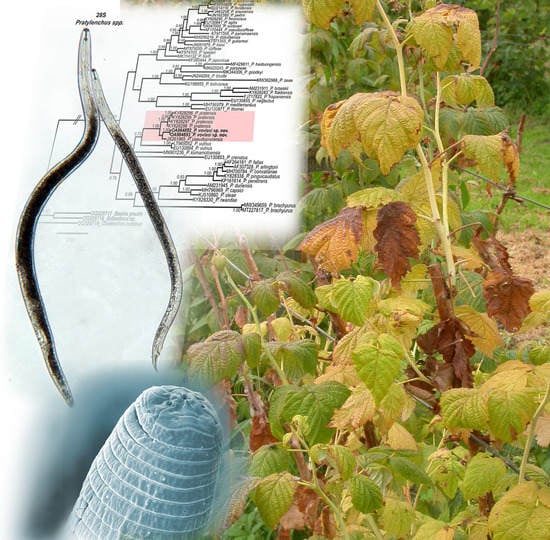
Pratylenchus vovlasi sp. Nov. (Nematoda: Pratylenchidae) on Raspberries in North Italy with a Morphometrical and Molecular Characterization †
Abstract
1. Introduction
2. Results
2.1. Molecular Characterization
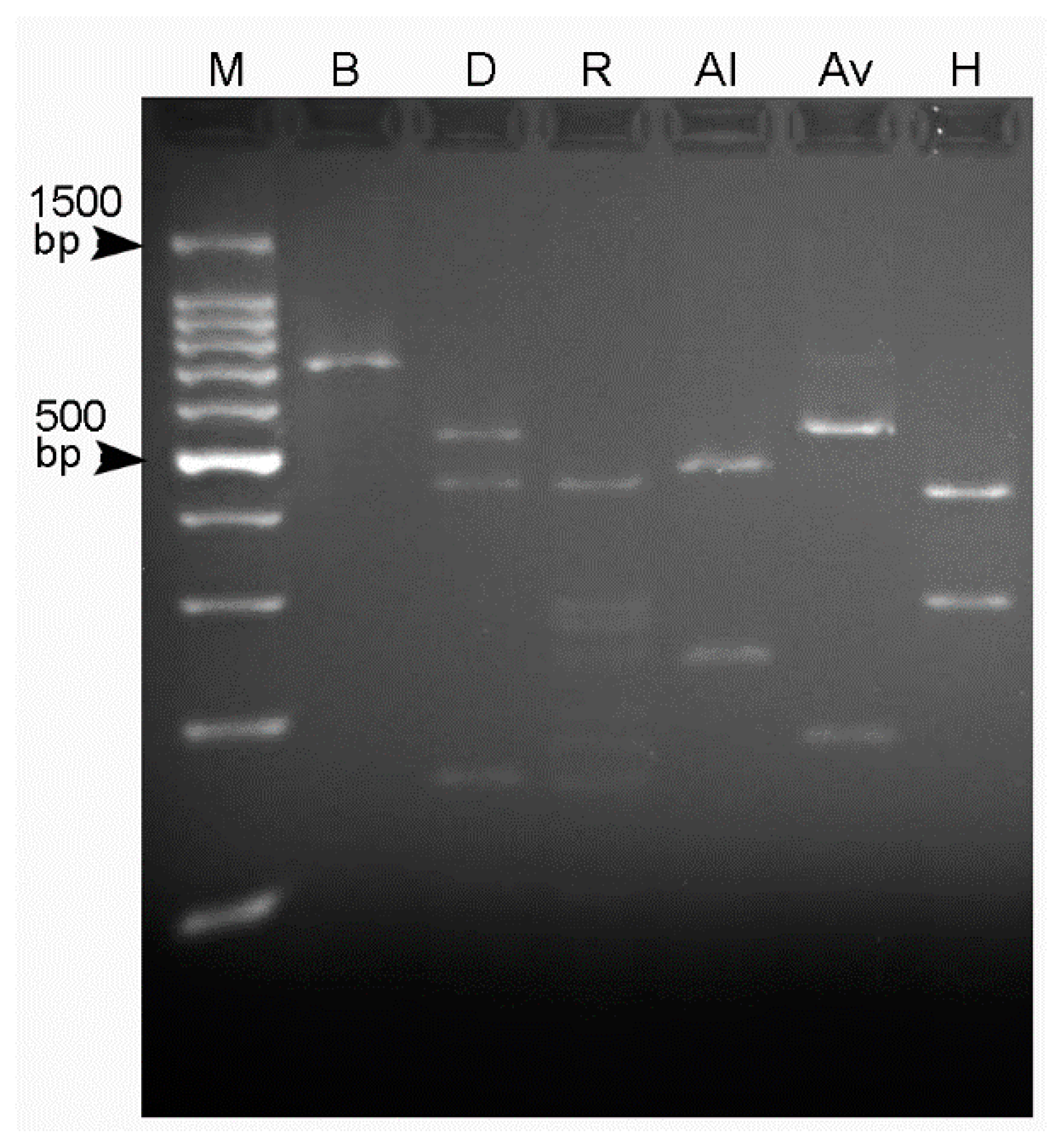
2.2. Restriction Profiles
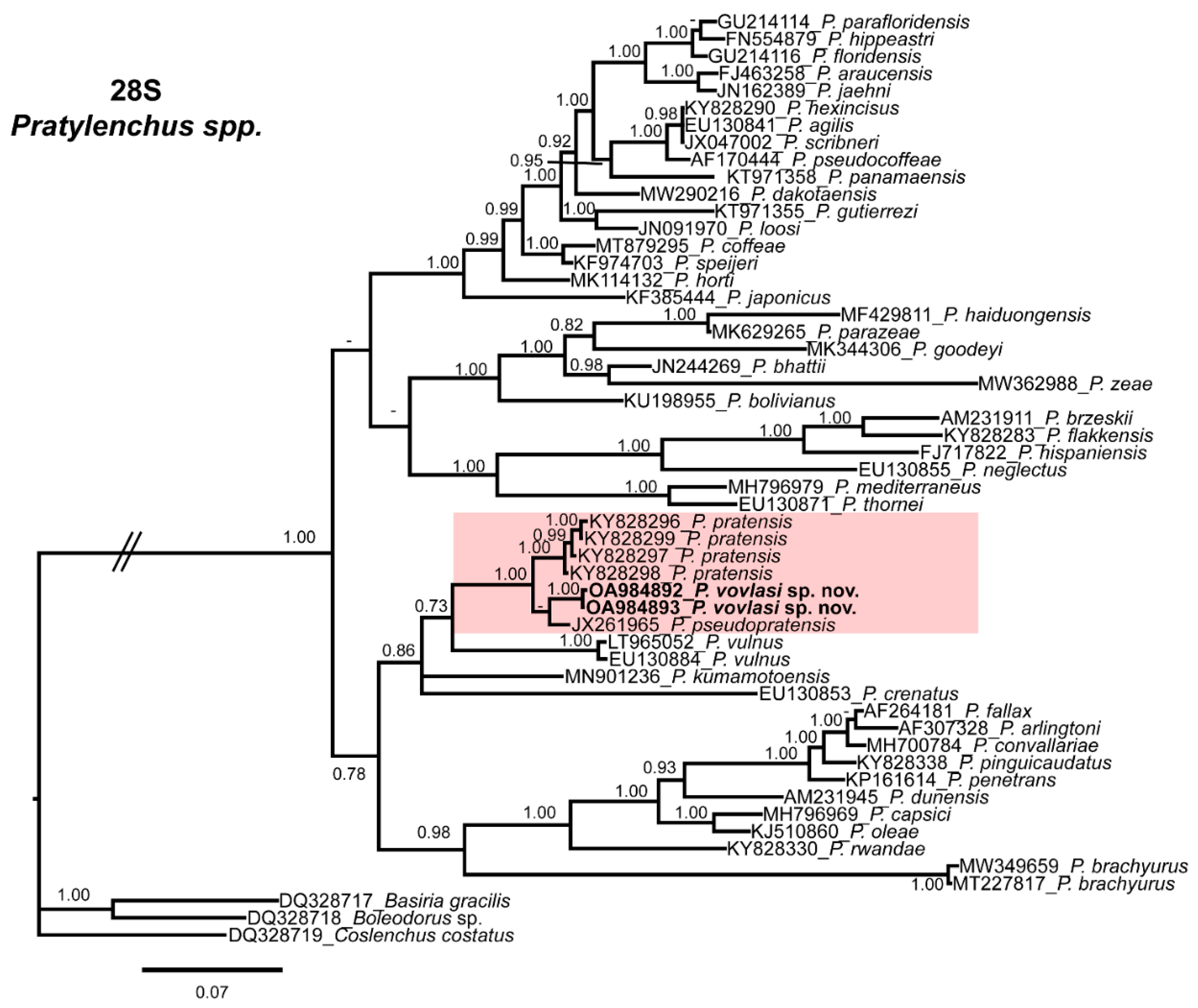
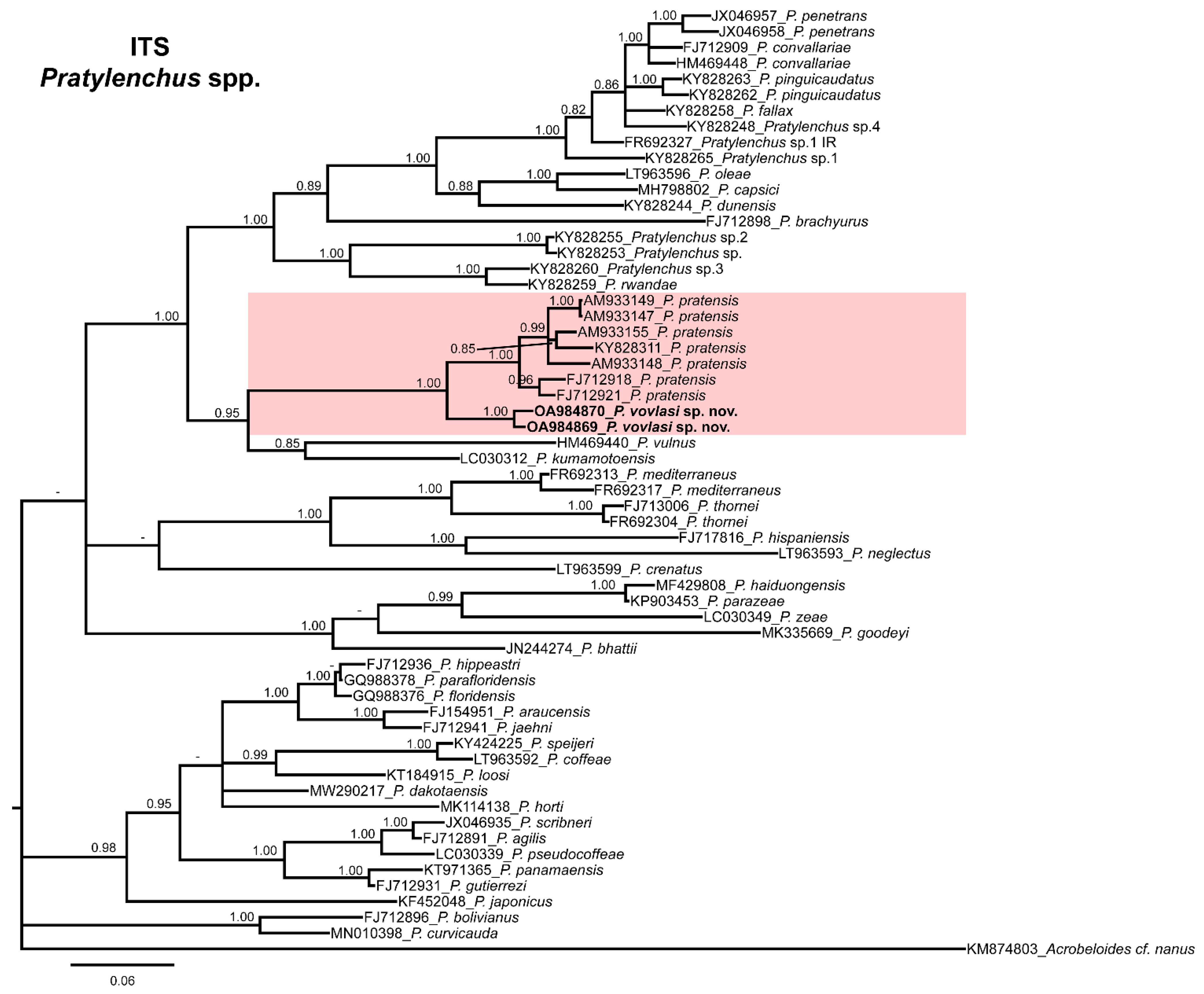
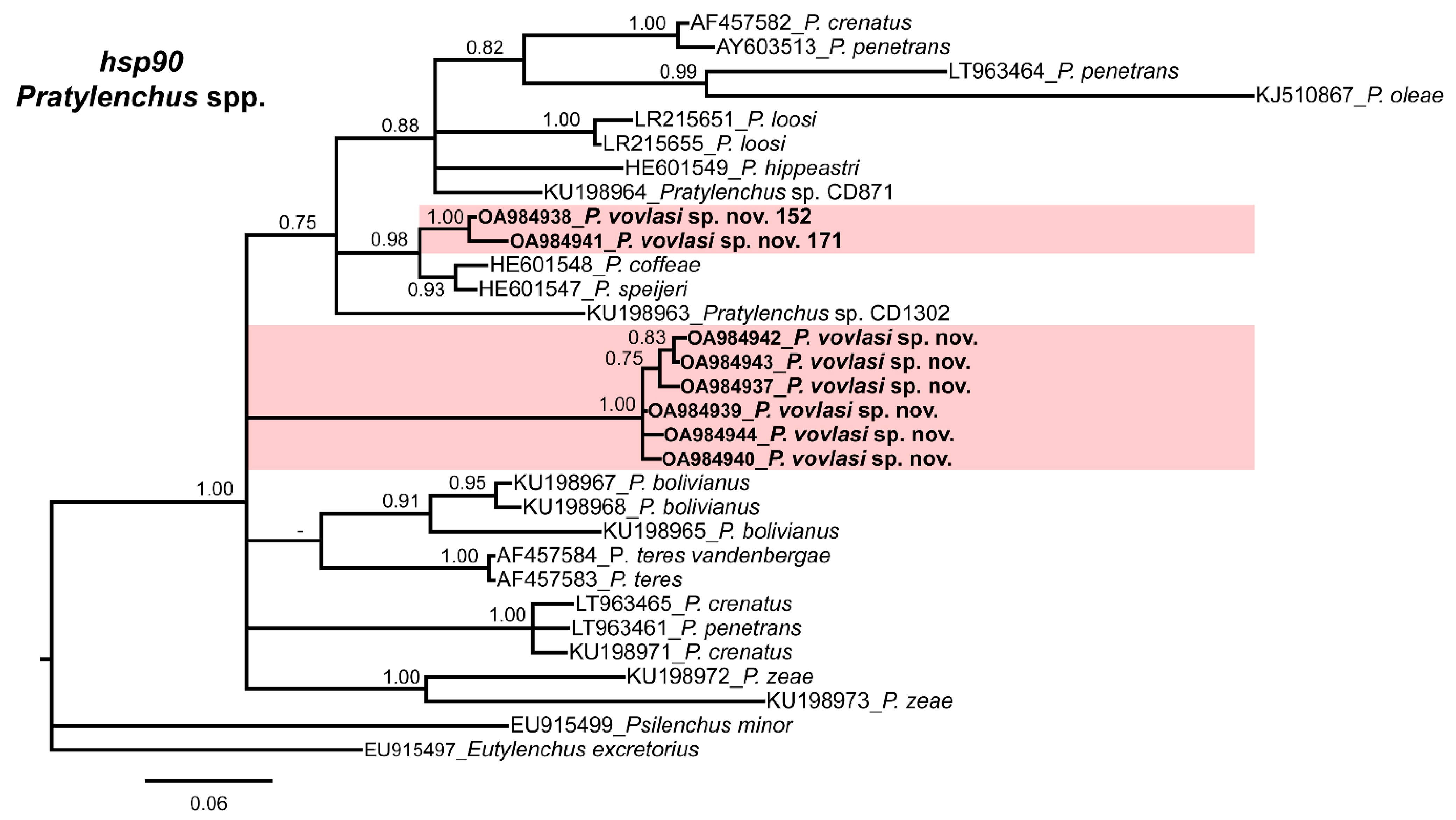
2.3. Phylogenetic Relationships
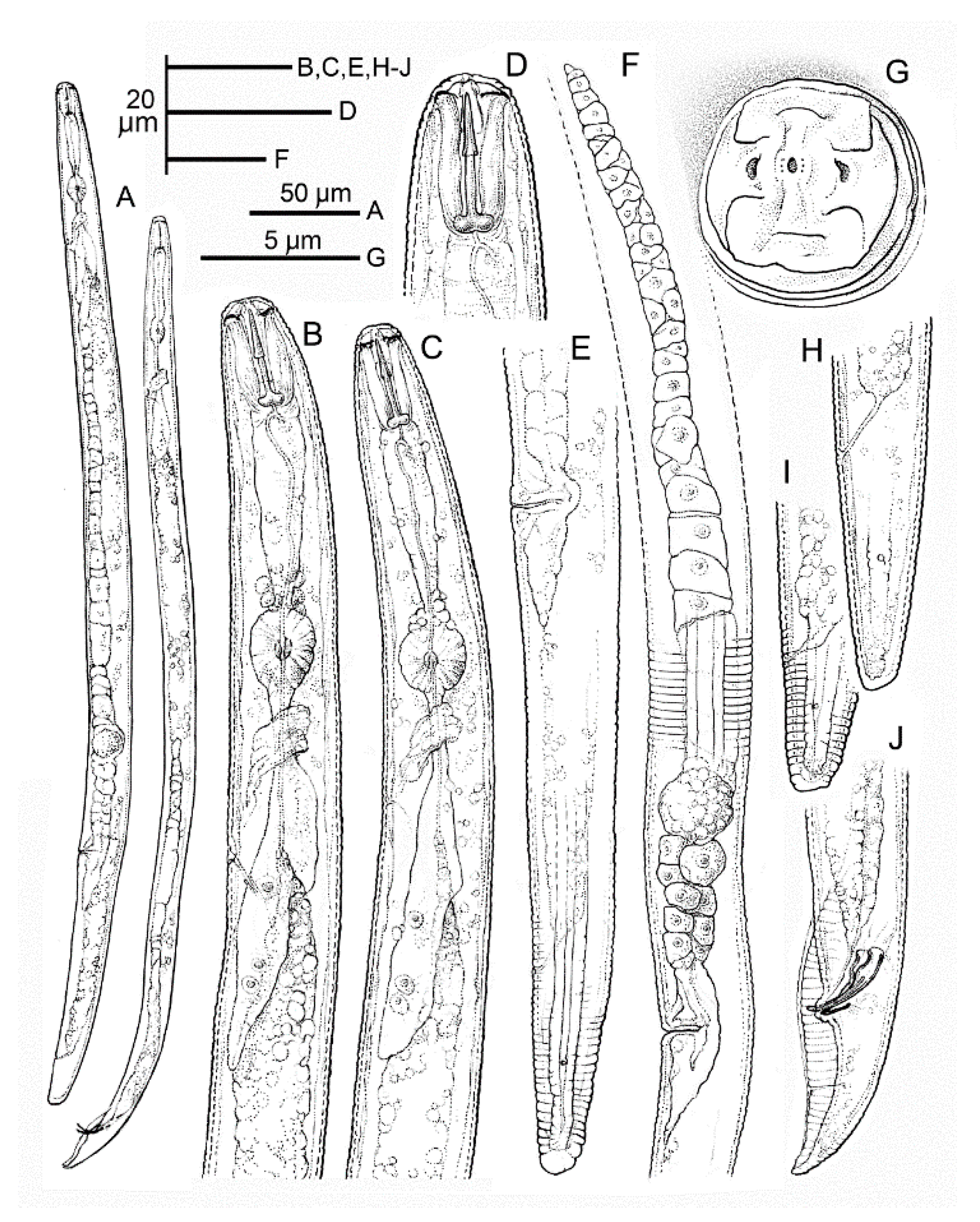
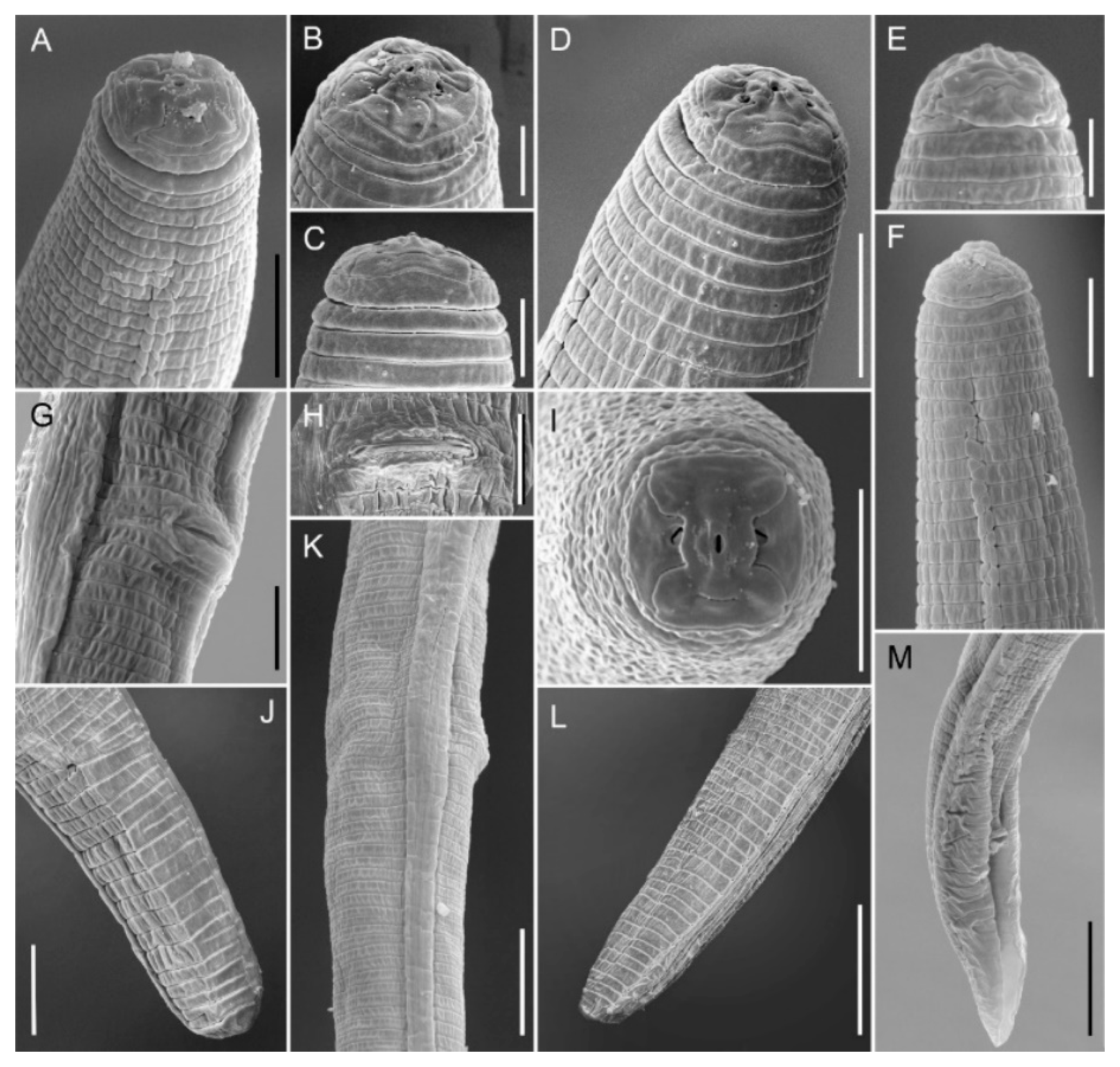
2.4. Morphology and Morphometry of Pratylenchus vovlasi sp. nov.
2.4.1. Description
2.4.2. Type Host and Locality
2.4.3. Type Material
2.4.4. Diagnosis and Relationships
2.4.5. Etymology
3. Discussion
4. Materials and Methods
4.1. Nematode Isolate and Morphological Studies
4.2. DNA Extraction, Polymerase Chain Reaction (PCR) and Sequencing
4.3. PCR-RFLP
4.4. Phylogenetic Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. 2009. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 2 April 2021).
- Skrovankova, S.; Sumczynski, D.; Mlcek, J.; Jurikova, T.; Sochor, J. Bioactive compounds and antioxidant activity in different types of berries. Int. J. Mol. Sci. 2015, 16, 24673–24706. [Google Scholar] [CrossRef]
- Tefera, T.; Tysnes, K.R.; Utaaker, K.S.; Robertson, L.J. Parasite contamination of berries: Risk, occurrence, and approaches for mitigation. Food Waterborne Parasitol. 2018, 10, 23–38. [Google Scholar] [CrossRef]
- Gigot, J.; Walters, T.; Zasada, I.A. Impact and occurrence of Phytophthora rubi and Pratylenchus penetrans in commercial red raspberry (Rubus ideaus) fields in North-western Washington. Int. J. Fruit Sci. 2013, 13, 357–372. [Google Scholar] [CrossRef]
- Poiras, L.; Cernet, A.; Buvol, A.; Poiras, N.; Iurcu-Straistraru, E. Preliminary analysis of plant parasitic nematodes associated with strawberry and raspberry crops in the Republic of Moldova. Oltenia. Studii şi comunicări. Ştiinţele Naturii 2014, 30, 98–104. [Google Scholar]
- Tsolova, E.; Koleva, L. Ecological characteristics of plant parasitic nematodes in conventional and organic production of raspberries. J. Mt. Agric. Balk. 2015, 18, 727–739. [Google Scholar]
- Zasada, I.A.; Weiland, J.E.; Han, Z.; Walters, T.W.; Moore, P. Impact of Pratylenchus penetrans on establishment of red raspberry. Plant Dis. 2015, 99, 939–946. [Google Scholar] [CrossRef] [PubMed]
- Kroese, D.R.; Weiland, J.E.; Zasada, I.A. Distribution and longevity of Pratylenchus penetrans in the red raspberry production system. J. Nematol. 2016, 48, 241–247. [Google Scholar] [CrossRef]
- Mohamedova, M.; Samaliev, H. Phytonematodes associated with red raspberry (Rubus idaeus L.) in Bulgaria. J. Entomol. Zool. Stud. 2018, 6, 123–127. [Google Scholar]
- Subbotin, S.A.; Ragsdale, E.J.; Mullens, T.; Roberts, P.A.; Mundo-Ocampo, M.; Baldwin, J.G. A phylogenetic framework for root lesion nematodes of the genus Pratylenchus (Nematoda): Evidence from 18S and D2-D3 expansion segments of 28S ribosomal RNA genes and morphological characters. Mol. Phylogenet. Evol. 2008, 48, 491–505. [Google Scholar] [CrossRef]
- Corbett, D.C.M.; Clark, S.A. Surface feature in the taxonomy of Pratylenchus species. Rev. Nématol. 1983, 6, 85–98. [Google Scholar]
- Castillo, P.; Vovlas, N. Pratylenchus (Nematoda: Pratylenchidae): Diagnosis, Biology, Pathogenicity and Management; Brill Academic Publishers: Leiden, The Netherlands, 2007; p. 555. [Google Scholar]
- De Luca, F.; Troccoli, A.; Duncan, L.W.; Subbotin, S.A.; Waeyenberge, L.; Moens, M.; Inserra, R.N. Characterisation of a population of Pratylenchus hippeastri from bromeliads and description of two related new species, P. floridensis n. sp. and P. parafloridensis n. sp. from grasses in Florida. Nematology 2010, 12, 847–868. [Google Scholar] [CrossRef]
- Palomares-Rius, J.E.; Castillo, P.; Liébanas, G.; Vovlas, N.; Landa, B.B.; Navas-Cortes, J.A.; Subbotin, S.A. Description of Pratylenchus hispaniensis n. sp. from Spain and considerations on the phylogenetic relationship among selected genera in the family Pratylenchidae. Nematology 2010, 12, 429–451. [Google Scholar]
- De Luca, F.; Troccoli, A.; Duncan, L.W.; Subbotin, S.A.; Waeyenberge, L.; Coyne, D.L.; Brentu, F.C.; Inserra, R.N. Pratylenchus speijeri n. sp. (Nematoda: Pratylenchidae), a new root-lesion nematode pest of plantain in West Africa. Nematology 2012, 14, 987–1004. [Google Scholar] [CrossRef]
- Wang, H.; Zhuo, K.; Ye, W.; Liao, J. Morphological and molecular characterisation of Pratylenchus parazeae n. sp. (Nematoda: Pratylenchidae) parasitizing sugarcane in China. Eur. J. Plant Pathol. 2015, 143, 173–191. [Google Scholar] [CrossRef]
- Araya, T.Z.; Padilla, W.P.; Archidona-Yuste, A.; Cantalapiedra-Navarrete, C.; Liébanas, G.; Palomares-Rius, J.E.; Castillo, P. Root-lesion nematodes of the genus Pratylenchus (Nematoda: Pratylenchidae) from Costa Rica with molecular identification of P. gutierrezi and P. panamaensis topotypes. Eur. J. Plant Pathol. 2016, 145, 973–998. [Google Scholar] [CrossRef]
- Palomares-Rius, J.E.; Guesmi, I.; Horrigue-Raouani, N.; Cantalapiedra-Navarrete, C.; Liébanas, G.; Castillo, P. Morphological and molecular characterisation of Pratylenchus oleae n. sp. (Nematoda: Pratylenchidae) parasitizing wild and cultivated olives in Spain and Tunisia. Eur. J. Plant Pathol. 2014, 140, 53–67. [Google Scholar] [CrossRef]
- Fanelli, E.; Troccoli, A.; Capriglia, F.; Lucarelli, G.; Vovlas, N.; Greco, N.; De Luca, F. Sequence variation in ribosomal DNA and in the nuclear hsp90 gene of Pratylenchus penetrans (Nematoda: Pratylenchidae) populations and phylogenetic analysis. Eur. J. Plant Pathol. 2018, 152, 355–365. [Google Scholar] [CrossRef]
- Chen, B.; Piel, W.H.; Gui, L.; Bruford, E.; Monteiro, A. The HSP90 family of genes in the human genome: Insights into their divergence and evolution. Genomics 2005, 86, 627–637. [Google Scholar] [CrossRef] [PubMed]
- Fanelli, E.; Troccoli, A.; Tarasco, E.; De Luca, F. Molecular characterization and functional analysis of the Hb-hsp90-1 gene in relation to temperature changes in Heterorhabditis bacteriophora. Front. Physiol. 2021, 12, 12. [Google Scholar] [CrossRef] [PubMed]
- Skantar, A.M.; Carta, L.K. Molecular characterization and phylogenetic evaluation of the Hsp90 gene from selected nematodes. J. Nematol. 2004, 36, 466–480. [Google Scholar] [PubMed]
- Troccoli, A.; Subbotin, S.A.; Chitambar, J.J.; Janssen, T.; Waeyenberge, L.; Stanley, J.D.; Duncan, L.W.; Agudelo, P.; Múnera Uribe, G.; Franco, J.; et al. Characterisation of amphimictic and parthenogenetic populations of Pratylenchus bolivianus Corbett, 1983 (Nematoda: Pratylenchidae) and their phylogenetic relationships with closely related species. Nematology 2016, 18, 651–678. [Google Scholar] [CrossRef]
- Coolen, W.A. Methods for extraction of Meloidogyne spp. and other nematodes from roots and soil. In Root-Knot Nematodes (Meloidogyne Species). Systematics, Biology and Control; Lamberti, F., Taylor, C.E., Eds.; Academic Press: New York, NY, USA, 1979; pp. 317–329. [Google Scholar]
- Seinhorst, J.W. Killing nematodes for taxonomic study with hot f.a. 4:1. Nematologica 1966, 12, 175. [Google Scholar] [CrossRef]
- Abolafia, J.; Liébanas, G.; Peña-Santiago, R. Nematodes of the order Rhabditida from Andalucia Oriental, Spain. The subgenus Pseudacrobeles Steiner, 1938, with description of a new species. J. Nematode Morphol. Syst. 2002, 4, 137–154. [Google Scholar]
- De Luca, F.; Fanelli, E.; Di Vito, M.; Reyes, A.; De Giorgi, C. Comparison of the sequences of the D3 expansion of the 26S ribosomal genes reveals different degrees of heterogeneity in different populations and species of Pratylenchus from the Mediterranean region. Eur. J. Plant Pathol. 2004, 111, 949–957. [Google Scholar] [CrossRef]
- Joyce, S.A.; Burnell, A.M.; Powers, T.O. Characterization of Heterorhabditis isolates by PCR amplification of segments of mtDNA and rDNA genes. J. Nematol. 1994, 26, 260–270. [Google Scholar] [PubMed]
- Joyce, S.A.; Reid, A.; Driver, F.; Curran, J. Application of polymerase chain reaction (PCR) methods to the identification of entomopathogenic nematodes. In Genetics of Entomopathogenic Nematodes-Bacterium Complexes, Proceedings of Symposium and Workshop; Burnell, A.M., Ehlers, R.U., Masson, J.P., Eds.; S. Patrick’s College, Maynooth, Co.: Kildrare, Ireland; EC DG XII: Luxembourg, 1994; pp. 178–187. [Google Scholar]
- Skantar, A.M.; Carta, L.K. Multiple Displacement Amplification (MDA) of total genomic DNA from Meloidogyne spp. and comparison to crude DNA extracts in PCR of ITS1, 28 S D2-D3 rDNA and Hsp90. Nematology 2005, 7, 285–293. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. FigTree v1.4.2, a Graphical Viewer of Phylogenetic Trees. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 2 April 2021).

| Character | Holotype | Paratypes | |
|---|---|---|---|
| Females | Males | ||
| n | - | 16 | 9 |
| L | 463 | 510 ± 33.4 (459–577) | 481 ± 32.1 (411–518) |
| a | 21 | 23.8 ± 2.2 (20.0–26.5) | 28.3 ± 1.8 (26.0–31.9) |
| b | 5.8 | 6.5 ± 0.5 (5.8–7.3) | 6.0 ± 0.3 (5.6–6.4) |
| b’ | 4.4 | 4.7 ± 0.4 (4.3–5.5) | 4.5 ± 0.2 (4.1–4.9) |
| c | 22.4 | 22.3 ± 2.7 (18.2–27.2) | 17.9 ± 4.0 (9.6–22.0) |
| c’ | 1.7 | 1.8 ± 0.2 (1.4–2.3) | 2.6 ± 0.7 (2.0–4.3) |
| Lip region height | 2.0 | 2.3 ± 0.3 (2.0–2.8) | 2.3 ± 0.2 (2.0–2.7) |
| Lip region diameter | 8.5 | 8.1 ± 1.0 (6.6–9.0) | 7.4 ± 0.6 (6.3–8.0) |
| Stylet length | 14.5 | 15.0 ± 0.6 (14.3–16.3) | 14.0 ± 0.6 (13.5–15.0) |
| Stylet cone | 7.0 | 6.8 ± 0.6 (6.0–8.0) | 6.5 ± 0.4 (6.5–7.5) |
| Stylet knob width | 4.0 | 4.0 ± 0.6 (3.0–4.5) | 2.7 ± 0.4 (2.5–3.5) |
| DGO from stylet base | 2.0 | 2.3 ± 0.3 (2.0–2.7) | 1.7 ± 0.4 (1.5–2.5) |
| o | 13.8 | 15.2 ± 1.9 (13.6–18.4) | 11.7 ± 2.8 (9.9–16.3) |
| Anterior End to: | |||
| center of metacorpus | 48.0 | 54 ± 2.6 (48–58) | 52.0 ± 2.8 (47.5–55.5) |
| cardia | 80.0 | 79 ± 5.4 (70–85) | 80.0 ± 5.1 (73.5–90.0) |
| end of pharyngeal lobe | 105.0 | 110 ± 9.0 (97–125) | 108 ± 6.5 (99.5–120) |
| secretory/excretory pore | 75 | 79 ± 3.5 (72–84) | 77.5 ± 5.6 (65.5–83) |
| vulva | 347 | 397 ± 31.8 (347–462) | - |
| Pharyngeal overlap | 25 | 32 ± 6.7 (20–43) | 28.0 ± 4.3 (22.5–36.5) |
| Max body diameter | 22.0 | 22.0 ± 1.5 (20–24.5) | 17.0 ± 1.0 (15.5–18.5) |
| Anal body diameter | 12.0 | 13.0 ± 1.1 (11.5–15.0) | 11.0 ± 1.0 (9.5–12.5) |
| Anterior genital tract length | 224.0 | 250 ± 30.8 (203–313) | 192 ± 23 (163–232) |
| Spermatheca to vagina distance | 42.0 | 50.0 ± 9.6 (36.5–64.5) | - |
| Spermatheca length | 17.0 | 17.0 ± 2.5 (12.5–21.0) | - |
| Spermatheca width | 14.0 | 15.0 ± 1.9 (11.5–18.0) | - |
| Vulva to anus distance | 96 | 90.0 ± 10.0 (78.5–110) | - |
| V or T | 75 | 77.8 ± 2.0 (74–80) | 40.1 ± 5.0 (32–50) |
| G1 | 48 | 49.0 ± 6.1 (39–59) | - |
| PUS | 26 | 21.7 ± 2.4 (18.0–26.0) | - |
| Tail length | 20.7 | 23.5 ± 3.3 (18.0–30.0) | 26.5 ± 4.6 (19.5–31.5) |
| Number of tail annuli | 16 | 16 ± 2.0 (14–20) | - |
| Spicule length | - | - | 16.5 ± 1.4 (14.5–18.5) |
| Gubernaculum length | - | - | 5.0 ± 0.3 (5.6–6.4) |
| Species | Morphological Characteristics * | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A1–3 | B1–2 | C1–5 | D1–4 | E1–4 | F1–6 | G1–3 | H1–4 | I1–4 | J1–3 | K1–2 | |
| Lip Annuli | Presence of Males | Stylet Length (µm) | Shape of Spermatheca | Vulva Position (%) | PUS ** (µm) | Female Tail Shape | Female Tail Tip | Pharyngeal Overlap (µm) | Lateral Field | Lateral Field Structures | |
| Pratylenchus vovlasi sp. nov. | A2 | B2 | C2 | D2 | E2 | F3 | G1,2 | H1 | I2 | J1 | K1 |
| Pratylenchus bhattii | A2 | B2 | C2 | D2 | E1 | F2 | G2 | H1 | I2 | J1 | K1 |
| Pratylenchus mediterraneus | A2 | B2 | C2 | D2 | E2 | F3 | G2 | H1 | I3 | J1 | K1 |
| Pratylenchus kralli | A2 | B2 | C2 | D2 | E2 | F1 | G3 | H1 | I1 | J1 | K1 |
| Pratylenchus fallax | A2 | B2 | C3 | D2 | E2 | F3 | G3 | H2 | I2 | J1 | K1 |
| Pratylenchus convallariae | A2 | B2 | C3 | D2 | E2 | F6 | G2 | H2 | I3 | J1 | K1 |
| Pratylenchus penetrans | A2 | B2 | C3 | D2 | E3 | F4 | G2 | H1 | I3 | J1 | K1 |
| Pratylenchus pratensis | A2 | B2 | C2 | D4 | E2 | F3 | G3 | H2 | I1 | J1 | K1 |
| Pratylenchus pseudopratensis | A2 | B2 | C3 | D4 | E2 | F3 | G3 | H2 | I3 | J1 | K1 |
| Pratylenchus thornei | A2 | B2 | C2 | D2 | E2 | F3 | G2 | H1 | I3 | J1 | K1 |
| Pratylenchus vulnus | A2 | B2 | C2 | D3 | E2 | F6 | G3 | H3 | I2 | J1 | K1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Troccoli, A.; Fanelli, E.; Castillo, P.; Liébanas, G.; Cotroneo, A.; De Luca, F. Pratylenchus vovlasi sp. Nov. (Nematoda: Pratylenchidae) on Raspberries in North Italy with a Morphometrical and Molecular Characterization. Plants 2021, 10, 1068. https://doi.org/10.3390/plants10061068
Troccoli A, Fanelli E, Castillo P, Liébanas G, Cotroneo A, De Luca F. Pratylenchus vovlasi sp. Nov. (Nematoda: Pratylenchidae) on Raspberries in North Italy with a Morphometrical and Molecular Characterization. Plants. 2021; 10(6):1068. https://doi.org/10.3390/plants10061068
Chicago/Turabian StyleTroccoli, Alberto, Elena Fanelli, Pablo Castillo, Gracia Liébanas, Alba Cotroneo, and Francesca De Luca. 2021. "Pratylenchus vovlasi sp. Nov. (Nematoda: Pratylenchidae) on Raspberries in North Italy with a Morphometrical and Molecular Characterization" Plants 10, no. 6: 1068. https://doi.org/10.3390/plants10061068
APA StyleTroccoli, A., Fanelli, E., Castillo, P., Liébanas, G., Cotroneo, A., & De Luca, F. (2021). Pratylenchus vovlasi sp. Nov. (Nematoda: Pratylenchidae) on Raspberries in North Italy with a Morphometrical and Molecular Characterization. Plants, 10(6), 1068. https://doi.org/10.3390/plants10061068