Abstract
Angiosperm plants reproduce both sexually and asexually (by apomixis). In apomictic plants, the embryo and endosperm develop without fertilization. Modern maize seems to have a broken apomixis-triggering mechanism, which still works in Tripsacum and in Tripsacum–maize hybrids. For the first time, maize lines characterized by pronounced and inheritable high-frequency maternal parthenogenesis were generated 40 years ago, but there are no data on gene expression in parthenogenic maize proembryos. Here we examined for the first time gene expression in parthenogenic proembryos isolated from unpollinated embryo sacs (ESs) of a parthenogenic maize line (AT-4). The DNA-methylation genes (dmt103, dmt105) and the genes coding for the chromatin-modifying enzymes (chr106, hdt104, hon101) were expressed much higher in parthenogenic proembryos than in unpollinated ESs. The expression of the fertilization-independent endosperm (fie1) genes was found for the first time in parthenogenic proembryos and unpollinated ESs. In parthenogenic proembryos, the Zm_fie2 gene was expressed up to two times higher than it was expressed in unpollinated ESs.
1. Introduction
Angiosperm plants reproduce both sexually and asexually (by apomixis) [1,2,3]. In sexual plants, in the absence of fertilization neither embryo nor endosperm can develop. In apomictic plants, mainly polyploid species, the embryo and endosperm develop under fertilization-free conditions (fertilization-independent seed (fis) and endosperm (fie)). Apomixis is of major interest for the fixation of genotypes obtained by using heterosis (hybrid vigor). Control of apomixis would make it possible to fix highly productive and environmentally adaptable genotypes for modern agriculture. An understanding of how apomixis acts at the molecular level could enable apomixis to be controlled.
Apomixis has been observed in the wild ancestor of maize, Tripsacum dactyloides L., and in Tripsacum–maize hybrids [4,5]. Hybrids with 38 chromosomes (2n = 20 maize + 18 Tripsacum) and with fertilization-free seed development were very interesting as a model [4,5,6,7]. Some researchers speculated that apomictic reproduction in Tripsacum is controlled by nine chromosomes [8]; others showed that one chromosome is responsible for apomixis-like development [9,10]. Which genes govern the switch from sexual to apomictic reproduction in Tripsacum–maize hybrids still remains unclear; there are data that the responsible genes are those coding for the chromatin-modifying enzymes (CMEs) [7].
It has been suggested that apomixis may result from temporal or spatial deregulation of the transcription programs that control sexual reproduction [11,12,13,14]. In particular, the CME genes are differently expressed in Tripsacum–maize hybrids and in sexual maize [7,15,16]. The CME genes have clear qualitative expression differences before and after pollination in maize and in Tripsacum–maize hybrids [7]. In contrast to sexual maize, the expression of three DNA methyltransferases (dmt102, dmt105, dmt103) and one chromatin-modulating factor (chr106) is suppressed in Tripsacum-maize hybrids at all developmental stages. Two genes [histone linker (hon101) and histone deacetylase (hdt104)] are expressed heterochronically [7].
The fertilization-independent development of the maize seed and endosperm is correlated with the methylation of the genes controlling endosperm development and the imprinting phenomenon [17,18,19,20]. Although gene imprinting (parents’ allele-dependent gene expression) is observed in the maize embryo and endosperm, it has not been found in adult plant tissues [21,22,23]. On the other hand, in the maize embryo, the genes are more methylated than in the endosperm [24].
The Zm_fie genes belong to the Рolycomb Repressive Complex 2 (PRC2) genes, which regulate the early stages in Drosophila and Arabidopsis development [25,26]. In particular, the Zm_fie1 gene is not expressed in the maize sperm, egg, or central cell before fertilization but is expressed in the endosperm at 2 days after pollination (DAP), with maximum activity at 10–11 DAP [25]. On the contrary, the Zm_fie2 gene is expressed in the egg and central cells before pollination and is probably repressed during maize endosperm development [25,26]. As suggested by phylogenetic analysis of the maize fie genes, the fie1 and fie2 genes arose through the duplication of one of the ancestral paralog genes during maize genome evolution [25].
Locus-specific imprinting in maize is observed in the genes fie, meg1 (maternally expressed gene 1), and nrp1 (no-apical-meristem-related protein 1) [25,27,28], which are expressed in the female gametophyte without fertilization.
Modern maize seems to have a broken apomixis-triggering mechanism. Forty years ago, however, Russian breeders from Saratov State University generated the first parthenogenic maize line (AT-1) with pronounced and inheritable high-frequency maternal parthenogenesis in the ESs [29]. The AT-1 line and its derivatives AT-3 [30,31] and AT-4 [32] are polyembryonic, with parthenogenic embryo and endosperm development. There are no data on gene expression in parthenogenic maize proembryos.
Previously, we examined for the first time the expression of the CME genes in the female gametophytes of the AT-3 line and have found no large differences between the ESs of the parthenogenic line AT-3 and the ordinary line DHH-1 before and after pollination, except that the expression of the hon101 and hdt104 genes was higher in DHH-1 [33]. However, there are no data on maize gene expression in parthenogenic proembryos or in unpollinated ESs. It is interesting to note that although the development of parthenogenic embryos in maize ESs has been described [29,30], we did not find any homologous Arabidopsis fis genes in the maize database (data not shown).
The maize CME genes are possibly responsible for apomictic-like egg-cell development in Tripsacum–maize hybrids [5,7,23]. Those findings suggest that the CMEs are implicated in the fate of the precursor cells of the maize ESs. We chose six CME loci (dmt102, dmt103, dmt105, chr106, hdt104, and hon101) to search for qualitative differences in gene expression between fertilization-free proembryos and ESs of the parthenogenic maize line AT-4. The reason was that only these genes differ qualitatively in expression between sexual maize B73 and apomictic Tripsacum-maize hybrid [7].
Here we observed for the first time the expression of the CME and Zm_fie genes in parthenogenic proembryos and ESs isolated from the unpollinated maize line AT-4.
2. Results and Discussion
2.1. Microscopy of ESs
In total, 50 frozen and fixed AT-4 ESs were analyzed 8 days after the appearance of pistillate stigmas (DAAPS). A typical ES contained one egg, two synergides, two polar nuclei, and an antipodal complex (Figure 1A). There were deviations from the typical ES structure, and parthenogenic proembryos developed under pollination-free conditions (8 DAAPS). At 8 DAAPS, the number of ESs with 2- to 16-cell parthenogenic proembryos was up to 18%. Figure 1B shows 8-cell proembryos at the early proembryo stage.
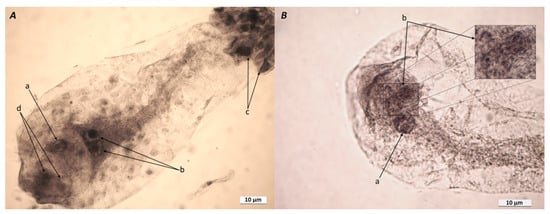
Figure 1.
(A) Typical embryo sac (ES) isolated from AT-4 ovules at 8 DAAPS. Notes: a, egg cell; b, two polar nuclei of the central cell; c, antipodal complex; d, two synergides. (B) Unpollinated ES isolated from AT-4 ovules at 8 DAAPS. Notes: a, central cell nucleus; b, 8-cell parthenogenic proembryo and an enlarged fragment of it. The ESs (A and B) were isolated from acetic-alcohol-fixed ovules, washed, and placed in glycerol–acetocarmine (5:1, v/v) for 24 h.
There also were ESs with additional cells and nuclei in the egg apparatus, synergide-like cells, and ESs with two-celled proembryos or additional egg cells (data not shown). The frequency of spontaneous proembryo formation in the AT-4 line was 10 times higher than the number of parthenogenic proembryos found earlier in the parthenogenic AT-3 line [33].
Approximately within 24 h after pollination (HAP), the zygote divides transversely, yielding a small terminal cell and a large basal cell. Forty-eight HAP, there occur additional rounds of cell division in the embryo, which generally result in a two- (or three-) celled proembryo and a two- (or three-) celled suspensor [34]. We observed 2- to 16-cell parthenogenic proembryos in the AT-4 ESs at 8 DAAPS, but we do not know exactly when a particular egg cell started dividing under pollination-free conditions. Microscopy showed that the egg cells in the AT-4 ESs were at different maturity stages and differed in their readiness for spontaneous division, because we observed 2- and 16-cell proembryos at 8 DAAPS.
Within 6–12 HAP, the pollen tube delivers two sperm cells to the ES, and within 10 s, one maize sperm attaches to the egg cell membrane, after which it fuses with it [35]. The first egg-cell division takes up to 48 HAP to produce a 2-cell proembryo [36,37] and 96 HAP to produce a late proembryo [35]. At present, we do not know how the parthenogenic proembryo and the embryo developed after pollination differs in the period of development. For 350,000 samples taken from the ears of the AT-3 maize line, no parthenogenic seed development was found (Yu.V. Smolkina, personal communication). The parthenogenic embryos disintegrated at 12–20 DAAPS, because they lacked support from the undeveloped endosperm.
Thus, at 8 DAAPS, the AT-4 line showed maternal parthenogenesis (spontaneous proembryo development from an unpollinated egg cell). The question arises: what changes in gene expression can accompany the development of parthenogenic proembryos? The DNA-methylation and CME genes, whose expression varies with apomictic development of the maize embryo [7], and the fie genes will be analyzed in the next sections.
2.2. Expression of DNA-Methylation Genes in ESs
To address the possible role of chromatin structure in the occurrence of apomixis-like proembryos in the AT-4 (unpollinated) ESs, we investigated the expression of the DNA-methylation genes in parthenogenic proembryos isolated from unpollinated ESs at 10 DAAPS, in ESs at 3 DAP, and in the embryo (7 DAP) and endosperm (10 DAP) isolated from AT-4 ESs. We wanted to determine whether there were any differences in the expression of the DNA-methylation genes (i) between the ESs with parthenogenic proembryos developed from unpollinated egg cells (10 DAAPS) and the unpollinated ESs at 7 and 10 DAAPS and (ii) between the parthenogenic proembryos at 10 DAAPS and the embryos and endosperm at 7 and 10 DAP.
The absence of qPCR products from the DNA-methylation genes dmt102, dmt103, and dmt105 in Tripsacum–maize ovules indicates apomictic development [7]. In our work, the expression patterns of the dmt103 and dmt105 genes in the parthenogenic proembryos were significantly higher than in unpollinated ESs (10 DAAPS; Figure 2A,B), except for the dmt102 gene (data not shown). In particular, in four biological samples consisting of five ESs with parthenogenic proembryos, the dmt103 gene was 1.6–3.3 times more expressed (average, 2.04 ± 0.65; p ≤ 0.05) than it was expressed in unpollinated ESs (Figure 2A). Yet, dmt103 expression in the ESs after pollination (3–10 DAP) gradually increased nonsignificantly from ESs (3 DAP) to embryos (7 DAP), becoming up to 1.5 times higher (p ≤ 0.05) than in unpollinated ESs (10 DAAPS) in the case of the endosperm (10 DAP). Surprisingly, dmt103 expression was higher in unpollinated ESs at 7 DAAPS (Figure 2A).
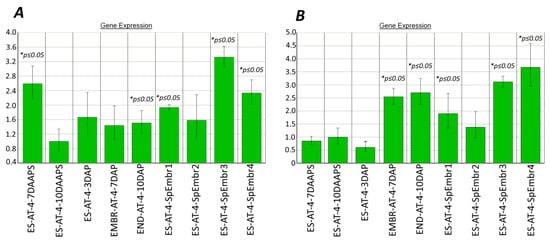
Figure 2.
Expression of DNA-methylation genes in the parthenogenic proembryos (10 DAAPS), ESs (7 and 10 DAAPS), and embryos and endosperm (7 and 10 DAP) of the AT-4 line. (А) dmt103; (B) dmt105. Control: unpollinated ESs (10 DAAPS). * Significant differences between target tissue and control cells (ESs at 10 DAAPS) are expressed as p ≤ 0.05. ES-AT-4-SpEmbr1-4: in one biological experiment, each of the four samples consisted of five ESs with parthenogenic proembryos. Data are presented from two independent biological experiments. ES-AT-4-7-10 DAAPS: in one biological experiment, each sample consisted of 12 unpollinated ESs. Data are presented from two independent biological experiments.
In four samples with parthenogenic proembryos, the dmt105 gene was 1.4–3.7 times more expressed (average, 2.44 ± 0.82) than it was expressed in unpollinated ESs (Figure 2B). Yet, dmt105 expression in the ESs after pollination (3–10 DAP) gradually increased, becoming more than two times higher than in unpollinated ESs (10 DAAPS). We speculate that the activity of dmt105 methyltransferase in parthenogenic embryos at 10 DAAPS may be higher than in unpollinated ESs at 10 DAAPS.
Microscopy showed that the parthenogenic proembryos in the AT-4 ESs at 8 DAAPS were at different division stages (Figure 1B). We used prefrozen (−20 °C) ears to isolate intact cooled ESs with visually observed parthenogenic proembryos and analyze the expression of the DNA-methylation genes. Unfortunately, it was impossible to recognize the development stage for parthenogenic proembryos in the intact ESs before RNA extraction. As can be seen from Figure 2, the expression levels for the dmt103 and dmt105 genes differed significantly (p ≤ 0.05), especially in samples 3 and 4 (parthenogenic proembryos). Therefore, the parthenogenic proembryos (numbered 1–4) from intact ESs possibly were at different division stages as well (Figure 2).
The DNA-methylation genes and the chr106 gene were broadly expressed during sexual development (sporogenesis; mature ESs at 3 DAP) in the B73 maize line but were totally absent during apomictic reproduction of a Tripsacum-maize hybrid (38C) [7]. Dmt102 was not expressed at any of the three stages in 38C. In this work, however, gene expression was detected in mature ESs, and it remained at the same level in parthenogenic proembryos and in reproductive embryos after pollination.
The expression of the dmt103 gene was observed in the B73 ovules with mature ESs, but at 3 DAP it dropped to the level of C38 hybrids [7]. In our work, dmt103 expression in unpollinated ESs was no different from that in embryos from pollinated ESs at 3 DAP; however, it increased in parthenogenic proembryos. The expression of the dmt103 gene was high in the ovules of the B73 line with mature ESs but lower in ovules with embryos at 3 DAP [7]. In our work, dmt105 expression did not differ between unpollinated ESs (7 DAAPS) and embryos from pollinated ESs (3 DAP). In parthenogenic proembryos, the expression of this gene was greatly increased (Figure 2B).
2.3. CME Gene Expression in ESs
2.3.1. Chromatin-Modulating Factor (chr106) Expression
In parthenogenic proembryo 1 (10 DAAPS), the chr106 gene was 1.4 times more expressed (p ≤ 0.05) than it was expressed in unpollinated ESs at 10 DAAPS. chr106 expression in sample 1 with proembryos was similar to that in embryos from pollinated ESs (7 DAP; Figure 3). We did not find any significant differences in chr106 expression between unpollinated ESs (7 and 10 DAAPS) and ESs with parthenogenic proembryos (sample 2) (Figure 3). In pollinated ESs (3 DAP), the chr106 gene was two times more expressed (p ≤ 0.05) than it was expressed in unpollinated ESs at 10 DAAPS.
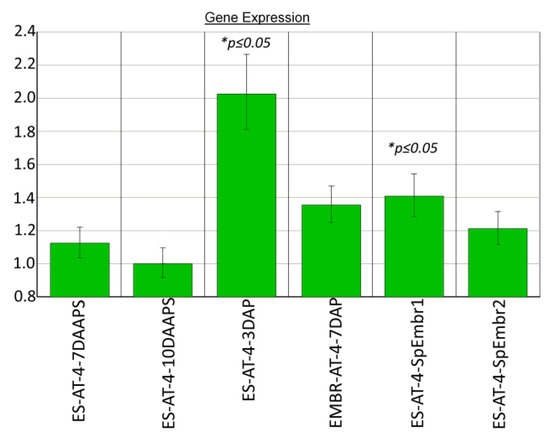
Figure 3.
Chromatin-modulating factor (chr106) expression in parthenogenic proembryos (10 DAAPS) and ESs (7 and 10 DAAPS) of the AT-4 line. Control: ESs at 10 DAAPS. ES-AT-4-7-10DAAPS: in one independent biological experiment, each sample consisted of 12 unpollinated ESs. Data are from four independent biological experiments.
2.3.2. Histone Deacetylase (hdt104) and Histone Linker (hon101) Expression in Proembryos and ESs
In parthenogenic proembryos (sample 1 but not sample 2) (10 DAAPS), the hon101 and hdt104 genes were 1.4 and 1.7 times more expressed (p ≤ 0.05), respectively, than they were expressed in unpollinated ESs at 10 DAAPS (Figure 4). Similarly, the hdt104 and hon101 genes were up to 2 and 2.4 times more expressed, respectively, in pollinated ESs at 3 DAP than they were expressed in unpollinated ESs at 10 DAAPS (Figure 4).
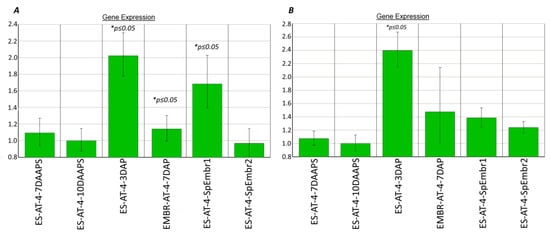
Figure 4.
Histone deacetylase (hdt104) (A) and histone linker (hon101) (B) expression in parthenogenic proembryos* (10 DAAPS) and ESs (7–10 DAAPS). Controls: ESs at 10 DAAPS for (A) and (B). *ES-AT-4, 7 and 10 DAAPS: in one biological experiment, each sample consisted of 12 unpollinated ESs. Data are from four independent biological experiments.
The parthenogenic embryos did not differ in chr106, hdt104, or hon101 expression in unpollinated ESs (10 DAAPS), as compared with embryos in pollinated ESs at 7 DAP in both independent experiments (Figure 3 and Figure 4).
Thus, except for the dmt102 gene, the expression levels of the dmt103, dmt105, chr106, hon101, and hdt104 genes were 1.4 to 3.7 times higher (p ≤ 0.05) in parthenogenic proembryos (10 DAAPS) than in unpollinated ESs (10 DAAPS). Of note, in the apomictic hybrid, the expression of the CME genes was repressed, as compared with that in the sexual B73 line [7].
The differences in the expression of the DNA-methylation (except dmt102) and CME genes at 10 DAAPS could clarify parthenogenic proembryo development in unpollinated AT-4 ESs (Figure 2 and Figure 3).
fie1 and fie2 Expression in ESs
The first nucleus division in the pollinated ESs began after 16–17 HAP, as soon as karyogamy in the central cell was complete [36]. Although we did not observe parthenogenic central cell development in 50 ESs isolated from AT-4 ovules, we believe that it does exist, because the AT-4 line is characterized by parthenogenic embryo and endosperm development [32].
We observed for the first time the expression of the Zm_fie1 gene in unpollinated AT-4 ESs at 3–10 DAAPS (p ≤ 0.05). We did not find any significant (p ≤ 0.05) differences in Zm_fie1 expression between unpollinated ESs (7 and 10 DAAPS) and parthenogenic proembryos (Figure 5A). Surprisingly, we observed higher-level Zm_fie1 expression in unpollinated ESs at 3 DAAPS (Figure 5A). In the endosperm from pollinated ESs (7–10 DAP), the Zm_fie1 gene was expressed three times more than it was expressed in unpollinated ESs at 10 DAAPS (Figure 5A). This finding is in agreement with the earlier data [25,26]. In particular, the Zm_fie1 gene is expressed in the maize endosperm at 2 DAP, with maximum activity at 10–11 DAP. It is interesting to note that Zm_fie1 expression in the embryo from pollinated ESs at 7 and 10 DAP was two times higher than in unpollinated ESs (10 DAAPS) (Figure 5A).
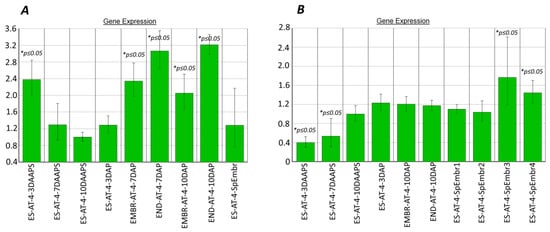
Figure 5.
fie1 (A) and fie2 (B) expression in ESs with parthenogenic proembryos* (10 DAAPS) and in unpollinated ESs (7 and 10 DAAPS). Control: ESs at 10 DAAPS. *ES-AT-4-7-10DAAPS: in one biological experiment, each sample consisted of 12 unpollinated ESs. Data are from two independent biological experiments.
We observed for the first time increased Zm_fie2 expression in parthenogenic proembryos, especially in samples 3 and 4 (Figure 5B). In these samples, fie2 expression at 10 DAAPS increased by 1.8 and 1.4 times, respectively (p ≤ 0.05), as compared with unpollinated ESs (10 DAAPS; Figure 5B). As expected, the Zm_fie2 gene was more expressed in the endosperm and embryo in 10 DAP (Figure 5B).
3. Materials and Methods
3.1. Plant Material
The diploid maize (Zea mays L.) line АТ-4 was produced by pollinating the tetraploid line KrP-1 with the diploid parthenogenic line AT-1. In the hybrid’s offspring, diploid plants (dihaploids) emerged, which were selected and self-pollinated [32]. The AT-1 line is the product of crossing the Stock 6 line [38] with the Brown Tester line [29]. The purple color of the AT-1 grains, leaves, and stems is determined by the A, B, PL, and R genes [38].
Seeds were planted by using tractor-mounted seeders (SKS-6-10, Russia) on May 14, 2020, in three 4 × 5 m plots. The planting density was 5–10 plants/m2. The AT-4 line was grown in the fields of the Russian Research, Design, and Technology Institute of Sorghum and Maize (Rossorgo Institute; Saratov, Russia) without watering or herbicide treatment. Weeds were removed on June 5, first by tractor and then by hand.
The maize ears were isolated with parchment bags before pistil filaments developed (late July–early August) and were grown without pollination for 3–10 days. The unpollinated ears were collected at 3, 7, and 10 DAAPS, placed in containers with ice, transported to the laboratory, and frozen at −20 °C. For ES and proembryo isolation, the ears were harvested at 7 and 10 DAAPS. For obtaining a sexual embryo and endosperm, preisolated pistils were pollinated manually with pollen collected from other AT-4 plants. The ears were collected at 3, 7, and 10 DAP, placed in containers with ice, transported to the laboratory, and frozen at −20 °C. ESs, parthenogenic proembryos, embryos, and endosperms were isolated from prefrozen (−20 °C) ears under an MBI-9 microscope (Russia) by using steel dissecting needles and were placed into Eppendorf tubes with a cooled RNA-isolation buffer.
3.2. Expression of CME, DNA-Methylation, and Fie Genes in Female Gametophyte Tissues
ESs, proembryos, embryos, and endosperms were isolated from freshly collected and prefrozen ears at 7 and 10 DAAPS and at 7 and 10 DAP. mRNA was extracted from 5 proembryos and 12 ESs (50 µg at 10 and 7 DAAPS, respectively), 5 embryos (50 µg at 7 and 10 DAP), and 2 endosperm samples (or 9 mg at 7 DAP) by using a commercial NucleoSpin RNA Plant kit (catalog no. 740949.50; Macherey Nagel, Dueren, North Rhine-Westphalia, Germany) with DNase. The total RNA, harvested from 12 proembryos (10 DAAPS) and 5 embryos (7 DAP), was 2.5 ng/µL. For cDNA synthesis, 1 µg of total RNA was subjected to reverse transcription with reverse transcriptase (catalog no. EP0442; Thermo Fisher, Waltham, MA, USA) and with an oligo (dT) primer (catalog no. AM5730G; Thermo Fisher). The cDNA concentration was measured on a Qubit fluorimeter (Thermo Fisher Scientific, Singapore) by using a Qubit DNA HS assay kit, (Q32850; Thermo Fisher Scientific, Waltham, MA, USA). For all samples, the cDNA concentration was made equal (2 ng) by diluting the high concentration. Quantitative PCR (qPCR) was done on an Applied Biosystems PCR amplifier (Applied Biosystems, Foster City, CA, USA) by using a set of reagents for real-time PCR in the presence of the SYBR green dye and the ROX reference dye (catalog no. M-435; Syntol, Russia) and specific primers (see Supplemental Data 1) handpicked by using the Primer3Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi, accession date was 24 March 2020) and Primer-BLAST programs. The primers were chosen so that the DNA and mRNA PCR products could differ in size (see Supplemental Data 1). Target gene expression profiles were determined relative to the expression level of the GAPDH gene as an endogenous control. For each data point, qPCR was done in four technical replicates for one cDNA sample. Two to four independent biological experiments were conducted (see figure notes). The relative gene expression levels were calculated by the 2−ΔΔCT method [39]. Following Weaver et al. [40], we discarded the difference <1.3 times.
3.3. Microscopy
The isolated ESs were stained with a glycerol–acetocarmine solution, as described by Volokhina et al. [33]. The ESs were analyzed with a DM2500 light fluorescence microscope (Leica Microsystems, Wetzlar, Heese, Germany) at the Simbioz Center for the Collective Use of Research Equipment in the Field of Physical–Chemical Biology and Nanobiotechnology (Institute of Biochemistry and Physiology of Plants and Microorganisms, Russian Academy of Sciences, Saratov).
4. Conclusions
Although apomixis has been observed in Tripsacum (the wild ancestor of maize) and in Tripsacum–maize hybrids [4,5], modern maize has a broken apomixis mechanism. The fertilization-independent development of the embryo and endosperm was discovered 40 years ago for the AT-1 and AT-3 maize lines [29,30] and for the recently generated AT-4 line [32]. However, there are no data on the genes controlling parthenogenic egg and central cell division. It is believed that the fertilization-independent development of the maize embryo and endosperm is correlated with the methylation of the genes controlling endosperm development and the imprinting phenomenon [17,18,19,20]. In particular, the DNA-methylation genes (dmt102, dmt103, and dmt105) and the genes coding for the CMEs (hdt104, chr106, and hon101) have clear qualitative expression differences in ESs before and after pollination between maize and Tripsacum–maize hybrids [7]. For this reason, we examined for the first time the expression of the DNA-methylation and CME genes in the parthenogenic proembryos of the AT-4 line. The expression of the DNA-methylation (except dmt102 gene) and CME genes differs between parthenogenic proembryos and unpollinated ESs and could be the reason for the development of parthenogenetic proembryos in the AT-4 line.
We observed for the first time the expression of the Zm_fie1 gene in unpollinated AT-4 ESs with or without parthenogenic proembryos. Zm_fie2 expression in parthenogenic proembryos was up to two times higher than in unpollinated ESs (10 DAAPS).
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/plants10050964/s1, Supplemetary Data 1-Primers for qPCR.
Author Contributions
M.C. conceived and designed the experiments, supervised the study, and wrote the manuscript; I.V., Y.M., V.F., and Y.G. performed the experiments; O.G. isolated maize proembryos and ESs; M.C., I.V., Y.M., and Y.G. analyzed the data; M.C., Y.G., Y.M. and I.V. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Russian Foundation for Basic Research (grant no. 18-016-00155) and by the 2021–2023 Basic Research Program of the State Academies of Sciences (no. 121031700141-7).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
We thank A.Yu. Kolesova for doing the microscopy and providing the AT-4 seeds. We thank reviewers for useful comments.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| CME | chromatin-modifying enzymes |
| DAAPS | days after the appearance of pistillate stigmas |
| DAP | days after pollination |
| ES | embryo sac |
| Fie | fertilization-independent endosperm |
| Fis | fertilization-independent seed |
| HAP | hours after pollination |
References
- Khokhlov, S.S.; Malysheva, N.A. Distribution and forms of apomixis in cereals. In Apomixis and selection; Nauka: Moscow, Russia, 1970; pp. 21–55. (in Russian) [Google Scholar]
- Nogler, G.A. Gametophytic Apomixis. In Embryology of Angiosperms; Johri, B.M., Ed.; Springer: Berlin/Heidelberg, Germany, 1984; pp. 475–518. [Google Scholar]
- Schmidt, A.; Schmid, M.W.; Grossniklaus, U. Plant germline formation: Common concepts and developmental flexibility in sexual and asexual reproduction. Development 2015, 142, 229–241. [Google Scholar] [CrossRef]
- Petrov, D. Apomixis and Its Significance for the Evolution and Selection; Nauka SB RAS: Novosibirsk, Russia, 1976. (In Russian) [Google Scholar]
- Grimanelli, D. Epigenetic regulation of reproductive development and the emergence of apomixis in angiosperms. Curr. Opin. Plant Biol. 2012, 15, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Kindiger, B.; Sokolov, V. Progress in the development of apomictic maize. Trends Agron. 1997, 1, 75–94. [Google Scholar]
- Garcia-Aguilar, M.; Michaud, C.; Leblanc, O.; Grimanelli, D. Inactivation of a DNA methylation pathway in maize reproductive organs results in apomixis-like phenotypes. Plant Cell 2010, 22, 3249–3267. [Google Scholar] [CrossRef] [PubMed]
- Belova, I.V.; Tarakanova, T.K.; Abdyrahmanova, E.A.; Sokolov, V.A.; Panikhin, P.A. Chromosome control of apomixis in maize–gamagrass hybrids. Russ. J. Genet. 2010, 46, 1055–1057. [Google Scholar] [CrossRef]
- Kindiger, B.; Bai, D.; Sokolov, V. Assignment of a gene (s) conferring apomixis in Tripsacum to a chromosome arm: Cytological and molecular evidence. Genome 1996, 39, 1133–1141. [Google Scholar] [CrossRef]
- Leblanc, O.; Pointe, C.; Hernandez, M. Cell cycle progression during endosperm development in Zea mays depends on parental dosage effects. Plant J. 2002, 32, 1057–1066. [Google Scholar] [CrossRef]
- Grimanelli, D.; García, M.; Kaszas, E.; Perotti, E.; Leblanc, O. Heterochronic expression of sexual reproductive programs during apomictic development in Tripsacum. Genetics 2003, 165, 1521–1531. [Google Scholar] [CrossRef]
- Koltunow, A.M.; Grossniklaus, U. Apomixis: A developmental perspective. Annu. Rev. Plant Biol. 2003, 54, 547–574. [Google Scholar] [CrossRef]
- Bicknell, R.A.; Koltunow, A.M. Understanding apomixis: Recent advances and remaining conundrums. Plant Cell 2004, 16, 228–246. [Google Scholar] [CrossRef]
- Bradley, E.; Carman, J.; Jamison, M.; Naumova, T. Heterochronic features of the female germline among several sexual diploid Tripsacum L. (Andropogoneae, Poaceae). Sex. Plant Reprod. 2007, 20, 9–17. [Google Scholar] [CrossRef]
- Lai, J.; Ma, J.; Swigon, Z.; Ramakrishna, W.; Linton, E.; Llaca, V.; Tanyolac, B.; Park, Y.; Jeong, O.; Bennetzen, J.L.; et al. Gene loss and movement in the maize genome. Genome Res. 2004, 14, 1924–1931. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Fu, J.; Gu, D.; Liu, W.; Liu, T.; Peng, Y.; Wang, J.; Wang, G. Genome-wide analysis of gene expression profiles during the kernel development of maize (Zea mays L.). Genomics 2008, 91, 378–387. [Google Scholar] [CrossRef] [PubMed]
- Haun, W.J.; Springer, N.M. Maternal and paternal alleles exhibit differential histone methylation and acetylation at maize imprinted genes. Plant J. 2008, 48, 903–912. [Google Scholar] [CrossRef]
- Köhler, C.; Lafon-Placette, C. Evolution and function of epigenetic processes in the endosperm. Front. Plant Sci. 2015, 6, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Eichten, S.R.; Springer, N.M. Minimal evidence for consistent changes in maize DNA methylation patterns following environmental stress. Front Plant Sci. 2015, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chettoor, A.M.; Phillips, A.R.; Clayton, C.; Dilkes, B.; Evans, M.M.S. Maternal gametophyte effects on seed development in maize. Genetics 2016, 204, 233–248. [Google Scholar] [CrossRef]
- Gehring, M.; Choi, Y.; Fischer, R.L. Imprinting and seed development. Plant Cell 2004, 16, 203–214. [Google Scholar] [CrossRef][Green Version]
- Penterman, J.; Zilberman, D.; Huh, J.H.; Ballinger, T.; Henikoff, S.; Fischer, R.L. DNA demethylation in the Arabidopsis genome. PNAS USA 2007, 104, 6752–6757. [Google Scholar] [CrossRef]
- García-Aguilar, M.; Gillmor, C.S. Zygotic genome activation and imprinting: Parent-of-origin gene regulation in plant embryogenesis. Curr. Opin. Plant Biol. 2015, 27, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Xia, H.; Zhang, Y.; Zhao, S.; Zhao, C.; Hou, L.; Li, C.; Li, A.; Ma, C.; Wang, X. Genome-wide high-resolution mapping of DNA methylation identifies epigenetic variation across embryo and endosperm in Maize (Zea may). BMC Genom. 2015, 16, 1–14. [Google Scholar] [CrossRef]
- Danilevskaya, O.N.; Hermon, P.; Hantke, S.; Muszynski, M.G.; Kollipara, K.; Ananiev, E. Duplicated fie genes in maize: Expression pattern and imprinting suggest distinct functions. Plant Cell 2003, 15, 425–438. [Google Scholar] [CrossRef]
- Hermon, P.; Kanok-orn, S.; Zou, J.; Dresselhaus, T.; Danilevskaya, O.N. Activation of the imprinted Polycomb Group Fie1 gene in maize endosperm requires demethylation of the maternal allele. Plant Mol. Biol. 2007, 64, 387–395. [Google Scholar] [CrossRef]
- Guo, M.; Rupe, M.A.; Danilevskaya, O.N.; Yang, X.; Hu, Z. Genome-wide mRNA pro ® ling reveals heterochronic allelic variation and a new imprinted gene in hybrid maize endosperm. Plant J. 2003, 36, 30–44. [Google Scholar] [CrossRef]
- Gutiérrez-Marcos, J.F.; Costa, L.M.; Biderre-Petit, C.; Khbaya, B.; O’Sullivan, D.M.; Wormald, M.; Perez, P.; Dickinson, H.G. Maternally expressed gene1 is a novel maize endosperm transfer cell-specific gene with a maternal parent-of-origin pattern of expression. Plant Cell 2004, 16, 1288–1301. [Google Scholar] [CrossRef]
- Tyrnov, V.; Enaleeva, N. Autonomous development of embryo and endosperm in maize. Dokl. Akad. Nauk 1983, 272, 722–725. (in Russian). [Google Scholar]
- Enaleeva, N.K.; Tyrnov, V.S. Cytological investigation of apomixis in AT-1 plants of corn. Maize Genet. Coop. Newsl. 1997, 71, 74–75. [Google Scholar]
- Enaleeva, N.; Ot’kalo, O.V.; Tyrnov, V.S. Phenotypic expression of the ig mutation in megagametophyte of the maize line embryonic marker. Russ. J. Genet. 1998, 34, 193–198. [Google Scholar]
- Kolesova, A.; Tyrnov, V. Embryological peculiarities of tetraploid parthenogenetic maize forms. Maize Genet. Coop. Newsl. 2012, 85, 65–66. [Google Scholar]
- Volokhina, I.; Gusev, Y.; Moiseeva, Y.; Fadeev, V.; Kolesova, A.; Gutorova, O.; Chumakov, M. Expression of genes coding for chromatin-modifying enzymes in maize embryo sacs before and after pollination. Plant Gene 2020, 22, 100221. [Google Scholar] [CrossRef]
- Wu, C.; Diggle, P.K.; Friedman, W.E. Female gametophyte development and double fertilization in Balsas teosinte, Zea mays subsp. parviglumis (Poaceae). Sex. Plant Reprod. 2011, 219–229. [Google Scholar] [CrossRef]
- Vernoud, V.; Hajduch, M.; Khaled, A.S.; Depège, N.; Rogowsky, P.M. Maize embryogenesis. Maydica 2005, 50, 469–483. [Google Scholar]
- Mòl, R.; Matthys-Rochon, E.; Dumas, C. The kinetics of cytological events during double fertilization in Zea mays L. Plant J. 1994, 5, 197–206. [Google Scholar] [CrossRef]
- Chen, J.; Strieder, N.; Krohn, N.G.; Cyprys, P.; Sprunck, S.; Engelmann, J.C.; Dresselhaus, T. Zygotic genome activation occurs shortly after fertilization in maize. Plant Cell 2017, 29, 2106–2125. [Google Scholar] [CrossRef] [PubMed]
- Coe, E.H. A Line of Maize with high haploid frequency. Am. Nat. 1959, 93, 381–382. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using realtime quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Weaver, S.; Dube, S.; Mir, A.; Qin, J.; Sun, G.; Ramakrishnan, R.; Jones, R.C.; Livak, K.J. Taking qPCR to a higher level: Analysis of CNV reveals the power of high throughput qPCR to enhance quantitative resolution. Methods 2010, 50, 271–276. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).