Genome Wide MeDIP-Seq Profiling of Wild and Cultivated Olives Trees Suggests DNA Methylation Fingerprint on the Sensory Quality of Olive Oil
Abstract
:1. Introduction
2. Results
2.1. Evolution of DNA Methylation Landscape across Olive Tree Tissues
2.2. Domestication Fingerprint on DNA Methylation Landscape
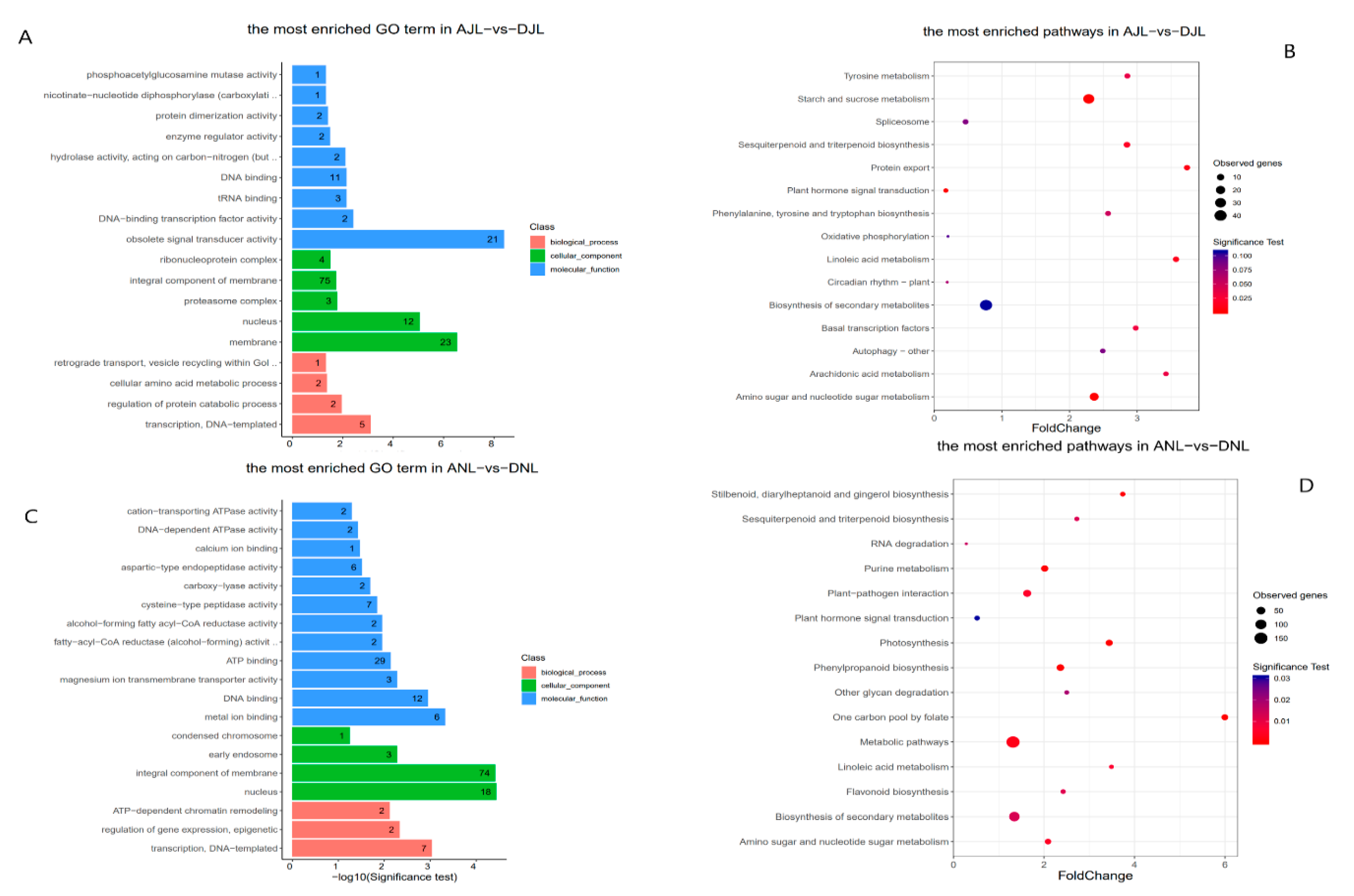
2.3. Functional Enrichment Analysis for Genes Associated with Differentially Methylated Regions
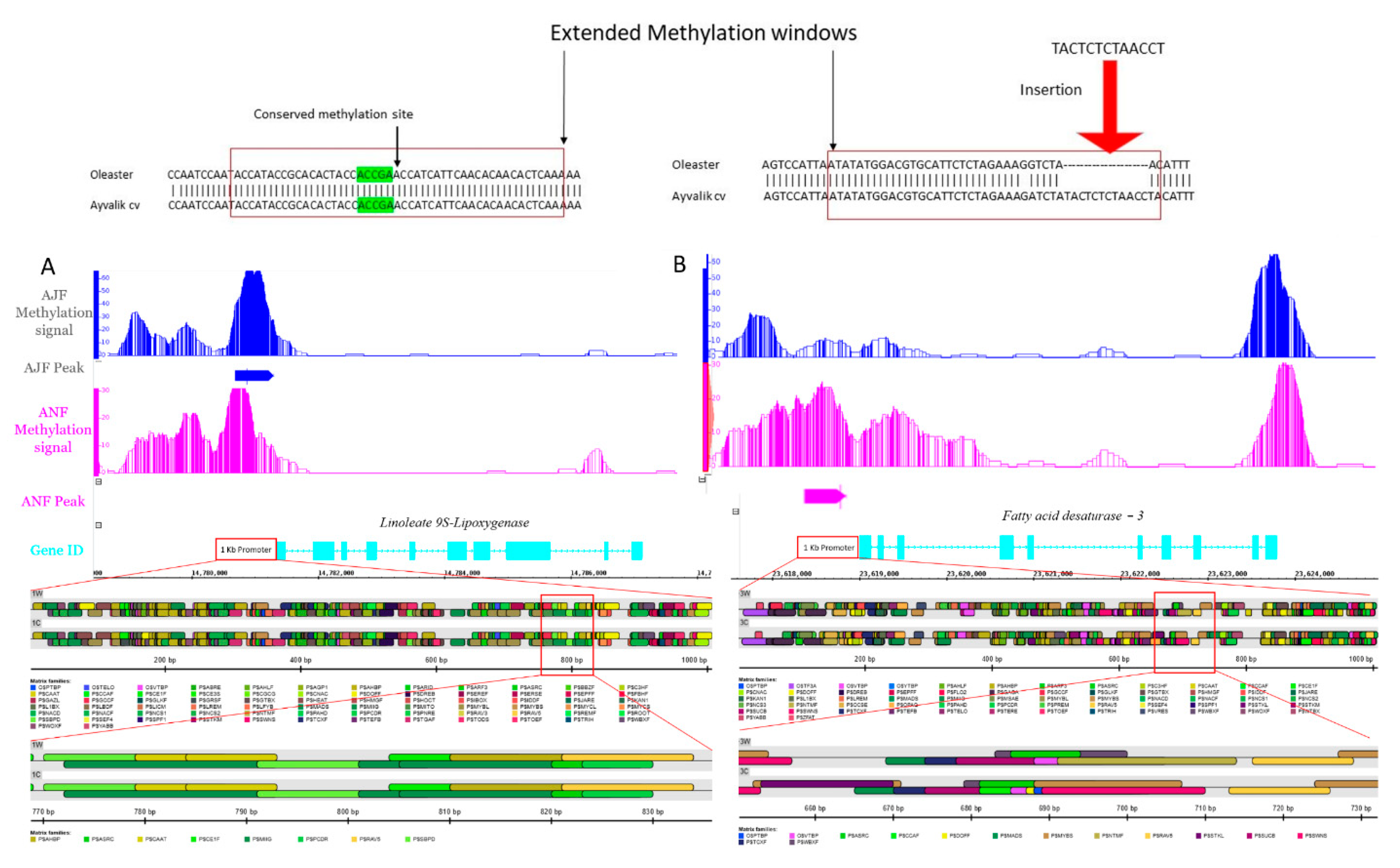
2.4. Comparative Methylated Promoter Analysis between Wild and Cultivated Olives
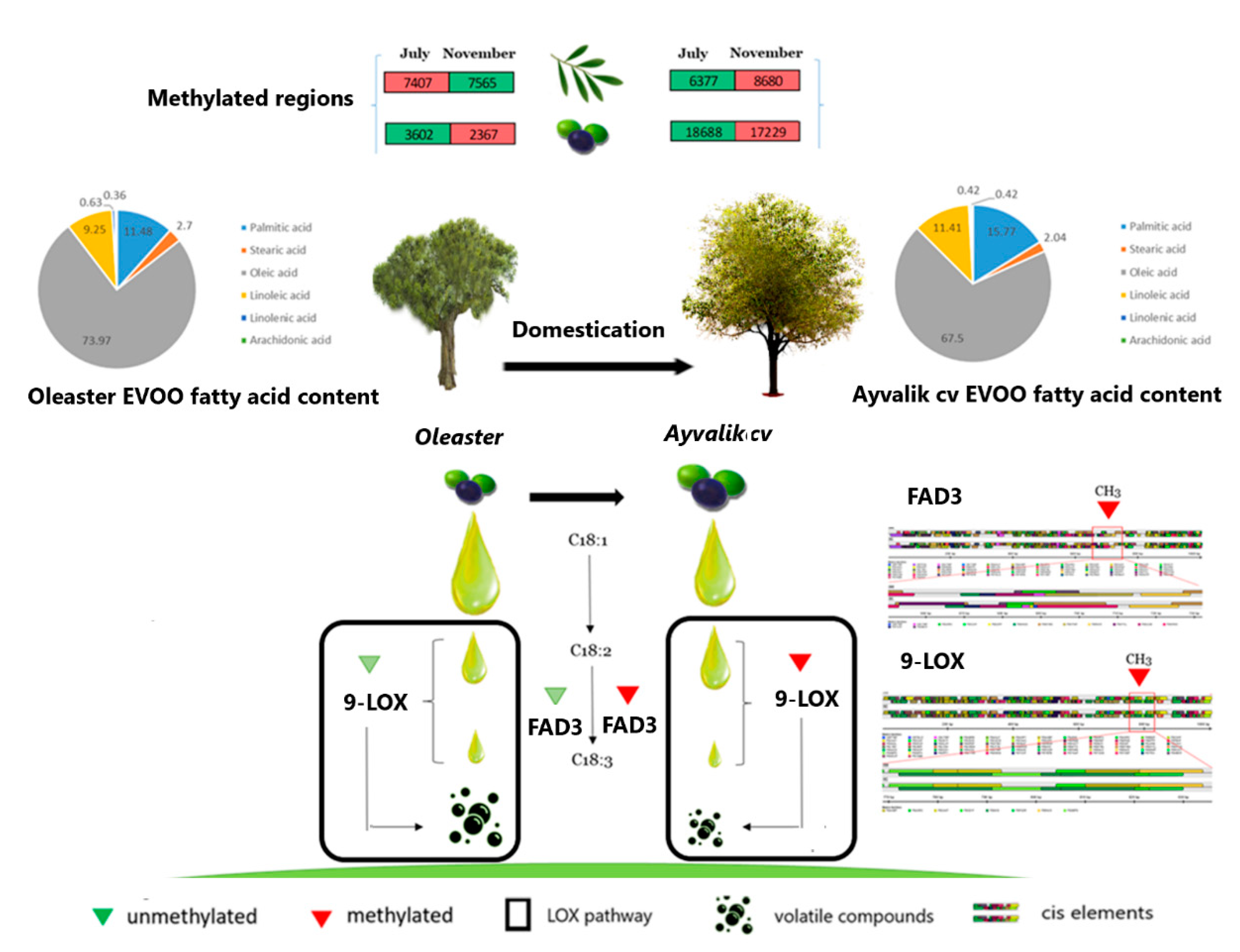
2.5. Metabolic Profiling of Ayvalik Cv and Wild Olea europaea ssp. europaea var. sylvestris
3. Discussion
3.1. Differential Methylation across Tissues
3.2. Global Level of Methylation during Fruit Ripening
3.3. Differential DNA Methylation Potential Role in the Sensory and Organoleptic Properties of Olive Oil
3.3.1. Genes Involved in Polyunsaturated Fatty Acids Metabolism and Their Derivatives Volatile Compounds
3.3.2. Differential Methylation in Putative Genes Involved in Phenolic Compound Metabolism in EVOO
3.4. Domestication Fingerprint on Cis-Elements Modulating Genes Activity Involved in Sensory Quality of Olive Oil
4. Materials and Methods
4.1. Sample Preparation: DNA Extraction and Quantification
4.2. Immunoprecipitated DNA Sequencing, Data Quality Control, and Read Alignment
4.3. Methylated Regions Calling, Annotation, and Functional Analysis
4.4. Comparative Promoter Analysis
4.5. Metabolic Analysis, Fatty Acid and Total Polyphenol Profiling
5. Conclusions
Remarks and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Pedan, V.; Popp, M.; Rohn, S.; Nyfeler, M.; Bongartz, A. Characterization of Phenolic Compounds and Their Contribution to Sensory Properties of Olive Oil. Molecules 2019, 24, 2041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stefani, M.; Rigacci, S. Beneficial properties of natural phenols: Highlight on protection against pathological conditions associated with amyloid aggregation. BioFactors 2014, 40, 482–493. [Google Scholar] [CrossRef]
- Luque-Muñoz, A.; Tapia, R.; Haidour, A.; Justicia, J.; Cuerva, J.M. Direct determination of phenolic secoiridoids in olive oil by ultra-high performance liquid chromatography-triple quadruple mass spectrometry analysis. Sci. Rep. 2019, 9, 15545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andjelkovic, M.; Acun, S.; Hoed, V.; Verhé, R.; Camp, J. Chemical Composition of Turkish Olive Oil–Ayvalik. JAOCS J. Am. Oil Chem. Soc. 2009, 86, 135–140. [Google Scholar] [CrossRef]
- Kelebek, H.; Selli, S.; Kola, O. Quantitative determination of phenolic compounds using LC-DAD-ESI-MS/MS in cv. Ayvalik olive oils as affected by harvest time. J. Food Meas. Charact. 2017, 11. [Google Scholar] [CrossRef]
- Dabbou, S.; Dabbou, S.; Selvaggini, R.; Urbani, S.; Taticchi, A.; Servili, M.; Hammami, M. Comparison of the Chemical Composition and the Organoleptic Profile of Virgin Olive Oil from Two Wild and Two Cultivated Tunisian Olea europaea. Chem. Biodivers. 2011, 8, 189–202. [Google Scholar] [CrossRef]
- Bouarroudj, K.; Tamendjari, A.; Larbat, R. Quality, composition and antioxidant activity of Algerian wild olive (Olea europaea L. subsp. Oleaster) oil. Ind. Crops Prod. 2016, 83. [Google Scholar] [CrossRef]
- Rodrigues, N.; Pinho, T.; Casal, S.; Peres, A.; Baptista, P.; Pereira, J. Chemical Characterization of Oleaster, Olea europaea var. sylvestris (Mill.) Lehr., Oils from Different Locations of Northeast Portugal. Appl. Sci. 2020, 10, 6414. [Google Scholar] [CrossRef]
- Espínola, F.; Vidal Castro, A.; Espínola, J.; Moya, M. Processing Effect and Characterization of Olive Oils from Spanish Wild Olive Trees (Olea europaea var. sylvestris). Molecules 2021, 26, 1304. [Google Scholar] [CrossRef]
- Ozkaya, M.; ErgÜLen, E.; Ülger, S.; Özilbey, N. Molecular Characterization of Some Selected Wild Olive (Olea oleaster L.) Ecotypes Grown in Turkey. J. Agric. Sci. 2009, 15, 14–19. [Google Scholar] [CrossRef]
- Unver, T.; Wu, Z.; Sterck, L.; Turktas, M.; Lohaus, R.; Li, Z.; Yang, M.; He, L.; Deng, T.; Escalante, F.J.; et al. Genome of wild olive and the evolution of oil biosynthesis. Proc. Natl. Acad. Sci. USA 2017, 114, E9413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gugger, P.F.; Fitz-Gibbon, S.; PellEgrini, M.; Sork, V.L. Species-wide patterns of DNA methylation variation in Quercus lobata and their association with climate gradients. Mol. Ecol. 2016, 25, 1665–1680. [Google Scholar] [CrossRef] [PubMed]
- Artemov, A.V.; Mugue, N.S.; Rastorguev, S.M.; Zhenilo, S.; Mazur, A.M.; Tsygankova, S.V.; Boulygina, E.S.; Kaplun, D.; Nedoluzhko, A.V.; Medvedeva, Y.A.; et al. Genome-Wide DNA Methylation Profiling Reveals Epigenetic Adaptation of Stickleback to Marine and Freshwater Conditions. Mol. Biol. Evol. 2017, 34, 2203–2213. [Google Scholar] [CrossRef] [Green Version]
- Metzger, D.C.H.; Schulte, P.M. Persistent and plastic effects of temperature on DNA methylation across the genome of threespine stickleback (Gasterosteus aculeatus). Proc. Biol. Sci. 2017, 284, 20171667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angiolillo, A.; Mencuccini, M.; Baldoni, L. Olive genetic diversity assessed using amplified fragment length polymorphisms. Theor. Appl. Genet. 1999, 98, 411–421. [Google Scholar] [CrossRef]
- Besnard, G.; Bervillé, A. On chloroplast DNA variations in the olive (Olea europaea L.) complex: Comparison of RFLP and PCR polymorphisms. Theor. Appl. Genet. 2002, 104, 1157–1163. [Google Scholar] [CrossRef] [PubMed]
- Bronzini de Caraffa, V.; Giannettini, J.; Gambotti, C.; Maury, J. Genetic relationships between cultivated and wild olives of Corsica and Sardinia using RAPD markers. Euphytica 2002, 123, 263–271. [Google Scholar] [CrossRef]
- Lumaret, R.; Ouazzani, N.; Michaud, H.; Vivier, G.; Deguilloux, M.F.; Di Giusto, F. Allozyme variation of oleaster populations (wild olive tree) (Olea europaea L.) in the Mediterranean Basin. Heredity 2004, 92, 343–351. [Google Scholar] [CrossRef]
- Baldoni, L.; Tosti, N.; Ricciolini, C.; Belaj, A.; Arcioni, S.; Pannelli, G.; Germana, M.A.; Mulas, M.; Porceddu, A. Genetic Structure of Wild and Cultivated Olives in the Central Mediterranean Basin. Ann. Bot. 2006, 98, 935–942. [Google Scholar] [CrossRef]
- Breton, C.; Tersac, M.; Bervillé, A. Genetic diversity and gene flow between the wild olive (oleaster, Olea europaea L.) and the olive: Several Plio-Pleistocene refuge zones in the Mediterranean basin suggested by simple sequence repeats analysis. J. Biogeogr. 2006, 33, 1916–1928. [Google Scholar] [CrossRef]
- Besnard, G.; Khadari, B.; Navascués, M.; Fernández-Mazuecos, M.; El Bakkali, A.; Arrigo, N.; Baali-Cherif, D.; Brunini-Bronzini de Caraffa, V.; Santoni, S.; Vargas, P.; et al. The complex history of the olive tree: From Late Quaternary diversification of Mediterranean lineages to primary domestication in the northern Levant. Proc. Biol. Sci. 2013, 280, 20122833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gros-Balthazard, M.; Besnard, G.; Sarah, G.; Holtz, Y.; Leclercq, J.; Santoni, S.; Wegmann, D.; Glémin, S.; Khadari, B. Evolutionary transcriptomics reveals the origins of olives and the genomic changes associated with their domestication. Plant J. 2019, 100, 143–157. [Google Scholar] [CrossRef]
- Jiménez-Ruiz, J.; Ramírez-Tejero, J.A.; Fernández-Pozo, N.; Leyva-Pérez, M.d.l.O.; Yan, H.; Rosa, R.d.l.; Belaj, A.; Montes, E.; Rodríguez-Ariza, M.O.; Navarro, F.; et al. Transposon activation is a major driver in the genome evolution of cultivated olive trees (Olea europaea L.). Plant Genome 2020, 13, e20010. [Google Scholar] [CrossRef] [Green Version]
- Kelly, T.K.; De Carvalho, D.D.; Jones, P.A. Epigenetic modifications as therapeutic targets. Nat. Biotechnol. 2010, 28, 1069–1078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, Z.D.; Meissner, A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoder, J.A.; Walsh, C.P.; Bestor, T.H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997, 13, 335–340. [Google Scholar] [CrossRef]
- Aran, D.; Toperoff, G.; Rosenberg, M.; Hellman, A. Replication timing-related and gene body-specific methylation of active human genes. Hum. Mol. Genet. 2011, 20, 670–680. [Google Scholar] [CrossRef]
- Dimond, J.L.; Nguyen, N.; Roberts, S.B. DNA methylation profiling of a cnidarian-algal symbiosis using nanopore sequencing. G3 Genes Genomes Genet. 2021. [Google Scholar] [CrossRef]
- Lu, Y.C.; Feng, S.J.; Zhang, J.J.; Luo, F.; Zhang, S.; Yang, H. Genome-wide identification of DNA methylation provides insights into the association of gene expression in rice exposed to pesticide atrazine. Sci. Rep. 2016, 6, 18985. [Google Scholar] [CrossRef] [Green Version]
- Zuo, J.; Wang, Y.; Zhu, B.; Luo, Y.; Wang, Q.; Gao, L. Comparative Analysis of DNA Methylation Reveals Specific Regulations on Ethylene Pathway in Tomato Fruit. Genes 2018, 9, 266. [Google Scholar] [CrossRef] [Green Version]
- Lang, Z.; Wang, Y.; Tang, K.; Tang, D.; Datsenka, T.; Cheng, J.; Zhang, Y.; Handa, A.K.; Zhu, J.-K. Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc. Natl. Acad. Sci. USA 2017, 114, E4511–E4519. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Ge, X.; Wang, J.; Tan, C.; King, G.J.; Liu, K. Genome-wide DNA methylation profiling by modified reduced representation bisulfite sequencing in Brassica rapa suggests that epigenetic modifications play a key role in polyploid genome evolution. Front. Plant Sci. 2015, 6, 836. [Google Scholar] [CrossRef] [Green Version]
- Manning, K.; Tör, M.; Poole, M.; Hong, Y.; Thompson, A.J.; King, G.J.; Giovannoni, J.J.; Seymour, G.B. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 2006, 38, 948–952. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Niu, Q.; Zhang, B.; Chen, K.; Yang, R.; Zhu, J.-K.; Zhang, Y.; Lang, Z. Downregulation of RdDM during strawberry fruit ripening. Genome Biol. 2018, 19, 212. [Google Scholar] [CrossRef] [PubMed]
- Schiller, D.; Contreras, C.; Vogt, J.; Dunemann, F.; Defilippi, B.G.; Beaudry, R.; Schwab, W. A dual positional specific lipoxygenase functions in the generation of flavor compounds during climacteric ripening of apple. Hortic. Res. 2015, 2, 15003. [Google Scholar] [CrossRef] [Green Version]
- Hwang, I.S.; Hwang, B.K. The pepper 9-lipoxygenase gene CaLOX1 functions in defense and cell death responses to microbial pathogens. Plant Physiol. 2010, 152, 948–967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Padilla, M.N.; Hernández, M.L.; Sanz, C.; Martínez-Rivas, J.M. Functional Characterization of Two 13-Lipoxygenase Genes from Olive Fruit in Relation to the Biosynthesis of Volatile Compounds of Virgin Olive Oil. J. Agric. Food Chem. 2009, 57, 9097–9107. [Google Scholar] [CrossRef]
- Nagpala, E.G.; Guidarelli, M.; Gasperotti, M.; Masuero, D.; Bertolini, P.; Vrhovsek, U.; Baraldi, E. Polyphenols Variation in Fruits of the Susceptible Strawberry Cultivar Alba during Ripening and upon Fungal Pathogen Interaction and Possible Involvement in Unripe Fruit Tolerance. J. Agric. Food Chem. 2016, 64, 1869–1878. [Google Scholar] [CrossRef] [PubMed]
- Bertin, N.; Gomez-Caravaca, A.; Cirilli, M.; Caruso, G.; Gennai, C.; Urbani, S.; Frioni, E.; Ruzzi, M.; Servili, M.; Gucci, R.; et al. The Role of Polyphenoloxidase, Peroxidase, and β-Glucosidase in Phenolics Accumulation in Olea europaea L. Fruits under Different Water Regimes. Front. Plant Sci. 2017, 8, 717. [Google Scholar] [CrossRef] [Green Version]
- Velázquez-Palmero, D.; Romero-Segura, C.; García-Rodríguez, R.; Hernández, M.L.; Vaistij, F.E.; Graham, I.A.; Pérez, A.G.; Martínez-Rivas, J.M. An Oleuropein β-Glucosidase from Olive Fruit Is Involved in Determining the Phenolic Composition of Virgin Olive Oil. Front. Plant Sci. 2017, 8, 1902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.; Liu, R.; Niu, Q.; Tang, K.; Zhang, B.; Zhang, H.; Chen, K.; Zhu, J.-K.; Lang, Z. Global increase in DNA methylation during orange fruit development and ripening. Proc. Natl. Acad. Sci. USA 2019, 116, 1430–1436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Messeguer, R.; Ganal, M.W.; Steffens, J.C.; Tanksley, S.D. Characterization of the level, target sites and inheritance of cytosine methylation in tomato nuclear DNA. Plant Mol. Biol. 1991, 16, 753–770. [Google Scholar] [CrossRef]
- Ruiz-García, L.; Cervera, M.T.; Martínez-Zapater, J.M. DNA methylation increases throughout Arabidopsis development. Planta 2005, 222, 301–306. [Google Scholar] [CrossRef]
- Zluvova, J.; Janousek, B.; Vyskot, B. Immunohistochemical study of DNA methylation dynamics during plant development. J. Exp. Bot. 2001, 52, 2265–2273. [Google Scholar] [CrossRef] [PubMed]
- Bartels, A.; Han, Q.; Nair, P.; Stacey, L.; Gaynier, H.; Mosley, M.; Huang, Q.Q.; Pearson, J.K.; Hsieh, T.-F.; An, Y.-Q.C.; et al. Dynamic DNA Methylation in Plant Growth and Development. Int. J. Mol. Sci. 2018, 19, 2144. [Google Scholar] [CrossRef] [Green Version]
- Fray, R.; Zhong, S. 11–Genome-wide DNA methylation in tomato. In Applied Plant Genomics and Biotechnology; Poltronieri, P., Hong, Y., Eds.; Woodhead Publishing: Oxford, UK, 2015; pp. 179–193. [Google Scholar]
- Brenet, F.; Moh, M.; Funk, P.; Feierstein, E.; Viale, A.J.; Socci, N.D.; Scandura, J.M. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS ONE 2011, 6, e14524. [Google Scholar] [CrossRef]
- Siedow, J.N. Plant Lipoxygenase: Structure and Function. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1991, 42, 145–188. [Google Scholar] [CrossRef]
- Morales, M.T.; Alonso, M.V.; Rios, J.J.; Aparicio, R. Virgin Olive Oil Aroma: Relationship between Volatile Compounds and Sensory Attributes by Chemometrics. J. Agric. Food Chem. 1995, 43, 2925–2931. [Google Scholar] [CrossRef]
- Angerosa, F.; Basti, C. Olive oil volatile compounds from the lipoxygenase pathway in relation to fruit ripeness. Ital. J. Food Sci. 2001, 13, 421–428. [Google Scholar]
- Kiritsakis, A.K. Flavor components of olive oil—A review. J. Am. Oil Chem. Soc. 1998, 75, 673–681. [Google Scholar] [CrossRef]
- Ryan, D.; Robards, K. Critical Review. Phenolic compounds in olives. Analyst 1998, 123, 31R–44R. [Google Scholar] [CrossRef]
- Servili, M.; Montedoro, G. Contribution of phenolic compounds to virgin olive oil. Eur. J. Lipid Sci. Technol. 2002, 104. [Google Scholar] [CrossRef]
- Tura, D.; Failla, O.; Pedò, S.; Gigliotti, C.; Bassi, D.; Serraiocco, A. Effects of Seasonal Weather Variability on Olive Oil Composition in Northern Italy. Acta Hortic. 2008, 791. [Google Scholar] [CrossRef] [Green Version]
- Boucheffa, S.; Tamendjari, A.; Rovellini, P.; Venturini, S. Composition and antioxidant activity of some Algerian wild extra virgin olive oils. Riv. Ital. Delle Sostanze Grasse 2014, 91, 177–185. [Google Scholar]
- Hannachi, H.; Nasri, N.; Elfalleh, W.; Tlili, N.; Ferchichi, A.; Msallem, M. Fatty Acids, Sterols, Polyphenols, and Chlorophylls of Olive Oils Obtained from Tunisian Wild Olive Trees (Olea europaea L. var. Sylvestris). Int. J. Food Prop. 2012, 16. [Google Scholar] [CrossRef]
- Amiot, M.J.; Fleuriet, A.; Macheix, J.J. Importance and evolution of phenolic compounds in olive during growth and maturation. J. Agric. Food Chem. 1986, 34, 823–826. [Google Scholar] [CrossRef]
- Ryan, D.; Robards, K.; Lavee, S. Changes in phenolic content of olive during maturation. Int. J. Food Sci. Technol. 1999, 34, 265–274. [Google Scholar] [CrossRef]
- Alagna, F.; Mariotti, R.; Panara, F.; Caporali, S.; Urbani, S.; Veneziani, G.; Esposto, S.; Taticchi, A.; Rosati, A.; Rao, R.; et al. Olive phenolic compounds: Metabolic and transcriptional profiling during fruit development. BMC Plant Biol. 2012, 12, 162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monteleone, E.; Caporale, G.; Carlucci, A.; Pagliarini, E. Optimisation of extra virgin olive oil quality. J. Sci. Food Agric. 1998, 77, 31–37. [Google Scholar] [CrossRef]
- Romani, A.; Mulinacci, N.; Pinelli, P.; Vincieri, F.F.; Cimato, A. Polyphenolic Content in Five Tuscany Cultivars of Olea europaea L. J. Agric. Food Chem. 1999, 47, 964–967. [Google Scholar] [CrossRef]
- Rotondi, A.; Bendini, A.; Cerretani, L.; Mari, M.; Lercker, G.; Gallina Toschi, T. Effect of Olive Ripening Degree on the Oxidative Stability and Organoleptic Properties of Cv. Nostrana di Brisighella Extra Virgin Olive Oil. J. Agric. Food Chem. 2004, 52, 3649–3654. [Google Scholar] [CrossRef]
- Talhaoui, N.; Taamalli, A.; Caravaca, A.M.G.; Fernández-Gutiérrez, A.; Segura Carretero, A. Phenolic compounds in olive leaves: Analytical determination, biotic and abiotic influence, and health benefits. Food Res. Int. 2015, 77. [Google Scholar] [CrossRef]
- Gutierrez-Rosales, F.; Romero, M.P.; Casanovas, M.; Motilva, M.J.; Mínguez-Mosquera, M.I. Metabolites Involved in Oleuropein Accumulation and Degradation in Fruits of Olea europaea L.: Hojiblanca and Arbequina Varieties. J. Agric. Food Chem. 2010, 58, 12924–12933. [Google Scholar] [CrossRef] [PubMed]
- Koudounas, K.; Banilas, G.; Michaelidis, C.; Demoliou, C.; Rigas, S.; Hatzopoulos, P. A defence-related Olea europaea β-glucosidase hydrolyses and activates oleuropein into a potent protein cross-linking agent. J. Exp. Bot. 2015, 66, 2093–2106. [Google Scholar] [CrossRef] [Green Version]
- Koudounas, K.; Thomopoulou, M.; Michaelidis, C.; Zevgiti, E.; Papakostas, G.; Tserou, P.; Daras, G.; Hatzopoulos, P. The C-Domain of Oleuropein β-Glucosidase Assists in Protein Folding and Sequesters the Enzyme in Nucleus. Plant Physiol. 2017, 174, 1371–1383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zha, L.; Liu, S.; Liu, J.; Jiang, C.; Yu, S.; Yuan, Y.; Yang, J.; Wang, Y.; Huang, L. DNA Methylation Influences Chlorogenic Acid Biosynthesis in Lonicera japonica by Mediating LjbZIP8 to Regulate Phenylalanine Ammonia-Lyase 2 Expression. Front. Plant Sci. 2017, 8, 1178. [Google Scholar] [CrossRef] [Green Version]
- Gutiérrez-Velázquez, M.V.; Almaraz-Abarca, N.; Herrera-Arrieta, Y.; Ávila-Reyes, J.A.; González-Valdez, L.S.; Torres-Ricario, R.; Uribe-Soto, J.N.; Monreal-García, H.M. Comparison of the phenolic contents and epigenetic and genetic variability of wild and cultivated watercress (Rorippa nasturtium var. aquaticum L.). Electron. J. Biotechnol. 2018, 34, 9–16. [Google Scholar] [CrossRef]
- Yang, D.; Huang, Z.; Jin, W.; Xia, P.; Jia, Q.; Yang, Z.; Hou, Z.; Zhang, H.; Ji, W.; Han, R. DNA methylation: A new regulator of phenolic acids biosynthesis in Salvia miltiorrhiza. Ind. Crops Prod. 2018, 124, 402–411. [Google Scholar] [CrossRef]
- Taiwo, O.; Wilson, G.A.; Morris, T.; Seisenberger, S.; Reik, W.; Pearce, D.; Beck, S.; Butcher, L.M. Methylome analysis using MeDIP-seq with low DNA concentrations. Nat. Protoc. 2012, 7, 617–636. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef] [Green Version]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef] [Green Version]
- Walter, W.; Sánchez-Cabo, F.; Ricote, M. GOplot: An R package for visually combining expression data with functional analysis. Bioinformatics 2015, 31, 2912–2914. [Google Scholar] [CrossRef] [PubMed]
- Sterck, L.; Billiau, K.; Abeel, T.; Rouzé, P.; Van de Peer, Y. ORCAE: Online resource for community annotation of eukaryotes. Nat. Methods 2012, 9, 1041. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cartharius, K.; Frech, K.; Grote, K.; Klocke, B.; Haltmeier, M.; Klingenhoff, A.; Frisch, M.; Bayerlein, M.; Werner, T. MatInspector and beyond: Promoter analysis based on transcription factor binding sites. Bioinformatics 2005, 21, 2933–2942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Smith, J.S. A gas chromatography-flame ionization detection method for detection of fusaproliferin in corn. J. Agric. Food Chem. 2007, 55, 3211–3216. [Google Scholar] [CrossRef]
- Soman, A.; Qiu, Y.; Chan Li, Q. HPLC-UV method development and validation for the determination of low level formaldehyde in a drug substance. J. Chromatogr. Sci. 2008, 46, 461–465. [Google Scholar] [CrossRef] [Green Version]
- Karkoula, E.; Skantzari, A.; Melliou, E.; Magiatis, P. Direct Measurement of Oleocanthal and Oleacein Levels in Olive Oil by Quantitative 1H NMR. Establishment of a New Index for the Characterization of Extra Virgin Olive Oils. J. Agric. Food Chem. 2012, 60, 11696–11703. [Google Scholar] [CrossRef] [PubMed]
- Sáez-Laguna, E.; Guevara, M.-Á.; Díaz, L.-M.; Sánchez-Gómez, D.; Collada, C.; Aranda, I.; Cervera, M.-T. Epigenetic Variability in the Genetically Uniform Forest Tree Species Pinus pinea L. PLoS ONE 2014, 9, e103145. [Google Scholar] [CrossRef] [Green Version]
- Liang, L.; Chang, Y.; Lu, J.; Wu, X.; Liu, Q.; Zhang, W.; Su, X.; Zhang, B. Global Methylomic and Transcriptomic Analyses Reveal the Broad Participation of DNA Methylation in Daily Gene Expression Regulation of Populus trichocarpa. Front. Plant Sci. 2019, 10, 243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lieben, L. Bad Karma reduces palm oil yields. Nat. Rev. Genet. 2015, 16, 625. [Google Scholar] [CrossRef]

| Compound | C18:1 | C18:2 | C183 | Oleocanthal | Oleacein | Aole agl | diAOle agl | Alig agl | diALig agl | Total Polyphenol |
|---|---|---|---|---|---|---|---|---|---|---|
| Ayvalik cv EVOO | 69.3 | 10.8 | 0.7 | 4.9 | 4.2 | 3.1 | 0.8 | 2.7 | ND | 227 |
| wild EVOO | 71.07 | 9.08 | 0.98 | 74 | 76 | 216 | 462 | 81 | 521 | 1034 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Badad, O.; Lakhssassi, N.; Zaid, N.; El Baze, A.; Zaid, Y.; Meksem, J.; Lightfoot, D.A.; Tombuloglu, H.; Zaid, E.H.; Unver, T.; et al. Genome Wide MeDIP-Seq Profiling of Wild and Cultivated Olives Trees Suggests DNA Methylation Fingerprint on the Sensory Quality of Olive Oil. Plants 2021, 10, 1405. https://doi.org/10.3390/plants10071405
Badad O, Lakhssassi N, Zaid N, El Baze A, Zaid Y, Meksem J, Lightfoot DA, Tombuloglu H, Zaid EH, Unver T, et al. Genome Wide MeDIP-Seq Profiling of Wild and Cultivated Olives Trees Suggests DNA Methylation Fingerprint on the Sensory Quality of Olive Oil. Plants. 2021; 10(7):1405. https://doi.org/10.3390/plants10071405
Chicago/Turabian StyleBadad, Oussama, Naoufal Lakhssassi, Nabil Zaid, Abdelhalim El Baze, Younes Zaid, Jonas Meksem, David A Lightfoot, Huseyin Tombuloglu, El Houcine Zaid, Turgay Unver, and et al. 2021. "Genome Wide MeDIP-Seq Profiling of Wild and Cultivated Olives Trees Suggests DNA Methylation Fingerprint on the Sensory Quality of Olive Oil" Plants 10, no. 7: 1405. https://doi.org/10.3390/plants10071405
APA StyleBadad, O., Lakhssassi, N., Zaid, N., El Baze, A., Zaid, Y., Meksem, J., Lightfoot, D. A., Tombuloglu, H., Zaid, E. H., Unver, T., & Meksem, K. (2021). Genome Wide MeDIP-Seq Profiling of Wild and Cultivated Olives Trees Suggests DNA Methylation Fingerprint on the Sensory Quality of Olive Oil. Plants, 10(7), 1405. https://doi.org/10.3390/plants10071405









