NF-YB-Mediated Active Responses of Plant Growth under Salt and Temperature Stress in Eucalyptus grandis
Abstract
1. Introduction
2. Results
2.1. Phylogenetic and Evolutionary Analysis of EgNF-YB
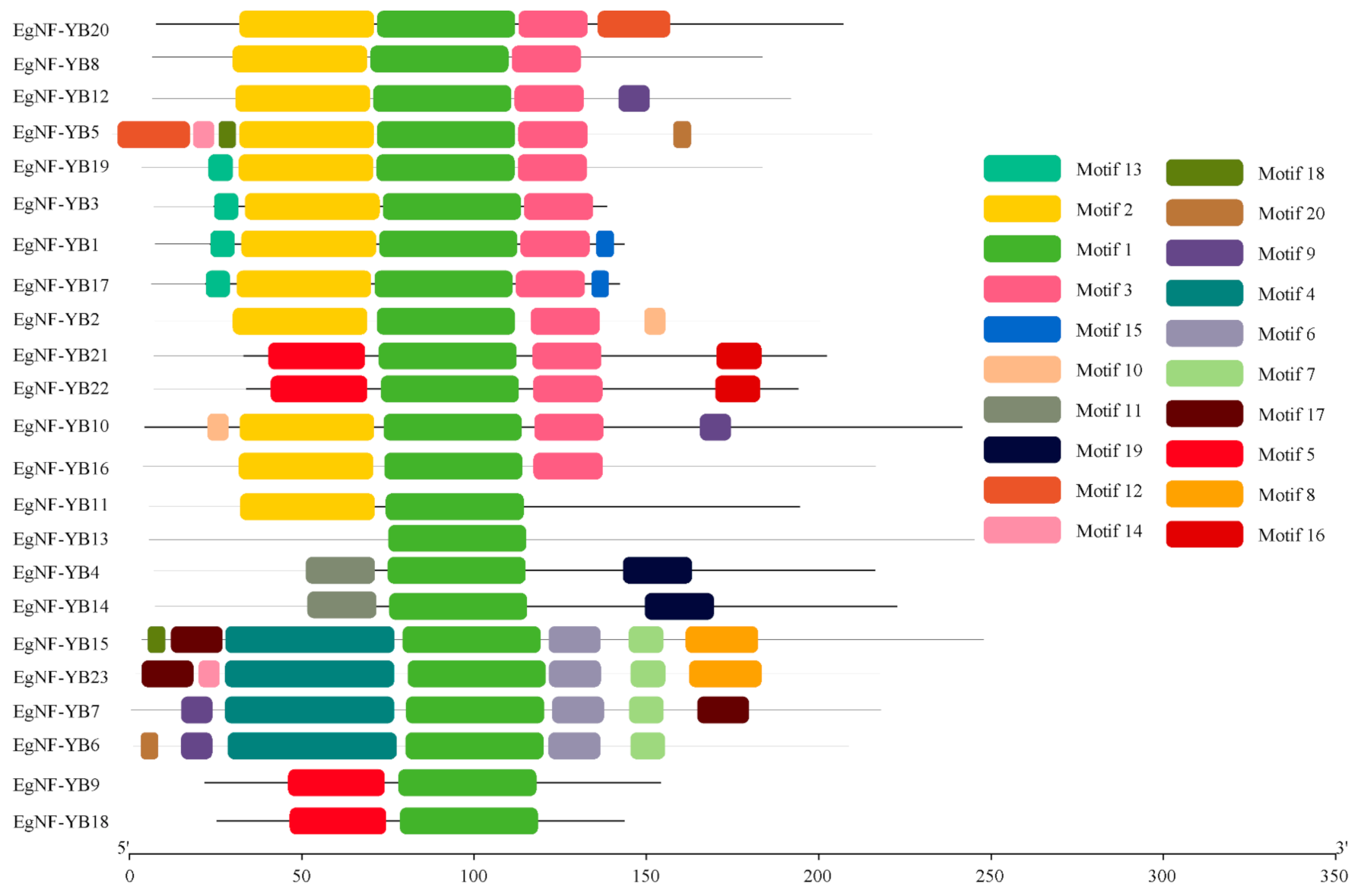
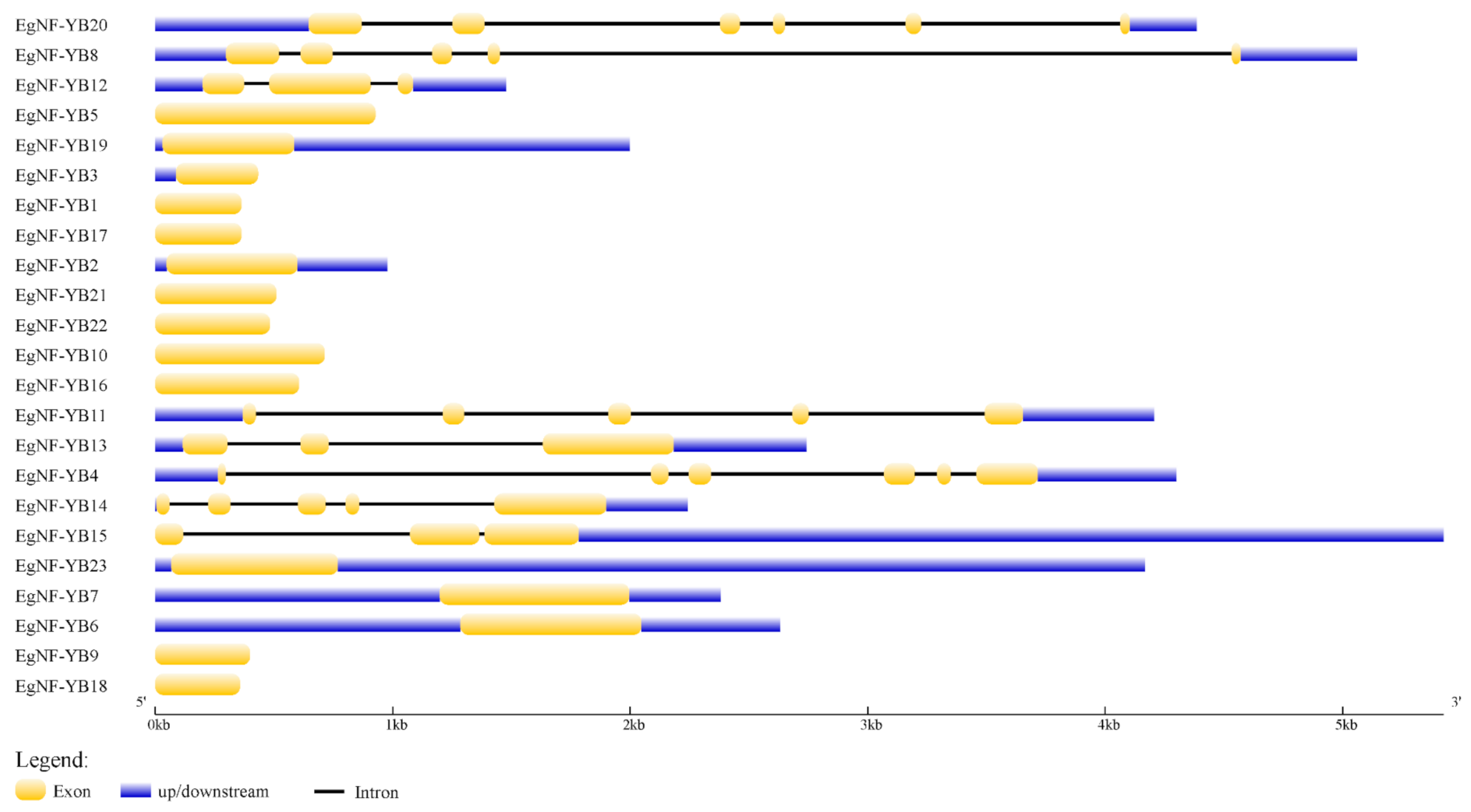
2.2. Classification and Structural Analysis of EgNF-YB
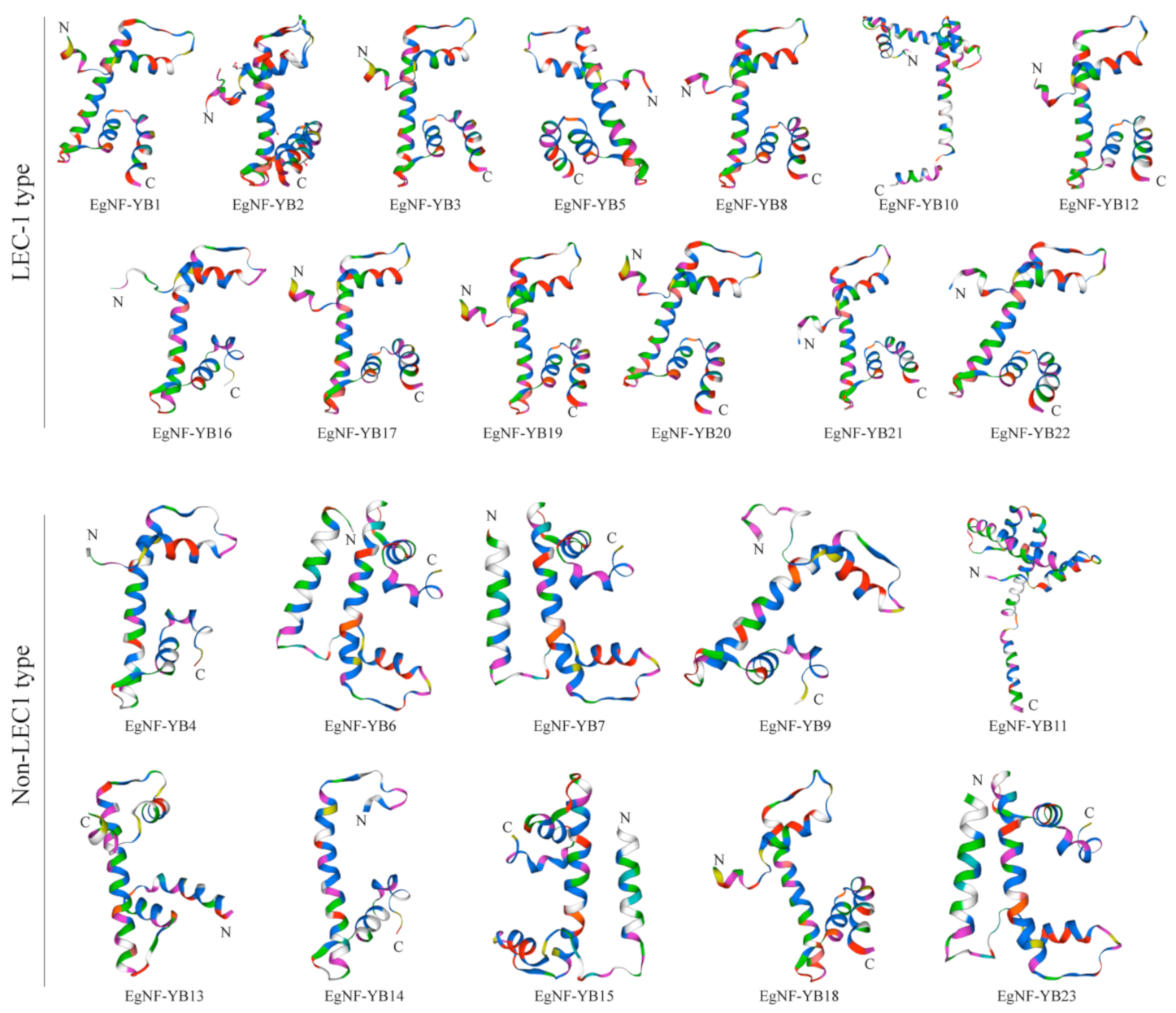
2.3. 3D Structure Analysis of EgNF-YBs
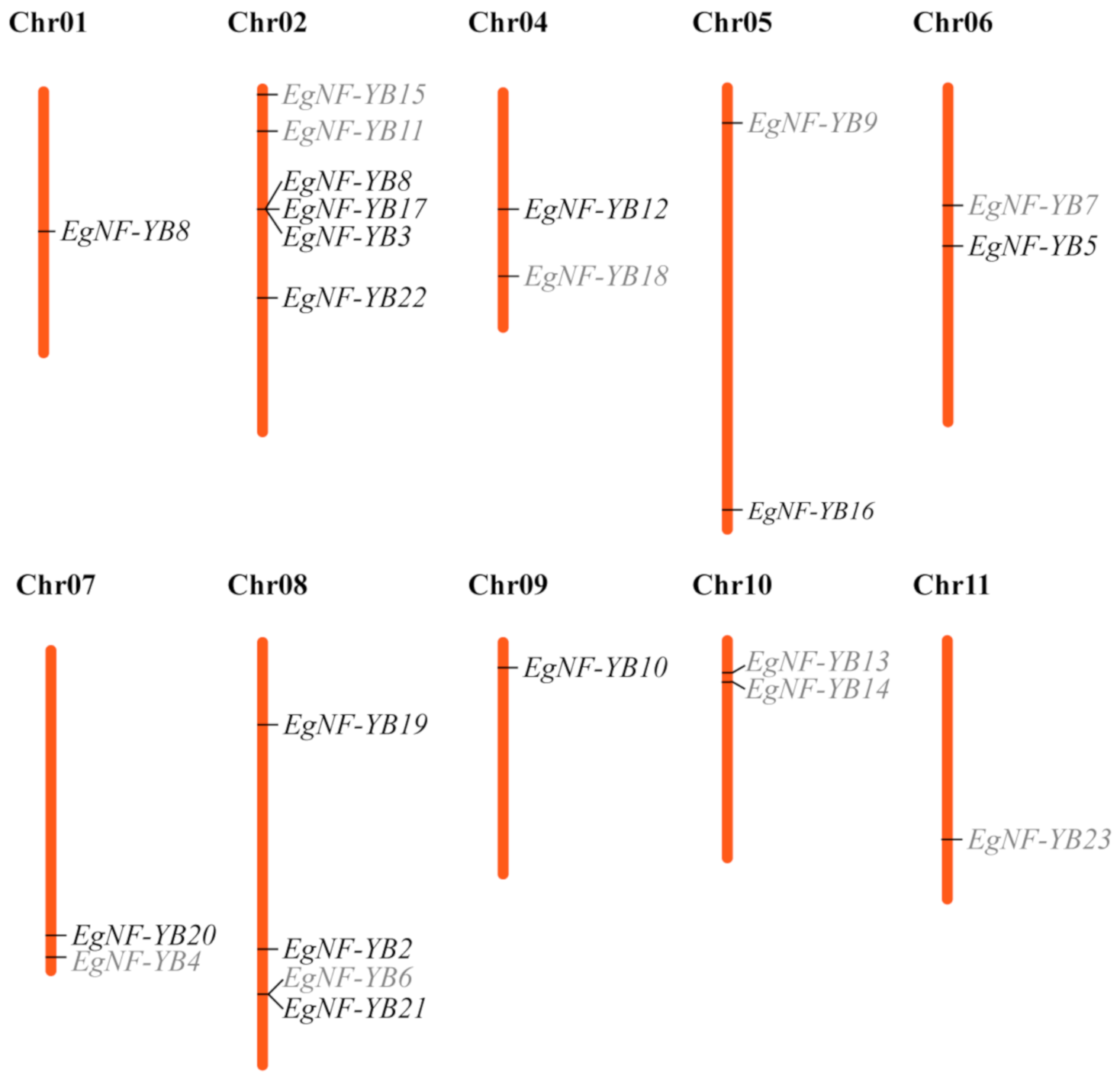
2.4. Chromosome Localization of EgNF-YB Genes
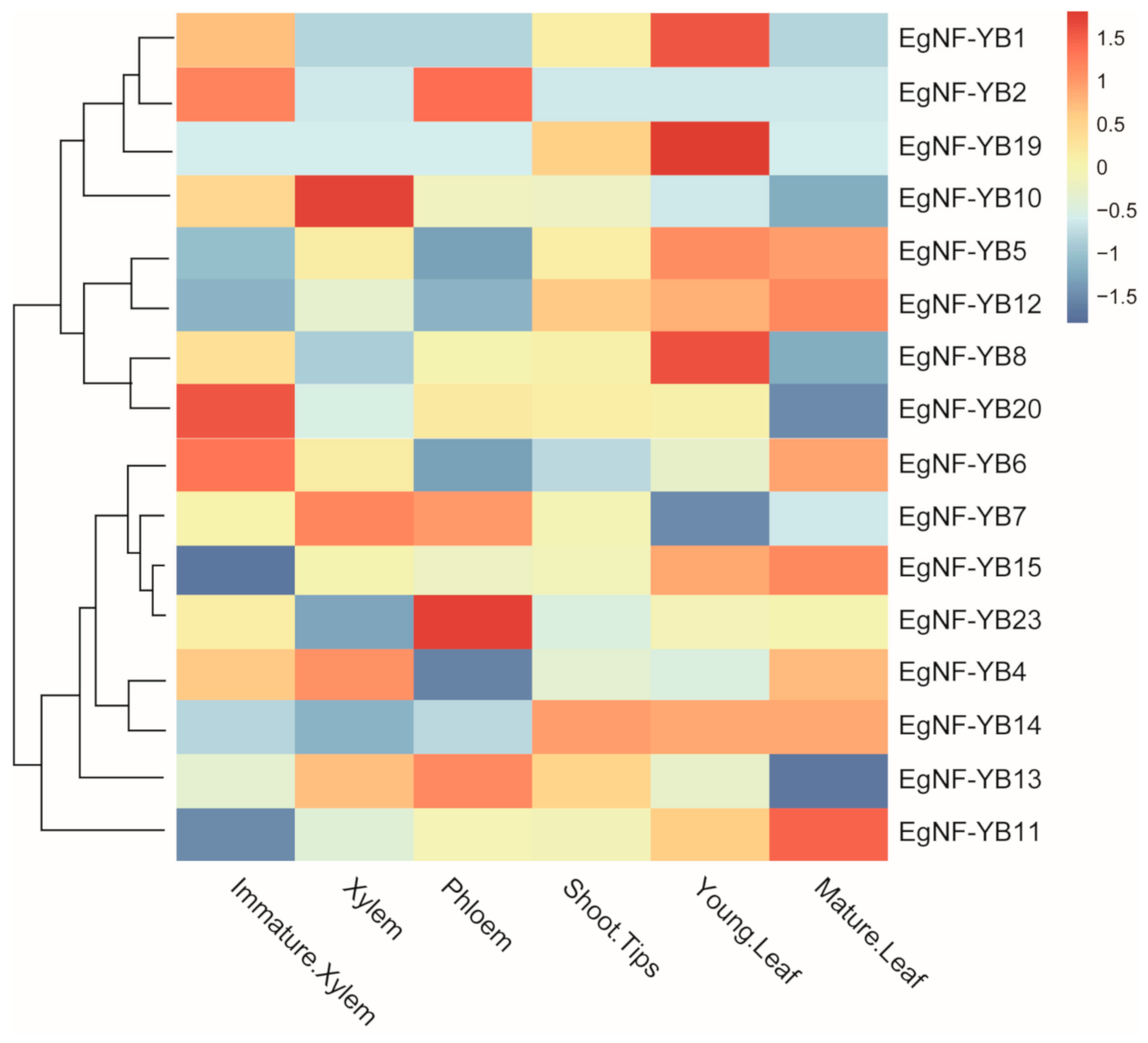
2.5. Expression Pattern of Eucalyptus EgNF-YB Genes in Various Tissues
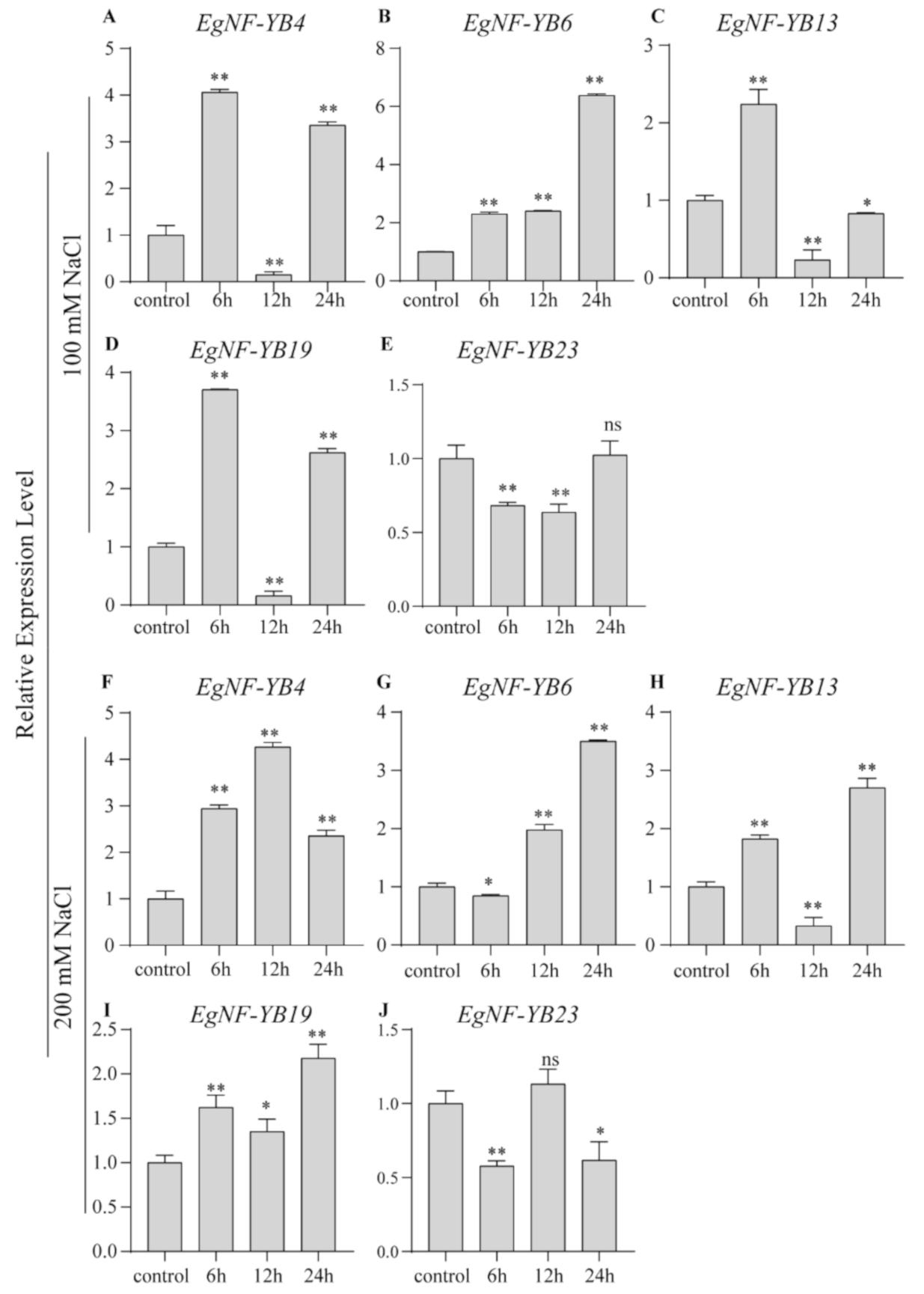
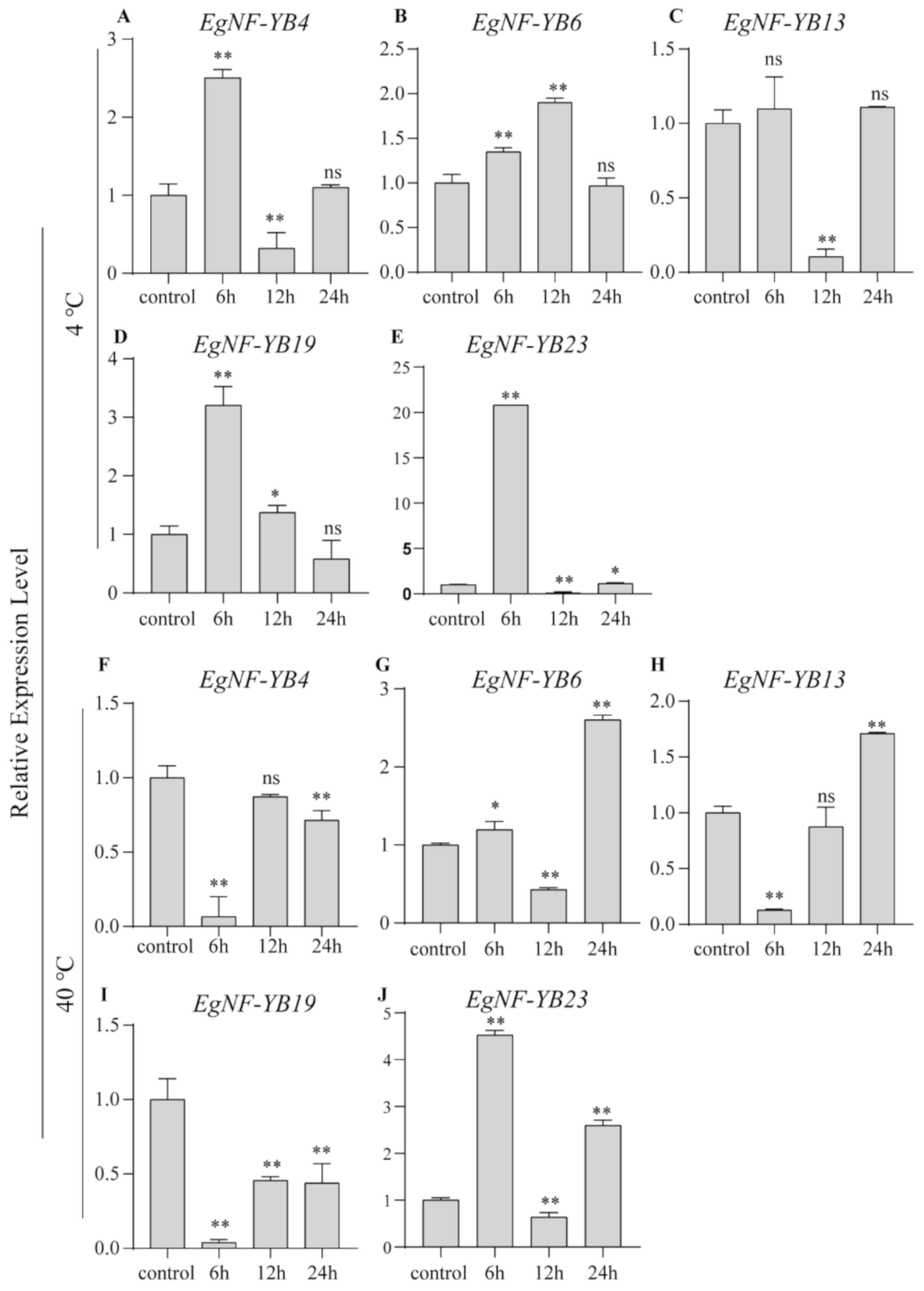
2.6. EgNF-YB Genes Response to Salinity and Extreme Temperature Stress
3. Discussion
4. Materials and Methods
4.1. Identification and Chromosomal Location of NF-YB Family Members in Eucalyptus
4.2. Gene Duplication and Phylogenetic Analysis
4.3. Analysis of NF-YB Gene Structure and Protein Conserved Motif
4.4. Tissue Expression Profile Analysis of NF-YB Genes
4.5. Plant Materials and Abiotic Treatment
4.6. RNA Extraction and Quantitative Real-Time PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, W.B.; Li, S.C.; Wang, Z.K. Review of structure and function of nuclear factor Y in plant. J. Northeast. Agric. Univ. 2012, 43, 12–17. [Google Scholar]
- Mantovani, R. The molecular biology of the CCAAT-binding factor NF-Y. Gene 1999, 239. [Google Scholar] [CrossRef]
- Paolo, B.; Valentina, B.; Daniele, M.; Isaia, F.L.; Enrico, T.; Carol, I. A balance between NF-Y and p53 governs the pro- and anti-apoptotic transcriptional response. Nucleic Acids Res. 2008, 36, 1415–1428. [Google Scholar]
- Zemzoumi, K.; Frontini, M.; Bellorini, M.; Mantovani, R. NF-Y histone fold α1 helices help impart CCAAT specificity. J. Mol. Biol. 1999, 286, 327–337. [Google Scholar] [CrossRef]
- Gusmaroli, G.; Tonelli, C.; Mantovani, R. Regulation of the CCAAT-binding NF-Y subunits in Arabidopsis thaliana. Gene 2001, 264, 173–185. [Google Scholar] [CrossRef]
- Dolfini, D.; Gatta, R.; Mantovani, R. NF-Y and the transcriptional activation of CCAAT promoters. Crit. Rev. Biochem. Mol. Biol. 2011, 47, 29–49. [Google Scholar] [CrossRef]
- Frontini, M.; Imbriano, C.; Manni, I.; Mantovani, R. Cell cycle regulation of NF-YC nuclear localization. Cell Cycle 2004, 3, 205–210. [Google Scholar] [CrossRef]
- Li, X.Y.; Mantovani, R.; Hooft van Huijsduijnen, R.; Andre, I.; Benoist, C.; Mathis, D. Evolutionary variation of the CCAAT-binding transcription factor NF-Y. Nucleic Acids Res. 1992, 20, 1087–1091. [Google Scholar] [CrossRef]
- Jennifer, M. CONSTANS companion: CO binds the NF-YB/NF-YC dimer and confers sequence-specific DNA binding. Plant Cell 2017, 29. [Google Scholar] [CrossRef]
- Lee, H.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc. Natl. Acad. Sci. USA 2003, 100, 2152–2156. [Google Scholar] [CrossRef]
- Riechmann, J.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.; Samaha, R. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Maere, S.; Bodt, S.D.; Raes, J.; Casneuf, T.; Montagu, M.V.; Kuiper, M.; Peer, Y.V.D. Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 5454–5459. [Google Scholar] [CrossRef]
- Petroni, K.; Kumimoto, R.W.; Gnesutta, N.; Calvenzani, V.; Fornari, M.; Tonelli, C.; Holt, B.F.; Mantovani, R. The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 2012, 24, 4777–4792. [Google Scholar] [CrossRef]
- Laloum, T.; Mita, S.D.; Gamas, P.; Baudin, M.; Niebel, A. Erratum to: CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 2013, 18, 157–166. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; Solano, R. Identification of plant transcription factor target sequences. Biochim. Biophys. Acta 2017, 1860, 21–30. [Google Scholar] [CrossRef]
- Ding, H.X.; Liu, F.; Zhang, L.j.; Yu, Y.X.; Wei, Q. Review of structure and function of nuclear factor Y in plant. Mol. Plant Breed. 2017, 15, 105–115. [Google Scholar]
- Yang, S.; Xiao, X.; Yang, L.H.; Qian, X.; Chen, X.J. Bioinformatics analysis of the expansin gene family in rice. Hereditas 2014, 36, 809–820. [Google Scholar]
- Lotan, T.; Ohto, M.-A.; Yee, K.M.; West, M.A.L.; Lo, R.; Kwong, R.W.; Yamagishi, K.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 1998, 93, 1195–1205. [Google Scholar] [CrossRef]
- Liang, M.; Hole, D.; Wu, J.; Blake, T.; Wu, Y. Expression and functional analysis of NUCLEAR FACTOR-Y, subunit B genes in barley. Planta 2012, 235, 779–791. [Google Scholar] [CrossRef]
- Hwang, Y.H.; Kim, S.K.; Lee, K.C.; Chung, Y.S.; Kim, J.K. Functional conservation of rice OsNF-YB/YC and Arabidopsis AtNF-YB/YC proteins in the regulation of flowering time. Plant Cell Rep. 2016, 35, 857–865. [Google Scholar] [CrossRef]
- Feng, Z.J.; He, G.H.; Zheng, W.J.; Lu, P.P.; Ming, C.; Gong, Y.M.; Ma, Y.Z.; Xu, Z.S. Foxtail millet NF-Y families: Genome-wide survey and evolution analyses identified two functional genes important in abiotic stresses. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef]
- Xu, Z.J.; Liu, Y.; Xu, L.; An, D.S. bioinformatics analysis of NF-YB gene family of transcription factor in zea mays. Mol. Plant Breed. 2019, 17, 3807–3816. [Google Scholar]
- Yang, M.Y.; Zhao, Y.J.; Shi, S.Y.; Du, X.M.; Gu, J.T.; Xiao, K. Wheat nuclear factor Y (NF-Y) B subfamily gene TaNF-YB3; I confers critical drought tolerance through modulation of the ABA-associated signaling pathway. Plant Cell Tissue Organ Cult. 2017, 128, 97–111. [Google Scholar] [CrossRef]
- Su, Y.S.; Dong, W.Y.; Fu, Y.H.; Bo, G.H.; Ming, G. Identification and characterization of NF-YB family genes in tung tree. Mol. Genet. Genom. 2015, 290, 2187–2198. [Google Scholar]
- Yan, D.H.; Fenning, T.; Tang, S.; Xia, X.; Yin, W. Genome-wide transcriptional response of Populus euphratica to long-term drought stress. Plant Sci. 2012, 195, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Brooker, M. A new classification of the genus Eucalyptus L’Hér. (Myrtaceae). Aust. Syst. Bot. 2000, 13, 79–148. [Google Scholar] [CrossRef]
- Coscolin, R.B.S.; Broetto, F.; Marchese, J.A.; Campohermoso, M.C.; Paladini, M.V. Effects of hydric deficiency on gas exchange parameters and metabolism of Eucalyptus grandis clones. Braz. J. Plant Physiol. 2010, 23, 255–262. [Google Scholar] [CrossRef]
- Myburg, A.A.; Grattapaglia, D.; Tuskan, G.A.; Hellsten, U.; Hayes, R.D.; Grimwood, J.; Jenkins, J.; Lindquist, E.; Tice, H.; Bauer, D.; et al. The genome of Eucalyptus grandis. Nature 2014, 510, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Kubo, T.; Serizawa, A. Identification, characterization and interaction of HAP family genes in rice. J. Recept. Res. 2008, 7, 279–289. [Google Scholar]
- Yuan, G.A.; Hui, Z.Q.; Xin, C.; Chu, L.J. GSDS: A gene structure display server. Hereditas 2007, 29, 1023–1026. [Google Scholar]
- Jeffares, D.C.; Penkett, C.J.; Baehler, J. Rapidly regulated genes are intron poor. Trends Genet. 2008, 24, 375–378. [Google Scholar] [CrossRef] [PubMed]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22. [Google Scholar] [CrossRef] [PubMed]
- Marco, B.; Stefan, B.; Andrew, W.; Konstantin, A.; Gabriel, S.; Tobias, S.; Florian, K.; Gallo, C.T.; Martino, B.; Lorenza, B. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar]
- Hackenberg, D.; Wu, Y.F.; Voigt, A.; Adams, R.; Schramm, P.; Grimm, B. Studies on differential nuclear translocation mechanism and assembly of the three subunits of the Arabidopsis thaliana transcription factor NF-Y. Mol. Plant 2012, 5, 876–888. [Google Scholar] [CrossRef]
- Hang, Z.; Di, W.; Kong, F.; Ke, L.; Zhang, H.; Gang, L. The Arabidopsis thaliana nuclear factor Y transcription factors. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef]
- Shun, Z.T.; Dong, Y.; Zhong, S.Z.; Wu, S.X.; Ning, T.Y.; Bing, S.X.; Juan, D.M.; Yang, Y.D. Bioinformatics analysis of the NF-YB gene family in rice. Mol. Plant Breed. 2018, 16, 5849–5861. [Google Scholar]
- Todeschini, A.L.; Georges, A.; Veitia, R.A. Transcription factors: Specific DNA binding and specific gene regulation. Trends Genet. 2014, 30, 211–219. [Google Scholar] [CrossRef]
- Maity, S.N.; Crombrugghe, B.D. Purification, characterization, and role of CCAAT-binding factor in transcription. Methods Enzymol. 1996, 273, 217–232. [Google Scholar]
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef]
- Lin, Y.X.; Cheng, Y.; Jin, J.; Jin, X.L.; Cheng, B.J. Genome duplication and gene loss affect the evolution of heat shock transcription factor genes in legumes. PLoS ONE 2014, 9, e102825. [Google Scholar] [CrossRef]
- Abe, H. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef]
- Stephenson, T.J.; McIntyre, C.L.; Collet, C.; Xue, G.P. Genome-wide identification and expression analysis of the NF-Y family of transcription factors in Triticum aestivum. Plant Mol. Biol. 2007, 65, 77–92. [Google Scholar] [CrossRef]
- Siefers, N.; Dang, K.K.; Kumimoto, R.W.; Bynum, W.E.; Tayrose, G.; Holt, B.F. Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 2009, 149, 625–641. [Google Scholar] [CrossRef]
- Yamamoto, A.; Kagaya, Y.; Toyoshima, R.; Kagaya, M.; Hattori, T. Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J. 2010, 58, 843–856. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-wide analysis of NAC transcription factor family in rice. Gene 2010, 465. [Google Scholar] [CrossRef]
- Kumimoto, R.W.; Adam, L.; Hymus, G.J.; Repetti, P.P.; Reuber, T.L.; Marion, C.M.; Hempel, F.D.; Ratcliffe, O.J. The nuclear factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta 2008, 228, 709–723. [Google Scholar] [CrossRef]
- Hussey, S.G.; Mizrachi, E.; Groover, A.; Berger, D.K.; Myburg, A.A. Genome-wide mapping of histone H3 lysine 4 trimethylation in Eucalyptus grandis developing xylem. BMC Plant Biol. 2015, 15. [Google Scholar] [CrossRef]
- Sato, H.; Mizoi, J.; Tanaka, H.; Maruyama, K.; Qin, F.; Osakabe, Y.; Morimoto, K.; Ohori, T.; Kusakabe, K.; Nagata, M.; et al. Arabidopsis DPB3-1, a DREB2A interactor, specifically enhances heat stress-induced gene expression by forming a heat stress-specific transcriptional complex with NF-Y subunits. Plant Cell 2014, 26, 4954–4973. [Google Scholar] [CrossRef]
- Sato, H.; Suzuki, T.; Takahashi, F.; Shinozaki, K.; Yamaguchi-Shinozaki, K. NF-YB2 and NF-YB3 Have Functionally Diverged and Differentially Induce Drought and Heat Stress-Specific Genes. Plant Physiol. 2019, 180, 1677–1690. [Google Scholar] [CrossRef]
- Ding, Y.; Shi, Y.; Yang, S. Molecular Regulation of Plant Responses to Environmental Temperatures. Mol Plant 2020, 13, 544–564. [Google Scholar] [CrossRef]
- Eddy, S.R. Profile hidden Markov models. Bioinformatics 1998, 14, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.L.; Jakovli, I.; Zou, H.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31. [Google Scholar] [CrossRef]
- Bailey, T.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In Proceedings of the International Conference on Intelligent Systems for Molecular Biology, Toronto, ON, Canada, 19–23 July 1994; pp. 28–36. [Google Scholar]
- Bailey, T.L.; Mikael, B.; Buske, F.A.; Martin, F.; Grant, C.E.; Luca, C.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Russell, H.; Rochak, N.; Hayes, R.D.; Joni, F.; Therese, M.; William, D.; Uffe, H.; Nicholas, P. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, J.-H.; Hu, A.-Q.; Zhang, J.-S.; Liao, W.-H.; Ma, H.-Y.; Wu, J.-Z.; Yu, Y.; Cao, S.-J. NF-YB-Mediated Active Responses of Plant Growth under Salt and Temperature Stress in Eucalyptus grandis. Plants 2021, 10, 1107. https://doi.org/10.3390/plants10061107
Dai J-H, Hu A-Q, Zhang J-S, Liao W-H, Ma H-Y, Wu J-Z, Yu Y, Cao S-J. NF-YB-Mediated Active Responses of Plant Growth under Salt and Temperature Stress in Eucalyptus grandis. Plants. 2021; 10(6):1107. https://doi.org/10.3390/plants10061107
Chicago/Turabian StyleDai, Jia-Hao, An-Qi Hu, Jia-Shuo Zhang, Wen-Hai Liao, Hua-Yan Ma, Jin-Zhang Wu, Yuan Yu, and Shi-Jiang Cao. 2021. "NF-YB-Mediated Active Responses of Plant Growth under Salt and Temperature Stress in Eucalyptus grandis" Plants 10, no. 6: 1107. https://doi.org/10.3390/plants10061107
APA StyleDai, J.-H., Hu, A.-Q., Zhang, J.-S., Liao, W.-H., Ma, H.-Y., Wu, J.-Z., Yu, Y., & Cao, S.-J. (2021). NF-YB-Mediated Active Responses of Plant Growth under Salt and Temperature Stress in Eucalyptus grandis. Plants, 10(6), 1107. https://doi.org/10.3390/plants10061107






