Abstract
Roots can produce mechanical and chemical alterations to building structures, especially in the case of underground historical artifacts. In archaeological sites, where vegetation plays the dual role of naturalistic relevance and potential threat, trees and bushes are under supervision. No customized measures can be taken against herbaceous plants lacking fast and reliable root identification methods that are useful to assess their dangerousness. In this study, we aimed to test the efficacy of DNA barcoding in identifying plant rootlets threatening the Etruscan tombs of the Necropolis of Tarquinia. As DNA barcode markers, we selected two sections of the genes rbcL and matK, the nuclear ribosomal internal transcribed spacer (nrITS), and the intergenic spacer psbA-trnH. All fourteen root samples were successfully sequenced and identified at species (92.9%) and genus level (7.01%) by GenBank matching and reference dataset implementation. Some eudicotyledons with taproots, such as Echium italicum L., Foeniculum vulgare Mill., and Reseda lutea L. subsp. lutea, showed a certain recurrence. Further investigations are needed to confirm this promising result, increasing the number of roots and enlarging the reference dataset with attention to meso-Mediterranean perennial herbaceous species. The finding of herbaceous plants roots at more than 3 m deep confirms their potential risk and underlines the importance of vegetation planning, monitoring, and management on archaeological sites.
1. Introduction
Vegetation plays a dual role on archaeological sites. Plants contribute significantly to the characterization of landscapes, enhancing their naturalistic, ecological, and cultural value [1,2,3,4,5,6,7,8]. However, vascular plants, especially trees, can seriously threaten the conservation of ancient monuments as they can directly colonize walls and damage structures by root expansion [1,9,10,11,12,13,14,15]. This risk can be highly relevant in the case of underground ruins as there might be a short distance between the buried archaeological structures and the vegetated ground level. Damages caused by roots have been reported for hypogeal tombs, including the Christian and Jewish catacombs [16], Mithraea, temples, and underground villas such as the Domus Aurea [17,18] and the Domus Tiberiana in Rome [19]. Roots with their growth may produce mechanical and chemical damages on foundations, mortars, plasters, walls, and frescoes [15,16,20,21], even dislodging large stones and weakening the mineral wall matrix and masonry texture by the release of chemical compounds [15]. Moreover, roots can be considered particularly detrimental in hypogea since they may favor water penetration (Figure 3E,F) and affect the internal microbial community [22,23]. Several studies evidenced that roots can modify the diversity and richness of the resident community into this fragile oligotrophic environment. Roots carrying exogenous rhizosphere microorganisms and organic carbon sources (as root litter and exudates) [24,25,26] can favor the growth and spread of detrimental heterotrophs.
The control of vegetation on archaeological sites has been addressed by several authors since the 1980s [1,11,17,27,28]. Particular attention has been paid to the classification of risk assessment tied to individual plant species and plant communities starting from species identification and information on relevant plant elements such as life form (according to Raunkiær), invasiveness, size, shape, and vigor of roots [29]. In this light, a hazard index (HI) ranging from 0 to 10 is assigned to each species. Recently, the ecological characteristics of the different plants in response to diverse micro-environmental (i.e., exposure and inclination) and micro-edaphic conditions (i.e., soil availability and composition) have been considered as additional parameters in the risk assessment [8,12].
Plant identification represents the basic step for the risk assessment, aimed to design a vegetation control plan with periodic monitoring and checks for undesired growth on archaeological sites [9]. Trees and bushes are given special attention on these sites, and their lignified roots can be identified through morpho-anatomical characters supported by the comparison with the nearby aboveground plant species [16,17]. However, morpho-anatomical identification is sometimes time-consuming and may be difficult when immature or ruined specimens lack one or more fundamental characters for their taxonomical identification [30]. In the case of primary roots, the identification is much more complex due to the substantial similarities of the stele organization among the different species. Moreover, the root system is affected by phenotypic plasticity so plants with identical genotypes adapt and modify their root system architectures based on the biotic and abiotic environmental factors [31]. Concerning the root penetration of herbaceous species and their architecture, very few data exist, limited to some important reviews [32,33]. Attempts to identify primary roots have been performed for the Etruscan tombs in Latium and Tuscany, where root penetration represents an enduring problem [2,23,34,35,36,37]. For example, cultures of root meristems, coupled with the analysis of the aboveground vegetation, were performed, but the limitations and difficulties of such methods were stressed [37].
In the last 15 years, DNA barcoding has become a primary tool for fast species identification. DNA sequence data from standard genome regions are routinely used in several applications: biomonitoring, invasive species identification, food fraud, forensics, etc. [38,39,40,41,42]. Although the nuclear Internal Transcribed Spacer (ITS) and the mitochondrial cox1 gene (Cytochrome c oxidase I) are universally used for fungi and animals, respectively, there is no strong consensus on which DNA regions should be used for plants (Fourth International Barcode of Life Conference, www.dnabarcodes2011.org, accessed on 6 January 2021). Two plastid coding regions, rbcL and matK, were suggested as a core-barcode for plants by the Plant Working Group of the Consortium for the Barcode of Life [43,44,45]. However, due to their differential discriminatory power across taxa, additional regions were also recommended, such as the plastid intergenic spacer psbA-trnH and the rapidly evolving Internal Transcribed Spacers (ITS) of nuclear ribosomal DNA [43,44,45,46,47,48,49]. As such, it is useful for a closer evaluation of the power and possible limits of this method.
We aimed to evaluate the power of the DNA barcode method in identifying higher plants starting from herbaceous roots and to test its application in the cultural heritage field, for the first time. The use of this method potentially has a great relevance when roots occur in underground layers and only roots are available for the identification of the plant species (i.e., when the aerial parts of a plant are not developed or visible). In the frame of an international cooperation project focused on biodeterioration and conservation of underground monuments, we analyzed the Necropolis of Monterozzi in Tarquinia (central Italy), where many painted hypogeal tombs are threatened from the penetration of rootlets and, consequently, are in need of conservation actions.
2. Results
2.1. DNA Marker Performances and Root Identification
We obtained readable sequences from all 14 root extracts (Table S1), belonging to eight genera. Overall, 78.57% of the samples were successfully sequenced for all the chosen molecular markers (Figure 3A). The matK target gene was more difficult to be amplified than others, as it was frequently necessary to repeat the amplifications using different primer sets. Nevertheless, 7.14% matK PCR resulted as negative. The highest incidence of successful sequencing was recorded with the psbA-trnH target (13 out of 14). As for drawbacks, using primers for ITS and rbcL regions (one time each), portions of fungal and mitochondrial genome were amplified (Figure 1 and Table 1). Figure 2B shows that technical factors, namely GenBank missing data and failure in PCR/sequencing, affected the contribution of each marker gene in the sample identification. Marker features such as the ability, or not, to return a single best match were also considered (recorded as “no contribution”, Figure 2B). In this light, it is possible to note each marker contribution in root identification at the species level. ITS was useful for the species identification in 85.71% of the cases, followed by psbA-trnH and matK with 64.29% and 35.71%, respectively. The rbcL sequences, not returning a sole best match, were not useful for species identification in 71.43% of the cases, while matK always returned a single species as the best match, even if this was not resolutive for Samples C1 and M1 (Table 1). A perfect match (100%) was recorded for Reseda crystallina Webb & Berthel. and an almost perfect match (99.83%) for Reseda lutea L.; however, as this match was in contrast with the results achieved by ITS and psbA-trnH sequences (Table 1), it was considered not reliable.
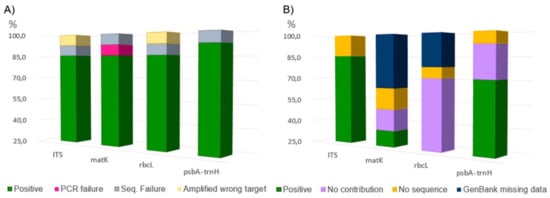
Figure 1.
Molecular marker performances (%). Sequencing performances, namely the frequencies on which were obtained high quality sequences (green) and technical affecting factors (A). Contribution to species identification, namely the frequencies on which Blast comparison returned a single reliable best match (green) and affecting factors (B). No statistical analysis was performed due to the number of samples.

Table 1.
Best BLASTn match results obtained for the 14 root samples processed. On the left, the tombs from which the samples were taken. For each sample and target gene, the following are reported in order: blast matches, percentage of identity (%), and accession number as found in GenBank. The most likely identifications are in bold. The order with which species names are reported within cells also considers parameters not shown, such as query coverage, alignment scores, and E value.
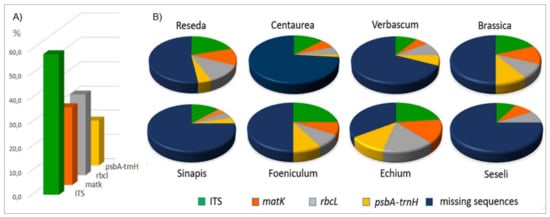
Figure 2.
GenBank sequence coverage of the eight plant genera identified by root sequencing, showing the coverage percentages by molecular marker (A) and genus (B).
As reported in Table 1, GenBank BLAST comparison was fruitful in identifying Foeniculum vulgare Mill. (21.4% of samples; Samples H2, CP01, and CP02), Echium italicum L. (14.3% of roots processed; Samples M2 and 02), Sinapis alba L. (Sample D4), and Reseda lutea (14.3% of samples; Samples C1 and M1). Further investigations are needed for the identification of the remaining root samples belonging to the genera Centaurea (Samples C2, LT01, and LT3, possibly Centaurea aspera L.), Brassica (Sample F1), Verbascum (Samples F3), and Seseli (Sample 01). The recurrence of some species was also evidenced.
2.2. Integrated Taxonomic Identification Method
BLASTn best matches (Table 1) were merged with local flora data to remove the alien species. This was the case of Reseda crystallina, which was previously considered not reliable despite the perfect match obtained (100%, matk, Samples M1 and C1) and now definitively discarded because does not belong to the Italian flora.
The genera of interest are not fully represented in the GenBank database (Figure 2 and Table S2). The highest number of sequences was recorded for the nuclear ITS, covering 58.02% of the local species, followed by rbcL, matk, and the intergenic psbA-trnH spacer with 33.33%, 32.10%, and 18.52%, respectively (Figure 2A). The poor representation of the local flora became more evident by genera. For example, 51.61% of Centaurea local species and subspecies are represented in GenBank with at least a single record each, and even lower is the occurrence for Seseli (33.33%) (Table S2). Local flora represented by at least two molecular markers per species ranges from 38.8% for Verbascum to 0% for Seseli. No sequences for the psbA-trnH intergenic spacer are present for Seseli (Figure 2B).
The six plants taken in the field, namely Centaurea aspera L. subsp. aspera, Reseda lutea L. subsp. lutea, Seseli tortuosum L. subsp. tortuosum, Verbascum sinuatum L., and two Diplotaxis (D. erucoides (L.) DC. and D. tenuifolia (L) DC.), were sequenced for all considered targets (except V. sinuatum ITS); all sequencing results and the relative GenBank accession number are shown in Table S3.
The Diplotaxis sp. were the only Brassicaceae species that we found along the visitors’ path. We also had root samples that showed matches with Sinapis alba and Brassica sp. The tombs where we initially collected the samples were later inaccessible (due to the COVID-19 restrictions). Due to the low number of samples processed starting from leaf extracts and the incomplete overlapping of the species considered, it was not possible to perform a statistical analysis to assess if the differences found in sequencing success yields (root vs. leaf sequencing) are significant.
The comparison of the root sequences with the new reference sequences allowed the identification of several species, e.g., S. tortuosum subsp. tortuosum (Tomb 5512), V. sinuatum (Tomb of the Sculptures), R. lutea subsp. lutea (the Moretti, M1, and Lotus flower, C1 tombs), and C. aspera subsp. aspera (Lotus Flower tomb, Samples C2, LT01, and LT3). Besides, no root matches were recorded with the species D. tenuifolia and D. erucoides, which commonly grow among the tombs in the aboveground area along the visitors’ path.
3. Discussion
There is a limited number of studies on roots’ identification by DNA barcode, mainly focused on the plant roots’ distribution and diversity in the belowground or aimed at authenticating medicinal plants [50,51,52,53,54]. On archeological sites, trees and bushes are commonly maintained under strict control, while no information and neither preventive measures nor guidelines are issued for herbaceous plants. This is mainly due to the general assumption that herbaceous plants are not dangerous, not deeply penetrating, and the lack of reliable and fast methods to identify plants starting from small, tiny roots. Indeed, the difficulty in identifying herbaceous roots allowed us to test the DNA barcoding efficiency.
Plant cells have three different genomes: nuclear, plastid, and mitochondrial [55]. Species, cell type, and age of the tissue affect the number of copies of the nuclear genome and the number of organelles, respectively [55]. Polysaccharides, polyphenolics, and secondary metabolites produced by plants could decrease the quality of their DNA extracts [56]. In this preliminary study, despite some difficulties, promising results were achieved with our protocol, leading to a successful four-marker sequencing in 11 out of 14 root samples (78.57%). Meanwhile, the negative outcomes can provide cues for improvement to be applied in the next step of this research.
Young, healthy, and tender tissues (better if from leaf meristems) are the ideal choice for good quality/quantity DNA extracts, due to the higher number of cells and the low deposition of starch and secondary metabolites [56]. Otherwise, in subterranean environments, a sufficient number of young fresh root samples is often not available, and this factor may affect the results.
Species discrimination with plant barcodes is typically lower than for animals and fungi, using cox1 and ITS barcodes, respectively [45]. This is in part due to the lower rate of nucleotide substitution in the plastid genome, but also tied, for example, to hybridization, polyploidy, and low levels of intraspecific gene flow for plastid markers [57].
It is well known that levels of species discrimination greatly vary among taxa, and several DNA barcoding studies on plants analyzed the discriminating power of molecular data within relatively homogeneous groups, such as families or genera [44,45]. Among plastid regions, rbcL is the best characterized gene because it is easily retrievable across terrestrial plants, suitable for high-quality bidirectional sequences, and easy to align [43,45]. Because of the best performing multi-locus combinations for species discrimination, rbcL was chosen as core-barcode with matK despite its modest discriminatory power [43]. In our study, rbcL sequences (500–650 bp) were never resolutive when used alone, but they were enough to identify the closest “species group” sharing the highest identity score. This information was useful in the field sampling to address the search for spontaneous species, which was not securely identifiable, allowing us to overcome the gap of missing sequences in GenBank. It was also highlighted that, when using rbcL primers, it is possible to amplify mitochondrial regions, a common event recorded in the Brassicaceae family [58].
Even though the matK gene showed high levels of discrimination power among angiosperm species [57,59], the main problem we had was due to the incomplete representation of the herbaceous local flora in GenBank.
The ITS target was the most useful DNA marker due to the number of sequences deposited in GenBank, with about 58.02% of congeneric species (and subspecies), and its recognized discriminatory power. It is characterized by an easy amplification, but as drawback the ITS of possible fungal endophytes can be amplified as well (recorded here as sequencing failure). Although the intergenic spacer psbA-trnH is demonstrated to be easily amplified and sequenced and useful for species identification (64.29% of samples) [60,61], it is poorly represented in GenBank (18.52% of congeneric species of the Latium flora). Its high sequence length variability, ranging from 152 to 851 bp in eudicotyledons, from 151 to 905 bp in monocotyledons, and from 283 to 1006 bp in gymnosperms [60], was useful in the lab practice to distinguish among different specimens just after an electrophoretic run (e.g., Foeniculum vulgare ca. 350 bp and Verbascum sinuatum ca. 600 bp).
In the light of these results, the four-target sequencing was useful to increase the identification rate and obtain more reliable results looking for consistent identity scores along with markers. Being the match scores tied to the specific fragment amplified (even within the same gene) and its length, a single perfect match does not provide a reliable identification. This is, for instance, what happened with the R. crystallina sequence match. Despite the full identity found in matK gene with this species, this result conflicted with the results achieved with the two other markers. Moreover, this species does not belong to the Italian flora. This evidence highlighted the importance of having more than a single discriminating marker for identification as well as the relevant contribution of the local floristic data.
Database improvement was, instead, crucial to achieve the 92.85% of identification at species and subspecies level (at the genus level for the remaining 7.15%), confirming the importance of comparisons with the aboveground vegetation. The improvement of the existing sequencing data on the autochthonous flora could be very useful, if not mandatory, to implement protection strategies for archaeological sites and underground buildings in general. Moreover, the enlargement of the reference database is necessary to assess the best marker barcodes for a faster and reliable identification.
From a conservation viewpoint, our results prove the herbaceous plants, typical of arid calcareous grasslands, can be a potential threat for hypogeal environments, as their roots were found more than 3 m deep (e.g., Sample M2). Indeed, all tombs of this study area are cut into a very porous (30–43% of porosity) yellowish limestone [62]. Being quite brittle, this stone does not offer great resistance to root penetration. Moreover, the xeric conditions that occur in summer in this Mediterranean site, may drive roots in search for water until they reach burial chambers, where the relative humidity is frequently between 90% and 100% [23]. The relevant deep growth is probably linked also to the fact that the most recurrent species, namely C. aspera, E. italicum, F. vulgare, R. lutea, S. tortuosum, and V. sinuatum, are biennial or perennial hemicryptophytes characterized by a vigorous root system. Other herbaceous annual species, such as Brassica sp. and S. alba, showed a well-developed root system as well. The recorded taxa belonged to the families Apiaceae, Asteraceae, Boraginaceae, Resedaceae, and Brassicaceae (eudicotyledons). Their vegetative growth varies [63], ranging from the medium C. aspera (30–60 cm high), R. alba (10–80 cm), S. tortuosum (20–70 cm), and S. alba (30–70 cm) to the medium-high E. italicum (35–100 cm) and F. vulgare (40–150 cm). Scant information is available about the architecture of their root system and behavior in drought conditions. A character shared by most of these species is the presence of taproots, probably able to penetrate more deeply than the adventitious roots of monocotyledons [32,33]. As roots were sampled at different depths and sites within the tombs, an accurate mapping of roots protrusion in different hypogea could provide useful information for conservation practices.
From an applicative side, there are several reasons to avoid large-scale interventions. It is well known that the vegetation generally benefits from policies designed to protect the archaeological site [64,65]. Moreover, the protection of the cultural heritage does not imply extensive and aggressive management routines (e.g., massive use of herbicide) especially when, as in our case, the archaeological area is also a site of naturalistic relevance. Vegetation affects the microclimate conditions of the sites and underground structures, decreasing the temperatures and increasing the humidity values. Recent studies in these Etruscan tombs showed some positive potential effects of plant cover in the stabilization of the local microclimate [66]. The negative counterpart is the role played by roots as carriers for rhizosphere microorganisms, water penetration, and organic carbon supply [22,24,26]. As previously stressed, being hypogea oligotrophic environments, these inputs could lead to a disequilibrium in the resident microbial communities and the spreading of further deteriogenous species [23,26]. Interestingly, the fungal strains sequenced by chance (Sample F1) showed the highest identity score with strains CCFEE 6623 and 6662 isolated previously from the Moretti tomb, deeply threatened by fungi [26].
4. Materials and Methods
4.1. Study Area
Due to its artistic and historic relevance, the Etruscan necropolis of Monterozzi in Tarquinia (Latium, Central Italy) (Figure 1A) has been included, together with those of Cerveteri, in the UNESCO World Heritage Site list since 2004. The tombs, dating from the 7th to the 3rd century BC, were dug in calcarenites banks (Macco stone) and lie at depths ranging from 2 to 8 m [23]. As with other hypogea, these tombs are characterized by a high humidity level, a stable temperature throughout the year, and limited air circulation [26,67]. The necropolis landscape (Figure 1B) is characterized by the presence of many tumuli (which gave the name to the area of Monterozzi). Most of them have been flattened by agricultural practices and others have been dismantled and partially rebuilt to protect the main chamber without considering the original shape [36,68].
The area falls within the Mediterranean macro-bioclimate, with a lower meso-Mediterranean thermotype and a lower subhumid ombrotype [23]. The presence of trees and shrubs is quite limited within the Monterozzi necropolis (Figure 1A,B), which is characterized mainly by ruderal synanthropic herbaceous vegetation, with annual and perennial herbaceous species typical of Mediterranean meadows. Despite the long anthropization and excavation history (since the 19th century), this area maintains a good level of naturalness, also linked to the low incidence of non-native species, and it is included among the protected area of SCI/SAC (IT6010028) of the European Directive 92/43/CEE “Habitat” for its naturalistic relevance. As commonly occurs in archaeological areas, the herbaceous vegetation is subject to periodic mowing.
4.2. Root Sampling, DNA Extraction, Amplification, and Sequence Comparison
Fourteen root samples arising from seven hypogeal tombs (Table S4) were aseptically collected between February and November 2019 (Figure 3C,D), placed in sterile bags, and stored at −20 °C until use. DNA was extracted from fresh root material (70–100 mg) using the Nucleospin Plant kit (Macherey-Nagel, Düren, Germany) following the manufacturer instructions. PCR reactions were performed using the BioMix (BioLine, Luckenwalde, Germany). The reaction solution was prepared with 12 μL of Biomix, 5 pmol of each primer, and about 30 ng of template DNA in a total volume of 25 μL. DNA barcoding analysis was performed using four different DNA markers: the plastid coding rbcL and matK genes and the noncoding psbA-trnH regions and the nuclear ITS. The different primer sets used and the annealing temperatures are listed in Table 2. Amplifications were carried out using the MyCycler™ Thermal Cycler (Bio-Rad Laboratories, Munich, Germany) applying the following protocol for plastid markers: an initial denaturation step for 2 min at 95 °C, 45 cycles at 95 °C for 30 s, annealing at 50 °C (or 53 °C as in Table 1) for 1 min 30 s, extension at 72 °C for 40 s, followed by a final extension at 72 °C for 5 min [52]. For ITS, PCR conditions were: initial denaturation for 3 min at 95 °C, 35 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, and extension for 32 s at 72 °C, with a final extension at 72 °C for 5 min. PCR amplicons were sequenced bidirectionally by Macrogen Spain (Madrid, Spain) and validated using CHROMASPRO v. 1.32 software (Technelysium, Southport, Queensland, Australia). All obtained sequences were searched through the GenBank database (BLASTn) and the best matches were recorded. PCR and sequencing were considered to have failed after four attempts. All sequences were deposited in GenBank (Table S2).

Figure 3.
Study area and root sampling: aerial view of the Monterozzi necropolis at Tarquinia (A); the landscape with tumuli (B); root sampling (C,D); and water droplets along roots, as highlighted by arrows (E,F).

Table 2.
Primer pairs and annealing temperatures used.
4.3. Integrated Taxonomic Identification Method
BLASTn best results (Table 2) were cross-referenced with our floristic data of the site and the checklist of the Italian flora [75]. In this way, it was possible to remove some matches at species level corresponding to plants not present in the local flora. To assess the matching reliability, we performed a search in the NCBI nucleotide database for all the congeneric species present in the Latium flora (https://www.ncbi.nlm.nih.gov/nucleotide/, accessed on 6 January 2021). For each congeneric species, we recorded the number of sequences found for each used DNA marker. Because the length of sequences could influence the best score, we reported the minimum and maximum sequence length. Due to the scarcity of genetic information when a plant was represented in the Latium flora as subspecies only, we also included the relative species, and plant data were recorded accordingly (Table S2). To implement the reference database, in summer 2020, after the COVID-19 lockdown and restrictions, we performed a recognition in the field (limited to the visitors’ path) looking for congeneric species. Six plant species were collected and identified according to the analytical keys in [63,76]; their leaves were processed for molecular purposes as previously described for root samples. The obtained sequences were used as additional reference material (Table S4). ClustalW was used to align/compare sequences of reference specimens with unidentified roots. The procedure of the identification workflow is resumed in Figure 4.
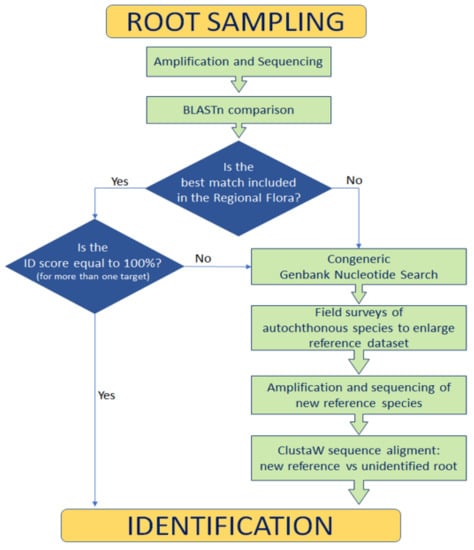
Figure 4.
Root identification workflow.
5. Conclusions
The collected data provide the first assessment of the efficiency of the DNA barcoding approach in the identification of plant rootlets for the preservation of cultural heritage. Despite the positive results, we highlighted the need for some improvements in the GenBank dataset and the selection of specific markers. The collected data also contribute to enhancing the role of herbaceous plant as risk factors for the conservation of hypogeal structures, in specific conditions of high rock porosity and xeric environmental conditions. Further studies are needed to assess species, depth, and risk frequencies, possibly leading, in the near future, to the design of customized control measures.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/plants10061138/s1, Table S1: Root sequences accession numbers, Table S2: Representation of congeneric Latium flora in the NCBI GenBank database, Table S3: Reference plant specimen and sequences accession numbers, Table S4: Tombs in study and samples taken.
Author Contributions
Conceptualization, G.C.; methodology, D.I., F.B. and G.C.; investigation, D.I., F.B. and S.L.; data curation, D.I., L.Z., S.L. and S.C.; writing—review and editing, D.I., L.Z., G.C. and S.C.; project administration, G.C. and F.B.; and funding acquisition, G.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research and the APC was funded by ITALIAN MINISTRY OF FOREIGN AFFAIRS AND INTERNATIONAL COOPERATION PROGRAM “Research of conservation environment and eco-friendly damage control of cultural heritage Korea and Italy”, grant number PGR01253. This work was also supported by the Grant to the DEPARTMENT OF SCIENCE, ROMA TRE UNIVERSITY (MIUR-323 Italy Departments of Excellence, Article 1, Comma 314–337, Law 232/2016).
Data Availability Statement
All data regarding this research are available. In supplementary material can be found the accession number of the all obtained sequences.
Acknowledgments
The authors thank Adele Cecchini (Ass. Amici Tombe di Tarquinia), the Direzione Regionale Musei Lazio (Daniela De Angelis, Director of the Cerveteri and Tarquinia UNESCO Site), and the Soprintendenza Archeologia Belle Arti e Paesaggio per l’Area Metropolitana di Roma, the Province of Viterbo e l’Etruria Meridionale (Daniele Federico Maras).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Caneva, G.; De Marco, G.; Pontrandolfi, M.A. Plant communities of the walls of Venosa Castle (Basilicata, Italy) as biodeteriogens and bioindicators. In Conservation of Stone and Other Materials; Thiel, M.J., Ed.; UNESCO: Paris, France, 1993; Volume 1, pp. 263–270. [Google Scholar]
- Caneva, G.; Benelli, F.; Bartoli, F.; Cicinelli, E. Safeguarding natural and cultural heritage on Etruscan tombs (La Banditaccia, Cerveteri, Italy). Rend Lincei Sci. Fis. 2018, 29, 891–907. [Google Scholar] [CrossRef]
- Celesti Grapow, L.; Caneva, G.; Pacini, A. La Flora dell’Anfiteatro Flavio (Roma). Webbia 2001, 56, 321–342. [Google Scholar]
- Corbetta, F.; Pavone, P.; Spampinato, G.; Tomaselli, V.; Trigilia, A. Studio della vegetazione dell’area archeologica della Neapolis (Siracusa, Sicilia) finalizzato alla conservazione dei manufatti architettonici. Fitosociologia 2002, 39, 3–24. [Google Scholar]
- Ceschin, S.; Caneva, G.; Kumbaric, A. Biodiversità ed emergenze floristiche nelle aree archeologiche romane. Webbia 2006, 61, 133–144. [Google Scholar] [CrossRef]
- Ceschin, S.; Cancellieri, L.; Caneva, G.; Battisti, C. Size area, patch heterogeneity and plant species richness across archaeological sites of Rome: Different patterns for different guilds. Vie Milieu 2012, 62, 165–171. [Google Scholar]
- Ceschin, S.; Kumbaric, A.; Caneva, G.; Zuccarello, V. Testing flora as bioindicator of buried structures in the archaeological area of Maxentius’s villa (Rome, Italy). J. Archaeol. Sci. 2012, 39, 1288–1295. [Google Scholar] [CrossRef]
- Cicinelli, E.; Benelli, F.; Bartoli, F.; Traversetti, L.; Caneva, G. Trends of plant communities growing on the Etruscan tombs (Cerveteri, Italy) related to different management practices. Plant. Biosyst. Int. J. Deal. all Asp. Plant Biol. 2020, 154, 158–164. [Google Scholar] [CrossRef]
- Caneva, G. A botanical approach to the planning of archaeological, parks in Italy. Conserv. Manag. Archaeol. Sites 1999, 3, 127–134. [Google Scholar] [CrossRef]
- Caneva, G.; Pacini, A.; Grapow, L.C.; Ceschin, S. The Colosseum’s use and state of abandonment as analysed through its flora. Int. Biodet. Biodeg. 2003, 51, 211–219. [Google Scholar] [CrossRef]
- Almeida, M.; Mouga, T.; Barracosa, P. The weathering ability of higher plants. The case of Ailanthus altissima (Miller) Swingle. Int. Biodet. Biodeg. 1994, 33, 333–343. [Google Scholar] [CrossRef]
- Motti, R.; Bonanomi, G. Vascular plant colonization of four castles in southern Italy: Effects of substrate bioreceptivity, local environmental factors, and current management. Int. Biodet. Biodeg. 2018, 133, 26–33. [Google Scholar] [CrossRef]
- Awang, A.; Nasir, M.R.M.; Mohamad, W.S.N.W. A Review on the Impacts of Plants Towards Heritage Buildings. J. Appl. Arts 2020, 2, 94–101. [Google Scholar]
- Dahmani, J.; Benharbit, M.; Fassar, M.; Hajila, R.; Zidane, L.; Magri, N.; Belahbib, N. Vascular plants census linked to the biodeterioration process of the Portuguese city of Mazagan in El Jadida, Morocco. J. King Saud Univ. Sci. 2020, 32, 682–689. [Google Scholar] [CrossRef]
- Trotta, G.; Savo, V.; Cicinelli, E.; Carboni, M.; Caneva, G. Colonization and damages of Ailanthus altissima (Mill.) Swingle on archaeological structures: Evidence from the Aurelian Walls in Rome (Italy). Int. Biodet. Biodeg. 2020, 153, 105054. [Google Scholar] [CrossRef]
- Caneva, G.; Galotta, G.; Cancellieri, L.; Savo, V. Tree roots and damages in the Jewish catacombs of Villa Torlonia (Roma). J. Cult. Heritage 2009, 10, 53–62. [Google Scholar] [CrossRef]
- Caneva, G. Tree roots and hypogeans conservation. Braun Blanquetia 1989, 3, 329–336. [Google Scholar]
- Caneva, G.; Ceschin, S.; De Marco, G. Mapping the risk of damage from tree roots for the conservation of archaeological sites: The case of the Domus Aurea, Rome. Conserv. Manag. Archaeol. Sites 2006, 7, 163–170. [Google Scholar] [CrossRef]
- Caneva, G.; Agostini, D.; Baldini, D. Domus Tiberiana and Horti Farnesiani (Rome): Further investigations on tree roots for the conservation of the archaeological site. In Proceedings of the 10th ICOMOS International Congress on Deterioration and Conservation of Stone, Stockholm, Sweden, 27 June–2 July 2004; Kwiatkowski, D., Löfvendahl, R., Eds.; Volume 2, pp. 955–961. [Google Scholar]
- Cuttler, D.F.; Richardson, I.B.K. Tree Root and Buildings; Kew Garden: London, UK, 1981; pp. 1–94. [Google Scholar]
- Biddle, P.G. Tree root damage to buildings. In Causes, Diagnosis, Remedy/Patterns of Soil Drying in Proximity to Trees on Clays Soils; Willowmead Publishing Ltd: Wantage, UK, 1998. [Google Scholar]
- Diaz–Herraiz, M.; Jurado, V.; Cuezva, S.; Laiz, L.; Pallecchi, P.; Tiano, P.; Sanchez-Moral, S.; Saiz-Jimenez, C. The actinobacterial colonisation of Etruscan paintings. Sci. Rep. 2013, 3, 1440. [Google Scholar] [CrossRef] [PubMed]
- Caneva, G.; Isola, D.; Lee, H.J.; Chung, Y.J. Biological Risk for Hypogea: Shared Data from Etruscan Tombs in Italy and Ancient Tombs of the Baekje Dynasty in Republic of Korea. Appl. Sci. 2020, 10, 6104. [Google Scholar] [CrossRef]
- Jasinska, E.J.; Knott, B.; McComb, A.J. Root Mats in Ground Water: A Fauna-Rich Cave Habitat. J. N. Am. Benthol. Soc. 1996, 15, 508–519. [Google Scholar] [CrossRef]
- Diaz-Herraiz, M.; Jurado, V.; Cuezva, S.; Laiz, L.; Pallecchi, P.; Tiano, P.; Sanchez-Moral, S.; Saiz-Jimenez, C. Deterioration of an Etruscan tomb by bacteria from the order Rhizobiales. Sci. Rep. 2015, 4, 3610. [Google Scholar] [CrossRef] [PubMed]
- Isola, D.; Zucconi, L.; Cecchini, A.; Caneva, G. Dark-pigmented biodeteriogenic fungi in etruscan hypogeal tombs: New data on their culture-dependent diversity, favouring conditions, and resistance to biocidal treatments. Fungal Biol. 2021. [Google Scholar] [CrossRef]
- Catizone, P. Il contenimento delle piante infestanti nelle aree di interesse archeologico. In Archeologia e Botanica; Mastroroberto, M., Ed.; L’Erma di Bretschneider Editore: Roma, Italy, 1990; pp. 59–64. [Google Scholar]
- Mishra, A.; Jain, K.K.; Garg, K. Role of higher plants in the deterioration of historic buildings. Sci. Total Environ. 1995, 167, 375–392. [Google Scholar] [CrossRef]
- Signorini, M.A. L’Indice di Pericolosità: Un contributo del botanico al controllo della vegetazione infestante nelle aree monumentali. Inf. Bot. Ital. 1996, 28, 7–14. [Google Scholar]
- Bruni, I.; De Mattia, F.; Martellos, S.; Galimberti, A.; Savadori, P.; Casiraghi, M.; Nimis, P.L.; Labra, M. DNA Barcoding as an Effective Tool in Improving a Digital Plant Identification System: A Case Study for the Area of Mt. Valerio, Trieste (NE Italy). PLoS ONE 2011, 7, e43256. [Google Scholar] [CrossRef] [PubMed]
- Bellini, C.; Pacurar, D.I.; Perrone, I. Adventitious Roots and Lateral Roots: Similarities and Differences. Annu. Rev. Plant Biol. 2014, 65, 639–666. [Google Scholar] [CrossRef] [PubMed]
- Cesari, M.G.; Rossi, W. Le radici minacciano le tombe dipinte di Tarquinia. Archeologia 1972, 3, 20–25. [Google Scholar]
- Kutschera, L. Root Atlas of Central European Weeds and Crop. Plants of Arable Land; DLG Verlag: Frankfurt am Main, Germany, 1960; p. 544. [Google Scholar]
- Kutschera, L.; Lichtenegger, E.; Sobotik, M.; Lichtenegger, E.; Haas, D. Würzeln. Bewurzelung von Pflanzen in verschidenen Lebensräumen. Stapfia 1997, 49, 1e331. [Google Scholar]
- Paribeni, M. Cause di Deperimento e Metodi di Conservazione Delle Pitture Murali Delle Tombe Sotterranee di Tarquinia. Scientific Report of C.N.R.—Istituto di Fisica Tecnica di Roma; Edizioni Sistema: Rome, Italy, 1970. [Google Scholar]
- Cecchini, A. Le Tombe Dipinte di Tarquinia. Vicenda Conservativa, Restauri e Tecnica di Esecuzione; Nardini Ed: Firenze, Italy, 2012; p. 102. [Google Scholar]
- Altieri, A.; Bartolini, M.; Pietrini, A.M. Relazione del Laboratorio di Indagini Biologiche: Tarquinia Necropoli dei Monterozzi—Consulenza Scientifica; ISCR Report: Rome, Italy, 2013. [Google Scholar]
- Adamowicz, S.J. International Barcode of Life: Evolution of a global research community. Genome 2015, 58, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Khaksar, R.; Carlson, T.; Schaffner, D.W.; Ghorashi, M.; Best, D.; Jandhyala, S.; Traverso, J.; Amini, S. Unmasking seafood mislabeling in U.S. markets: DNA barcoding as a unique technology for food authentication and quality control. Food Control 2015, 56, 71–76. [Google Scholar] [CrossRef]
- Pappalardo, A.M.; Ferrito, M. DNA barcoding species identification unveils mislabeling of processed flatfish products in southern Italy markets. Fish. Res. 2015, 164, 153–158. [Google Scholar] [CrossRef]
- Chimeno, C.; Morinière, J.; Podhorna, J.; Hardulak, L.; Hausmann, A.; Reckel, F.; Grunwald, J.E.; Penning, R.; Haszprunar, G. DNA Barcoding in Forensic Entomology—Establishing a DNA Reference Library of Potentially Forensic Relevant Arthropod Species. J. Forensic. Sci. 2019, 64, 593–601. [Google Scholar] [CrossRef]
- Weigand, H.; Beermann, A.J.; Čiampor, F.; Costa, F.O.; Csabai, Z.; Duarte, S.; Geiger, M.F.; Grabowski, M.; Rimet, F.; Rulik, B.; et al. DNA barcode reference libraries for the monitoring of aquatic biota in Europe: Gap-analysis and recommendations for future work. Sci. Total Environ. 2019, 678, 499–524. [Google Scholar] [CrossRef]
- CBOL Plant Working Group; Hollingsworth, P.M.; Forrest, L.L.; Spouge, J.L.; Hajibabaei, M.; Ratnasingham, S.; van der Bank, M.; Chase, M.W.; Cowan, R.S.; Erickson, D.L.; et al. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar]
- Hollingsworth, M.L.; Clark, A.A.; Forrest, L.L.; Richardson, J.; Pennington, R.T.; Long, D.G.; Cowan, R.; Chase, M.W.; Gaudeul, M.; Hollingsworth, P.M. Selecting barcoding loci for plants: Evaluation of seven candidate loci with species-level sampling in three divergent groups of land plants. Mol. Ecol. Resour. 2009, 9, 439–457. [Google Scholar] [CrossRef] [PubMed]
- Hollingsworth, P.M.; Graham, S.; Little, D.P. Choosing and Using a Plant DNA Barcode. PLoS ONE 2011, 6, e19254. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.-Y.; Qimike, A.; Yang, C.-F.; Chen, J.-M.; Wang, Q.-F. Testing four barcoding markers for species identification of Potamogetonaceae. J. Syst. Evol. 2011, 49, 246–251. [Google Scholar] [CrossRef]
- Guo, X.; Wang, X.; Su, W.; Zhang, G.; Zhou, R. DNA Barcodes for Discriminating the Medicinal Plant Scutellaria baicalensis (Lamiaceae) and Its Adulterants. Biol. Pharm. Bull. 2011, 34, 1198–1203. [Google Scholar] [CrossRef]
- De Mattia, F.; Gentili, R.; Bruni, I.; Galimberti, A.; Sgorbati, S.; Casiraghi, M.; Labra, M. A multi-marker DNA barcoding approach to save time and resources in vegetation surveys. Bot. J. Linn. Soc. 2012, 169, 518–529. [Google Scholar] [CrossRef]
- Zhang, C.Y.; Wang, F.Y.; Yan, H.F.; Hao, G.; Hu, C.M.; Ge, X.J. Testing DNA barcoding in closely related groups of Lysimachia L. (Myrsinaceae). Mol. Ecol. Resour. 2012, 12, 98–108. [Google Scholar] [CrossRef]
- Amritha, N.; Bhooma, V.; Parani, M. Authentication of the market samples of Ashwagandha by DNA barcoding reveals that powders are significantly more adulterated than roots. J. Ethnopharmacol. 2020, 256, 112725. [Google Scholar] [CrossRef]
- Jones, F.A.; Erickson, D.L.; Bernal, M.A.; Bermingham, E.; Kress, W.J.; Herre, E.A.; Muller-Landau, H.C.; Turner, B.L. The roots of diversity: Below ground species richness and rooting distributions in a tropical forest revealed by DNA barcodes and inverse modeling. PLoS ONE 2011, 6, e24506. [Google Scholar] [CrossRef]
- Kesanakurti, P.R.; Fazekas, A.J.; Burgess, K.S.; Percy, D.M.; Newmaster, S.G.; Graham, S.; Barrett, S.C.H.; Hajibabaei, M.; Husband, B.C. Spatial patterns of plant diversity below-ground as revealed by DNA barcoding. Mol. Ecol. 2011, 20, 1289–1302. [Google Scholar] [CrossRef] [PubMed]
- Park, I.; Yang, S.; Kim, W.J.; Noh, P.; Lee, H.O.; Moon, B.C. Authentication of Herbal Medicines Dipsacus asper and Phlomoides umbrosa Using DNA Barcodes, Chloroplast Genome, and Sequence Characterized Amplified Region (SCAR) Marker. Molecules 2018, 23, 1748. [Google Scholar] [CrossRef]
- Wallace, L.J.; Boilard, S.M.; Eagle, S.H.; Spall, J.L.; Shokralla, S.; Hajibabaei, M. DNA barcodes for everyday life: Routine authentication of Natural Health Products. Food Res. Int. 2012, 49, 446–452. [Google Scholar] [CrossRef]
- Lutz, K.A.; Wang, W.; Zdepski, A.; Michael, T.P. Isolation and analysis of high quality nuclear DNA with reduced organellar DNA for plant genome sequencing and resequencing. BMC Biotechnol. 2011, 11, 54–59. [Google Scholar] [CrossRef]
- Varma, A.; Padh, H.; Shrivastava, N. Plant genomic DNA isolation: An art or a science. Biotechnol. J. 2007, 2, 386–392. [Google Scholar] [CrossRef]
- Fazekas, A.J.; Burgess, K.S.; Kesanakurti, P.R.; Graham, S.; Newmaster, S.G.; Husband, B.C.; Percy, D.M.; Hajibabaei, M.; Barrett, S.C.H. Multiple Multilocus DNA Barcodes from the Plastid Genome Discriminate Plant Species Equally Well. PLoS ONE 2008, 3, e2802. [Google Scholar] [CrossRef]
- Cummings, M.P.; Nugent, J.M.; Olmstead, R.G.; Palmer, J.D. Phylogenetic analysis reveals five independent transfers of the chloroplast gene rbcL to the mitochondrial genome in angiosperms. Curr. Genet. 2003, 43, 131–138. [Google Scholar] [CrossRef][Green Version]
- Lahaye, R.; van der Bank, M.; Bogarin, D.; Warner, J.; Pupulin, F.; Gigot, G.; Maurin, O.; Duthoit, S.; Barraclough, T.G.; Savolainen, V. DNA barcoding the floras of biodiversity hotspots. Proc. Natl. Acad. Sci. USA 2008, 105, 2923–2928. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.; Liu, C.; Shi, L.; Liu, R.; Liang, D.; Li, H.; Cherny, S.; Chen, S. Utility of the trnH–psbA Intergenic Spacer Region and Its Combinations as Plant DNA Barcodes: A Meta-Analysis. PLoS ONE 2012, 7, e48833. [Google Scholar] [CrossRef] [PubMed]
- Uncu, A.T.; Uncu, A.O. Plastid trnH–psbA intergenic spacer serves as a PCR–based marker to detect common grain adulterants of coffee (Coffea arabica L.). Food Control. 2018, 91, 32–39. [Google Scholar] [CrossRef]
- Bugini, R.; Folli, L. Masonries and stone materials of Romanesque architecture (Northern Italy). Int. J. Mason. Res. Innov. 2021, 6, 45. [Google Scholar] [CrossRef]
- Pignatti, S. Flora d’Italia; Edagricole: Bologna, Italy, 1982; Volume 1–3. [Google Scholar]
- Cicinelli, E.; Salerno, G.; Caneva, G. An assessment methodology to combine the preservation of biodiversity and cultural heritage: The San Vincenzo al Volturno historical site (Molise, Italy). Biodivers. Conserv. 2018, 27, 1073–1093. [Google Scholar] [CrossRef]
- Lucchese, F.; Pignatti, E. La vegetazione delle aree archeologiche di Roma e della Campagna Romana. Quad. Bot. Ambient. Appl. 2009, 20, 3–89. [Google Scholar]
- Caneva, G.; Langone, S.; Bartoli, F.; Cecchini, A.; Meneghini, C. Vegetation Cover and Tumuli’s Shape as Affecting Factors of Microclimate and Biodeterioration Risk for the Conservation of Etruscan Tombs (Tarquinia, Italy). Sustainability 2021, 13, 3393. [Google Scholar] [CrossRef]
- Urzì, C.; De Leo, F.; Krakova, L.; Pangallo, D.; Bruno, L. Effects of biocide treatments on the biofilm community in Domitilla’s catacombs in Rome. Sci. Total Environ. 2016, 572, 252–262. [Google Scholar] [CrossRef]
- Marzullo, M. Grotte Cornetane: Materiale e Apparato Critico per lo Studio Delle Tombe Dipinte di Tarquinia; Ledizioni Ledi Publishing: Milano, Italy, 2016. [Google Scholar]
- De Vere, N.; Rich, T.C.; Trinder, S.A.; Long, C. DNA barcoding for plants. In Plant Genotyping; Batley, J., Ed.; Humana Press: New York, NY, USA, 2014; pp. 101–118. [Google Scholar]
- Ford, C.S.; Ayres, K.L.; Toomey, N.; Haider, N.; van AlphenStahl, J.; Kelly, L.J.; Wikström, N.; Hollingsworth, P.M.; Duff, R.J.; Hoot, S.B.; et al. Selection of candidate coding DNA barcoding regions for use on land plants. Bot. J. Linn. Soc. 2009, 159, 1–11. [Google Scholar] [CrossRef]
- Cuénoud, P.; Savolainen, V.; Chatrou, L.; Powell, M.; Grayer, R.J.; Chase, M.W. Molecular phylogenetics of Caryophyllales based on nuclear 18S rDNA and plastid rbcL, atpB, and matK DNA sequences. Am. J. Bot. 2002, 89, 132–144. [Google Scholar] [CrossRef]
- Fay, M.F.; Bayer, C.; Alverson, W.S.; de Bruijn, A.Y.; Chase, M.W. Plastid rbcL sequence data indicate a close affinity between Diegodendron and Bixa. Taxon 1998, 47, 43–50. [Google Scholar] [CrossRef]
- Newmaster, S.G.; Ragupathy, S. Testing plant barcoding in a sister species complex of pantropical Acacia (Mimosoideae, Fabaceae). Mol. Ecol. Res. 2009, 9, S172–S180. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press Inc: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Bartolucci, F.; Peruzzi, L.; Galasso, G.; Albano, A.; Alessandrini, A.; Ardenghi, N.M.G.; Astuti, G.; Bacchetta, G.; Ballelli, S.; Banfi, E.; et al. An updated checklist of the vascular flora native to Italy. Plant Biosyst. Int. J. Deal. All Asp. Plant. Biol. 2018, 152, 179–303. [Google Scholar] [CrossRef]
- Pignatti, S.; Guarino, R.; La Rosa, M. Flora d’Italia; Edagricole: Bologna, Italy, 2017. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).