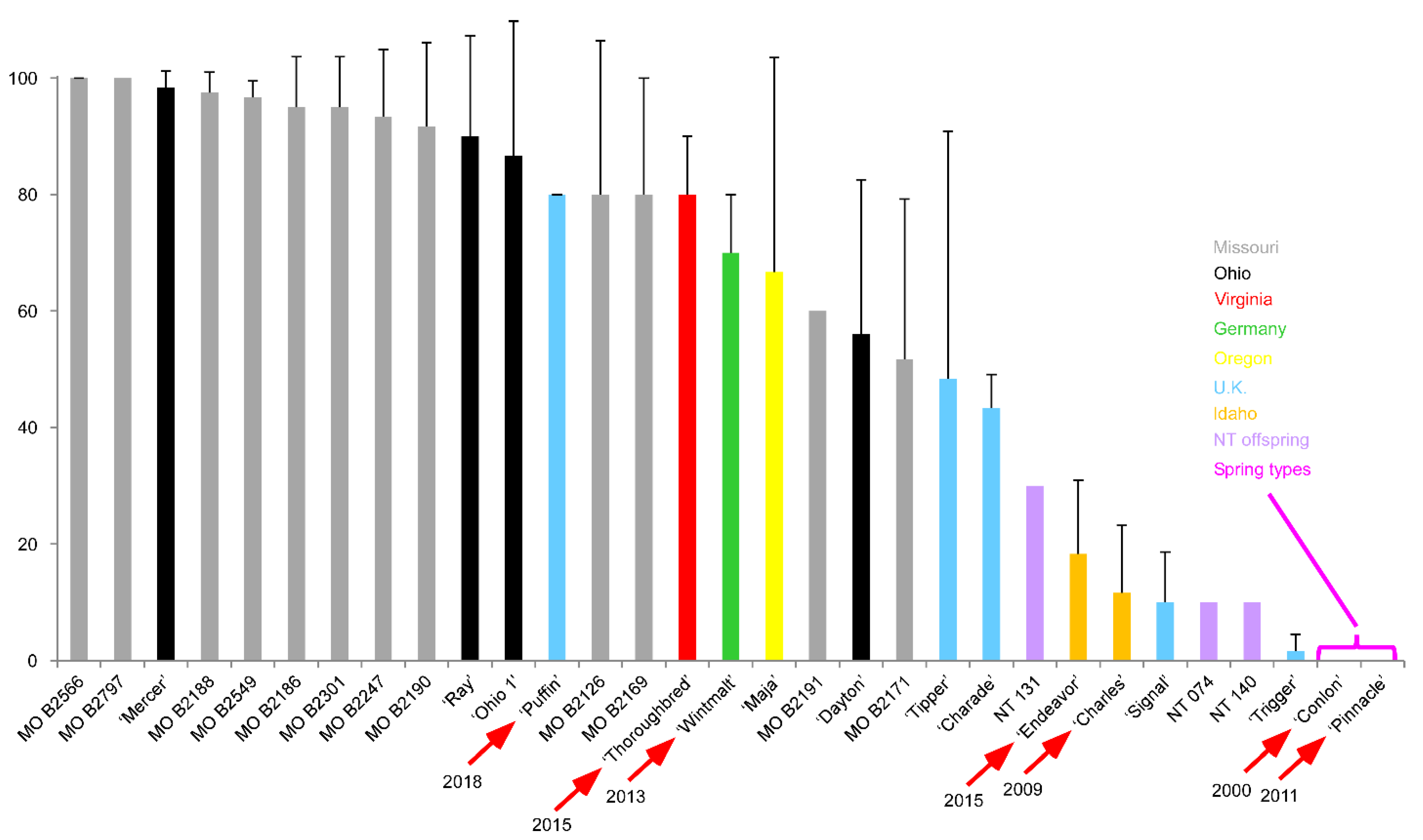
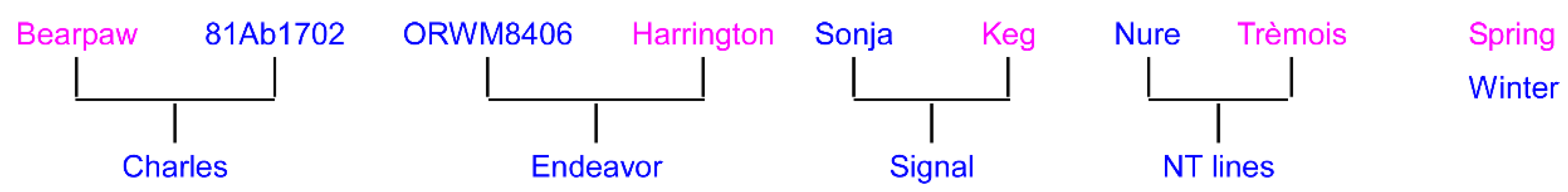
| Rank | Name | CIho No. | Pedigree |
|---|---|---|---|
| 1 | Kearney | 7580 | Selection from Composite Cross III (CIho 5530) |
| 2 | Dicktoo | 5529 | Selection #2 from unknown cross made pre 1917 at Dickinson, North Dakota |
| 3 | OAC * 4GH1 | 10,096 | Kenate/Wong |
| 4 | MO B893 | 9516 | Selection from Ludwig (CI 7525) |
| 5 | NE 62434 | 9581 | Selection from Korean landrace Cha-Dae-Maec (PI 157656, CI 7404) |
| 6 | NE 53417 | 9580 | Wong/Ludwig |
| 7 | Admire | 6377 | Selection from a Kansas farmer’s field |
| 8 | Kansas South Central | 6376 | Bulk seed from Kansas farmers’ fields |
| 9 | OAE 30GH10 | 10,097 | Kenate/Wong |
| 10 | Purdue B466A7-7-7-2 | 10,102 | Purdue 28156A3-2-2-2/Wisconsin H42-5-4-5-1-1/Kentucky1/ Purdue 400-17/Wong |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stockinger, E.J. The Breeding of Winter-Hardy Malting Barley. Plants 2021, 10, 1415. https://doi.org/10.3390/plants10071415
Stockinger EJ. The Breeding of Winter-Hardy Malting Barley. Plants. 2021; 10(7):1415. https://doi.org/10.3390/plants10071415
Chicago/Turabian StyleStockinger, Eric J. 2021. "The Breeding of Winter-Hardy Malting Barley" Plants 10, no. 7: 1415. https://doi.org/10.3390/plants10071415
APA StyleStockinger, E. J. (2021). The Breeding of Winter-Hardy Malting Barley. Plants, 10(7), 1415. https://doi.org/10.3390/plants10071415





