Genome-Wide Association and Prediction of Male and Female Floral Hybrid Potential Traits in Elite Spring Bread Wheat Genotypes
Abstract
1. Introduction
2. Results
2.1. Population Structure and Linkage Disequilibrium
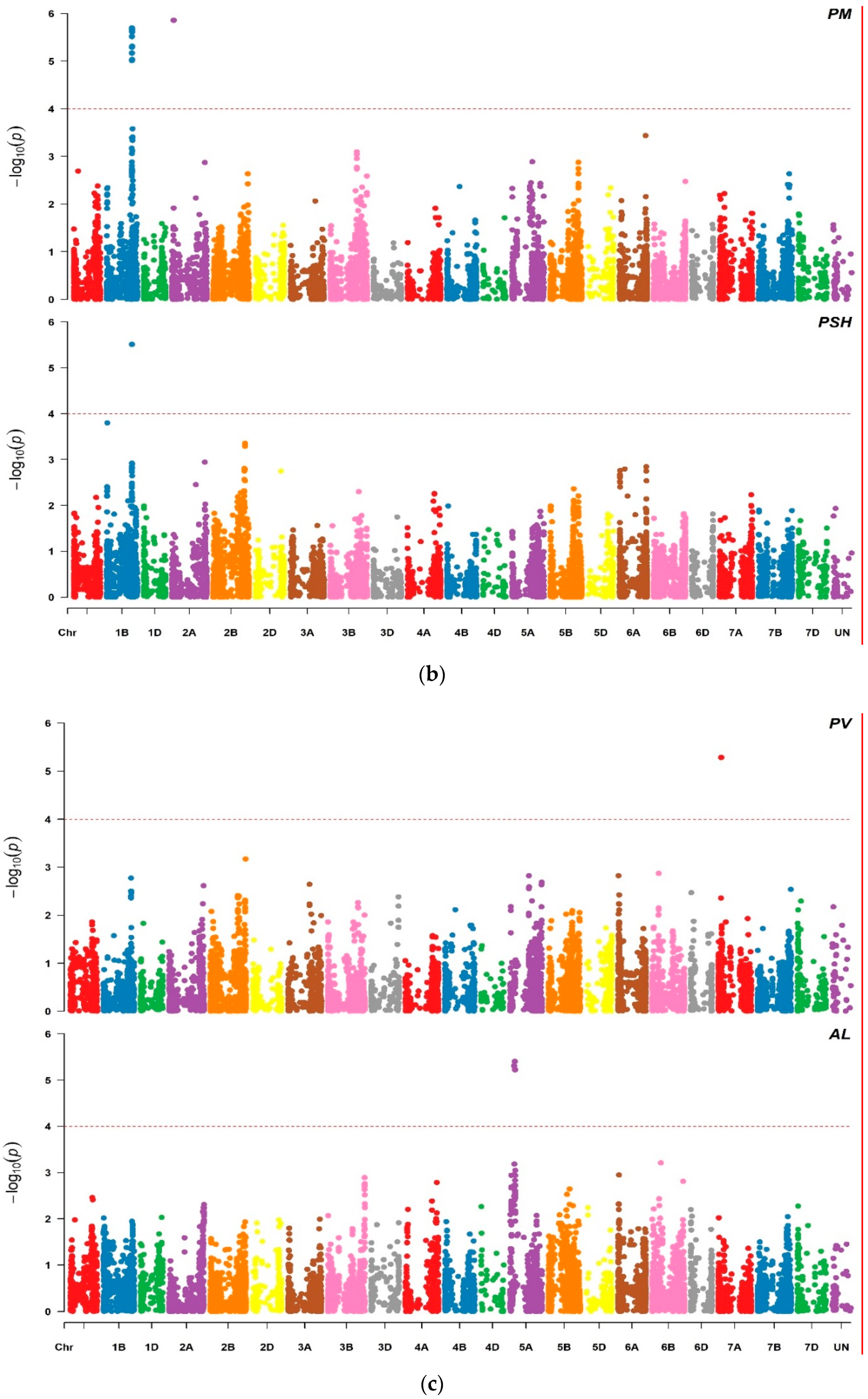
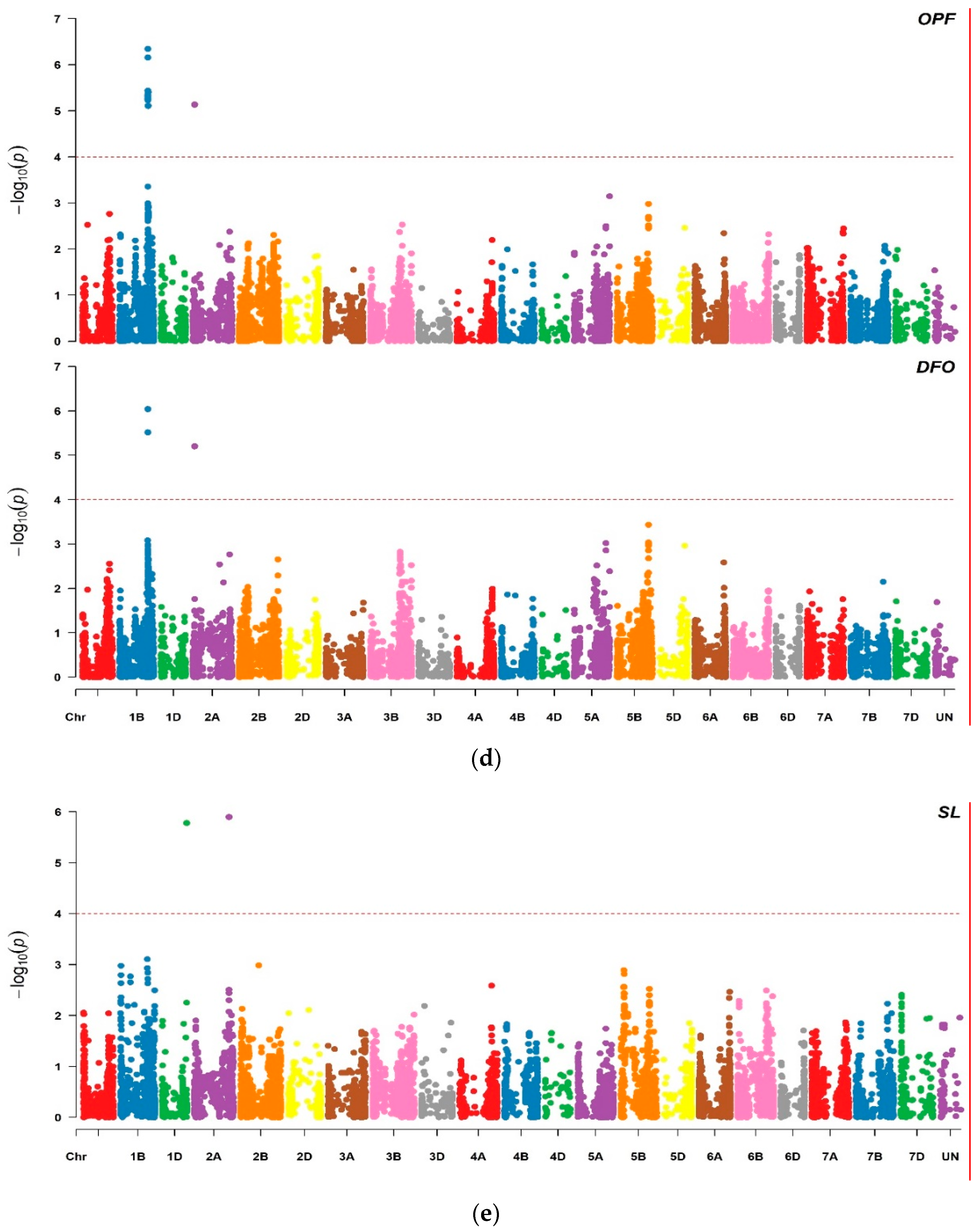
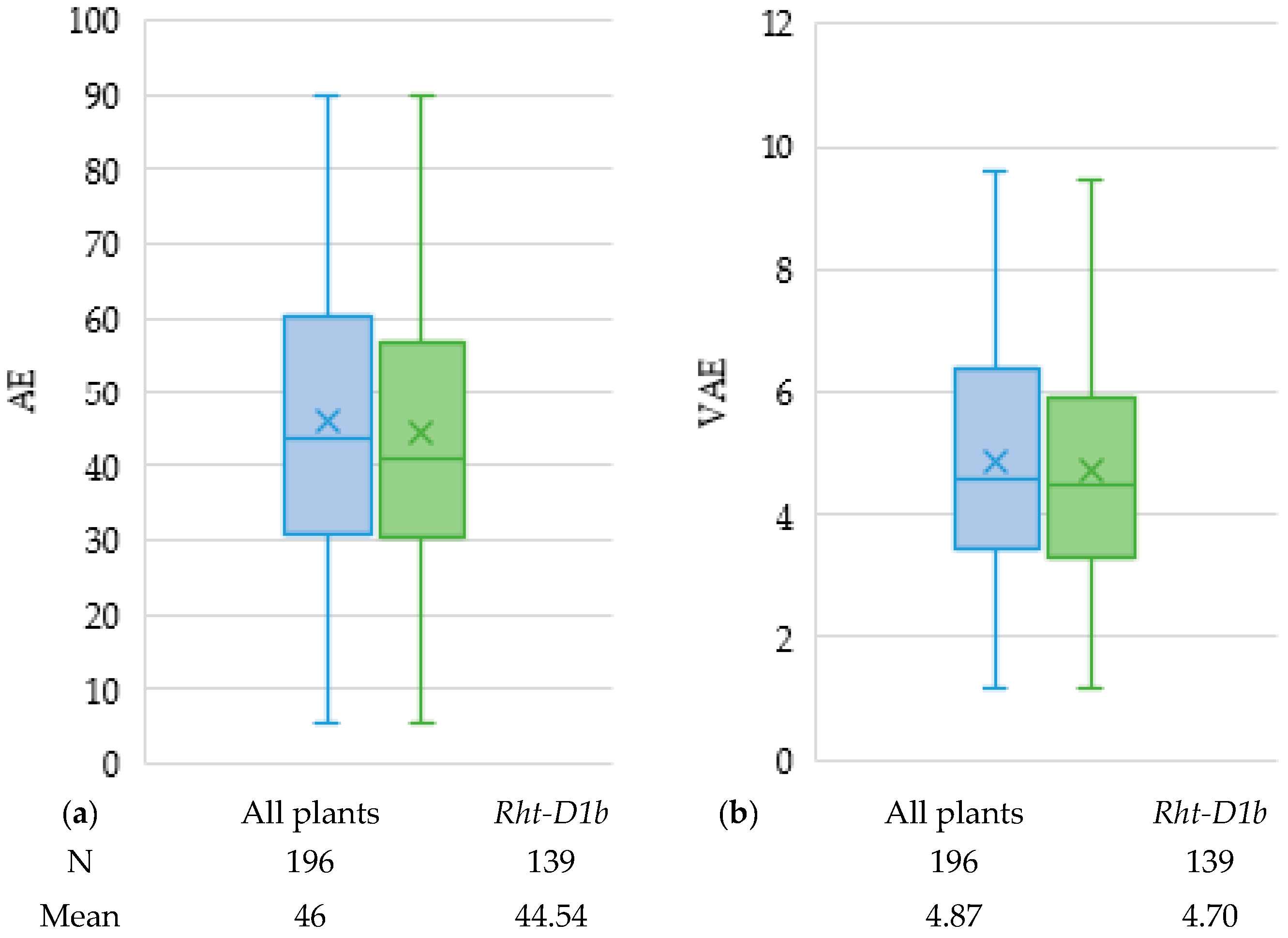
2.2. Marker Trait Associations
2.3. Genome-Wide Prediction of the Potential Floral Traits
3. Discussion
3.1. Marker Traits Associations
3.2. Genome-Wide Prediction of Hybrid Floral Traits
4. Materials and Methods
4.1. Plant Materials and Phenotyping
4.2. Genotyping by SNP Markers
4.3. Population Structure and Linkage Disequilibrium
4.4. Association Mapping
4.5. Genomic Prediction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 24 January 2021).
- Kempe, K.; Rubtsova, M.; Gils, M. Split-Gene System for Hybrid Wheat Seed Production. Proc. Natl. Acad. Sci. USA 2014, 111, 9097–9102. [Google Scholar] [CrossRef] [PubMed]
- Easterly, A.C.; Garst, N.; Belamkar, V.; Ibrahim, A.M.H.; Rudd, J.C.; Sarazin, J.; Baenziger, P.S. Evaluation of Hybrid Wheat Yield in Nebraska. Crop Sci. 2020, 60, 1210–1222. [Google Scholar] [CrossRef]
- El Hanafi, S.; Bendaou, N.; Kehel, Z.; Sanchez-Garcia, M.; Tadesse, W. Phenotypic Evaluation of Elite Spring Bread Wheat Genotypes for Hybrid Potential Traits. Euphytica 2020, 216, 168. [Google Scholar] [CrossRef]
- Boeven, P.H.G.; Longin, C.F.H.; Leiser, W.L.; Kollers, S.; Ebmeyer, E.; Würschum, T. Genetic Architecture of Male Floral Traits Required for Hybrid Wheat Breeding. Theor. Appl. Genet. 2016, 129, 2343–2357. [Google Scholar] [CrossRef]
- Beri, S.M.; Anand, S.C. Factors Affecting Pollen Shedding Capacity in Wheat. Euphytica 1971, 20, 327–332. [Google Scholar] [CrossRef]
- Lu, Q.; Lillemo, M.; Skinnes, H.; He, X.; Shi, J.; Ji, F.; Dong, Y.; Bjørnstad, Å. Anther Extrusion and Plant Height Are Associated with Type I Resistance to Fusarium Head Blight in Bread Wheat Line “Shanghai-3/Catbird”. Theor. Appl. Genet. 2013, 126, 317–334. [Google Scholar] [CrossRef]
- He, X.; Singh, P.K.; Dreisigacker, S.; Singh, S.; Lillemo, M.; Duveiller, E. Dwarfing Genes Rht-B1b and Rht-D1b Are Associated with Both Type I FHB Susceptibility and Low Anther Extrusion in Two Bread Wheat Populations. PLoS ONE 2016, 11, e0162499. [Google Scholar] [CrossRef] [PubMed]
- Crossa, J.; Pérez-Rodríguez, P.; Cuevas, J.; Montesinos-López, O.; Jarquín, D.; de Los Campos, G.; Burgueño, J.; González-Camacho, J.M.; Pérez-Elizalde, S.; Beyene, Y.; et al. Genomic Selection in Plant Breeding: Methods, Models, and Perspectives. Trends Plant Sci. 2017, 22, 961–975. [Google Scholar] [CrossRef]
- Hickey, J.M.; Chiurugwi, T.; Mackay, I.; Powell, W. Genomic Prediction Unifies Animal and Plant Breeding Programs to Form Platforms for Biological Discovery. Nat. Genet. 2017, 49, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Muqaddasi, Q.H.; Reif, J.C.; Li, Z.; Basnet, B.R.; Dreisigacker, S.; Röder, M.S. Genome-Wide Association Mapping and Genome-Wide Prediction of Anther Extrusion in CIMMYT Spring Wheat. Euphytica 2017, 213, 73. [Google Scholar] [CrossRef]
- Adhikari, A.; Basnet, B.R.; Crossa, J.; Dreisigacker, S.; Camarillo, F.; Bhati, P.K.; Jarquin, D.; Manes, Y.; Ibrahim, A.M.H. Genome-Wide Association Mapping and Genomic Prediction of Anther Extrusion in CIMMYT Hybrid Wheat Breeding Program via Modeling Pedigree, Genomic Relationship, and Interaction with the Environment. Front. Genet. 2020, 11, 586687. [Google Scholar] [CrossRef]
- Muqaddasi, Q.H.; Reif, J.C.; Röder, M.S.; Basnet, B.R.; Dreisigacker, S. Genetic Mapping Reveals Large-Effect QTL for Anther Extrusion in CIMMYT Spring Wheat. Agronomy 2019, 9, 407. [Google Scholar] [CrossRef]
- Song, X.; Feng, J.; Cui, Z.; Zhang, C.; Sun, D. Genome-Wide Association Study for Anther Length in Some Elite Bread Wheat Germplasm. Czech J. Genet. Plant Breed. 2018, 54, 109–114. [Google Scholar] [CrossRef]
- Muqaddasi, Q.H.; Pillen, K.; Plieske, J.; Ganal, M.W.; Röder, M.S. Genetic and Physical Mapping of Anther Extrusion in Elite European Winter Wheat. PLoS ONE 2017, 12, e0187744. [Google Scholar] [CrossRef] [PubMed]
- Muqaddasi, Q.H.; Lohwasser, U.; Nagel, M.; Börner, A.; Pillen, K.; Röder, M.S. Genome-Wide Association Mapping of Anther Extrusion in Hexaploid Spring Wheat. PLoS ONE 2016, 11, e0155494. [Google Scholar] [CrossRef] [PubMed]
- Tadesse, W.; Suleiman, S.; Tahir, I.; Sanchez-Garcia, M.; Jighly, A.; Hagras, A.; Thabet, S.; Baum, M. Heat-Tolerant QTLs Associated with Grain Yield and Its Components in Spring Bread Wheat under Heat-Stressed Environments of Sudan and Egypt. Crop Sci. 2019, 59, 199–211. [Google Scholar] [CrossRef]
- El Hanafi, S.; Backhaus, A.; Bendaou, N.; Sanchez-Garcia, M.; Al-Abdallat, A.; Tadesse, W. Genome-Wide Association Study for Adult Plant Resistance to Yellow Rust in Spring Bread Wheat (Triticum Aestivum L.). Euphytica 2021, 217, 87. [Google Scholar] [CrossRef]
- Skinnes, H.; Semagn, K.; Tarkegne, Y.; Marøy, A.G.; Bjørnstad, Å. The Inheritance of Anther Extrusion in Hexaploid Wheat and Its Relationship to Fusarium Head Blight Resistance and Deoxynivalenol Content. Plant Breed. 2010, 129, 149–155. [Google Scholar] [CrossRef]
- Buerstmayr, M.; Buerstmayr, H. Comparative Mapping of Quantitative Trait Loci for Fusarium Head Blight Resistance and Anther Retention in the Winter Wheat Population Capo × Arina. Theor. Appl. Genet. 2015, 128, 1519–1530. [Google Scholar] [CrossRef]
- Okada, T.; Jayasinghe, J.E.A.R.M.; Eckermann, P.; Watson-Haigh, N.S.; Warner, P.; Hendrikse, Y.; Baes, M.; Tucker, E.J.; Laga, H.; Kato, K.; et al. Effects of Rht-B1 and Ppd-D1 Loci on Pollinator Traits in Wheat. Theor. Appl. Genet. 2019, 132, 1965–1979. [Google Scholar] [CrossRef]
- Ollier, M.; Talle, V.; Brisset, A.L.; Le Bihan, Z.; Duerr, S.; Lemmens, M.; Goudemand, E.; Robert, O.; Hilbert, J.L.; Buerstmayr, H. QTL Mapping and Successful Introgression of the Spring Wheat-Derived QTL Fhb1 for Fusarium Head Blight Resistance in Three European Triticale Populations. Theor. Appl. Genet. 2020, 133, 457–477. [Google Scholar] [CrossRef] [PubMed]
- Langer, S.M.; Longin, C.F.H.; Würschum, T. Phenotypic Evaluation of Floral and Flowering Traits with Relevance for Hybrid Breeding in Wheat (Triticum Aestivum L.). Plant Breed. 2014, 133, 433–441. [Google Scholar] [CrossRef]
- Li, Y.; Xiao, J.; Wu, J.; Duan, J.; Liu, Y.; Ye, X.; Zhang, X.; Guo, X.; Gu, Y.; Zhang, L.; et al. A Tandem Segmental Duplication (TSD) in Green Revolution Gene Rht-D1b Region Underlies Plant Height Variation. New Phytol. 2012, 196, 282–291. [Google Scholar] [CrossRef]
- Miedaner, T.; Würschum, T.; Maurer, H.P.; Korzun, V.; Ebmeyer, E.; Reif, J.C. Association Mapping for Fusarium Head Blight Resistance in European Soft Winter Wheat. Mol. Breed. 2011, 28, 647–655. [Google Scholar] [CrossRef]
- ImageJ. Available online: https://imagej.nih.gov/ij/index.html (accessed on 30 March 2021).
- Ogbonnaya, F.C.; Seah, S.; Delibes, A.; Jahier, J.; López-Braña, I.; Eastwood, R.F.; Lagudah, E.S. Molecular-Genetic Characterisation of a New Nematode Resistance Gene in Wheat. Theor. Appl. Genet. 2001, 102, 623–629. [Google Scholar] [CrossRef]
- Würschum, T.; Liu, G.; Boeven, P.H.G.; Longin, C.F.H.; Mirdita, V.; Kazman, E.; Zhao, Y.; Reif, J.C. Exploiting the Rht Portfolio for Hybrid Wheat Breeding. Theor. Appl. Genet. 2018, 131, 1433–1442. [Google Scholar] [CrossRef]
- Okada, T.; Jayasinghe, J.E.A.R.M.; Eckermann, P.; Watson-Haigh, N.S.; Warner, P.; Williams, M.E.; Albertsen, M.C.; Baumann, U.; Whitford, R. Genetic Factors Associated with Favourable Pollinator Traits in the Wheat Cultivar Piko. Funct. Plant Biol. FPB 2020. [Google Scholar] [CrossRef]
- Cheng, H.; Qin, L.; Lee, S.; Fu, X.; Richards, D.E.; Cao, D.; Luo, D.; Harberd, N.P.; Peng, J. Gibberellin Regulates Arabidopsis Floral Development via Suppression of DELLA Protein Function. Development 2004, 131, 1055–1064. [Google Scholar] [CrossRef]
- Xu, K.; He, X.; Dreisigacker, S.; He, Z.; Singh, P.K. Anther Extrusion and Its Association with Fusarium Head Blight in CIMMYT Wheat Germplasm. Agronomy 2019, 10, 47. [Google Scholar] [CrossRef]
- Kubo, K.; Fujita, M.; Kawada, N.; Nakajima, T.; Nakamura, K.; Maejima, H.; Ushiyama, T.; Hatta, K.; Matsunaka, H. Minor Differences in Anther Extrusion Affect Resistance to Fusarium Head Blight in Wheat. J. Phytopathol. 2013, 161, 308–314. [Google Scholar] [CrossRef]
- Liu, S.; Anderson, J.A. Marker Assisted Evaluation of Fusarium Head Blight Resistant Wheat Germplasm. Crop Sci. 2003, 43, 760–766. [Google Scholar] [CrossRef]
- Impe, D.; Reitz, J.; Köpnick, C.; Rolletschek, H.; Börner, A.; Senula, A.; Nagel, M. Assessment of Pollen Viability for Wheat. Front. Plant Sci. 2020, 10, 1588. [Google Scholar] [CrossRef]
- De Vries, A.P. Some Aspects of Cross-Pollination in Wheat (Triticum Aestivum L.). 3. Anther Length and Number of Pollen Grains per Anther. Euphytica 1974, 23, 11–19. [Google Scholar] [CrossRef]
- Guo, Z.; Chen, D.; Schnurbusch, T. Variance Components, Heritability and Correlation Analysis of Anther and Ovary Size during the Floral Development of Bread Wheat. J. Exp. Bot. 2015, 66, 3099–3111. [Google Scholar] [CrossRef]
- Nguyen, V.; Fleury, D.; Timmins, A.; Laga, H.; Hayden, M.; Mather, D.; Okada, T. Addition of Rye Chromosome 4R to Wheat Increases Anther Length and Pollen Grain Number. Theor. Appl. Genet. 2015, 128, 953–964. [Google Scholar] [CrossRef]
- Kubo, K.; Kawada, N.; Fujita, M.; Hatta, K.; Shunsuke, O.; Nakajima, T. Effect of Cleistogamy on Fusarium Head Blight Resistance in Wheat. Breed. Sci. 2010, 60, 405–411. [Google Scholar] [CrossRef]
- Gilsinger, J.; Kong, L.; Shen, X.; Ohm, H. DNA Markers Associated with Low Fusarium Head Blight Incidence and Narrow Flower Opening in Wheat. Theor. Appl. Genet. 2005, 110, 1218–1225. [Google Scholar] [CrossRef]
- Sarinelli, J.M.; Murphy, J.P.; Tyagi, P.; Holland, J.B.; Johnson, J.W.; Mergoum, M.; Mason, R.E.; Babar, A.; Harrison, S.; Sutton, R.; et al. Training Population Selection and Use of Fixed Effects to Optimize Genomic Predictions in a Historical USA Winter Wheat Panel. Theor. Appl. Genet. 2019, 132, 1247–1261. [Google Scholar] [CrossRef]
- Joppa, L.R.; McNeal, F.H.; Welsh, J.R. Pollen and Anther Development in Cytoplasmic Male Sterile Wheat, (Triticum Aestivum L.) 1. Crop Sci. 1966, 6, 296–297. [Google Scholar] [CrossRef]
- SGS TraitGenetics. Available online: http://www.traitgenetics.com/ (accessed on 30 March 2021).
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for Association Mapping of Complex Traits in Diverse Samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Bi, I.V.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A Unified Mixed-Model Method for Association Mapping that Accounts for Multiple Levels of Relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2015; Available online: https://www.R-project.org/ (accessed on 28 February 2021).
- Endelman, J.B.; Jannink, J.L. Shrinkage Estimation of the Realized Relationship Matrix. G3 Genes Genomes Genet. 2012, 2, 1405–1413. [Google Scholar] [CrossRef] [PubMed]
- Endelman, J.B. Ridge Regression and Other Kernels for Genomic Selection with R Package rrBLUP. Plant Genome 2011, 4, 250–255. [Google Scholar] [CrossRef]
- Butler, D. Asreml: Asreml Fits the Linear Mixed Model; R Package Version 3.0. 2009. Available online: www.vsni.co.uk (accessed on 28 January 2021).
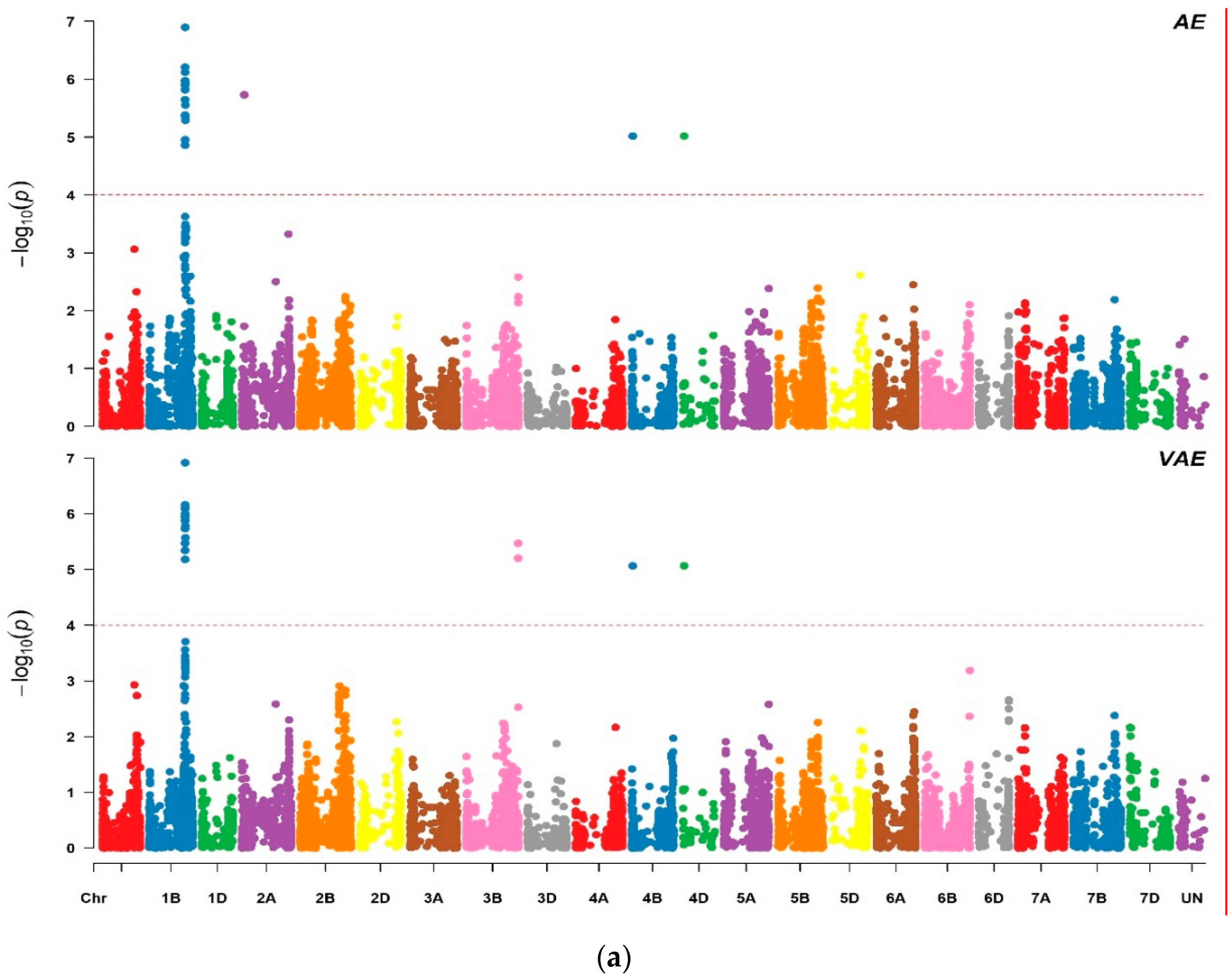
| Marker | Chr | Position (Mbp) a | AE | VAE | PM | OPF | DFO | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −Log10 (p) | Effect | %R2 b | −Log10 (p) | Effect | %R2 | −Log10 (p) | Effect | %R2 | −Log10 (p) | Effect | %R2 | −Log10 (p) | Effect | %R2 | |||
| Rht-D1b | 4D | - | 5.02 | 0.18 | 20.58 | 5.07 | 0.05 | 17.34 | |||||||||
| wsnp_Ex_rep_c103167_88182254 | 2A | 33.04 | 5.73 | −11.40 | 26.08 | 5.86 | −3.03 | 23.76 | 5.13 | −3.03 | 24.07 | 5.20 | −10.1 | 18.7 | |||
| Tdurum_contig30517_578 | 1B | 566.84 | 5.37 | 12.88 | 25.34 | 5.18 | 1.22 | 20.95 | 5.01 | 3.54 | 21.98 | ||||||
| Tdurum_contig30517_310 | 1B | 566.84 | 5.65 | 13.51 | 25.87 | 5.34 | 1.26 | 21.16 | 5.30 | 3.75 | 24.24 | ||||||
| BS00062724_51 | 1B | 567.71 | 6.13 | 14.17 | 26.21 | 6.10 | 1.38 | 22.39 | 5.64 | 3.69 | 22.79 | ||||||
| Kukri_rep_c103359_233 | 1B | 568.52 | 6.90 | 13.57 | 26.28 | 5.88 | 1.32 | 22.33 | 5.65 | 3.64 | 22.99 | 6.34 | 3.64 | 24.38 | 6.04 | 11.37 | 18.68 |
| wsnp_Ex_c13109_20725640 | 1B | 568.53 | 4.86 | −14.10 | 26.53 | 5.96 | −1.37 | 22.52 | 5.67 | −3.60 | 23.24 | 6.16 | −3.60 | 24.00 | |||
| wsnp_Ex_c23235_32471767 | 1B | 568.57 | 6.21 | 14.35 | 26.93 | 6.16 | 1.39 | 22.93 | 5.70 | 3.74 | 23.28 | 5.44 | 3.74 | 24.53 | |||
| Excalibur_c10496_651 | 1B | 568.97 | 5.38 | −12.80 | 24.75 | 5.74 | −1.33 | 21.64 | 5.17 | −3.01 | 21.88 | ||||||
| wsnp_Ex_c23230_32466568 | 1B | 568.97 | 4.96 | 12.68 | 25.56 | 5.48 | 1.23 | 21.62 | 5.30 | 3.37 | 22.39 | ||||||
| wsnp_Ex_rep_c107911_91350866 | 1B | 570.28 | 5.97 | 13.73 | 26.52 | 6.92 | 1.33 | 22.44 | 5.66 | 3.63 | 23.03 | 5.31 | 3.63 | 24.24 | |||
| wsnp_Ex_rep_c107911_91350930 | 1B | 570.28 | 5.90 | 13.57 | 26.28 | 5.88 | 1.32 | 22.33 | 5.65 | 3.64 | 22.99 | 5.34 | 3.64 | 24.38 | 5.52 | 11.36 | 18.67 |
| wsnp_Ex_c21198_30327016 | 1B | 570.29 | 5.98 | 13.79 | 26.47 | 6.00 | 1.35 | 22.66 | 5.52 | 3.61 | 22.98 | 5.27 | 3.61 | 24.24 | |||
| wsnp_Ku_rep_c89089_82009060 | 1B | 570.30 | 5.82 | −13.50 | 25.37 | 5.93 | −1.34 | 21.90 | 5.23 | −3.60 | 23.48 | ||||||
| IAAV3802 | 1B | 571.06 | 5.56 | −13.30 | 25.42 | 4.79 | −1.30 | 21.73 | 5.31 | −3.52 | 22.14 | 5.41 | −3.52 | 23.57 | |||
| wsnp_Ku_c13043_20902807 | 1B | 571.06 | 5.29 | 12.56 | 24.50 | 5.57 | 1.29 | 21.16 | 5.03 | 3.02 | 21.49 | ||||||
| Ex_c11930_509 | 1B | 571.18 | 5.93 | −3.52 | 24.10 | 5.98 | −1.36 | 22.67 | 5.62 | −3.52 | 23.22 | 5.11 | −3.52 | 24.10 | |||
| Excalibur_c20309_539 | 3B | 819.86 | 5.47 | 1.55 | 21.16 | ||||||||||||
| Kukri_rep_c110544_248 | 3B | 819.89 | 5.20 | 1.54 | 21.09 | ||||||||||||
| Marker | Trait | Chr | Physical Position (Mbp) a | −Log10 (p) | Effect | %R2 b |
|---|---|---|---|---|---|---|
| Excalibur_c10496_651 | PSH | 1B | 568.97 | 5.51 | −1.04 | 22.10 |
| BS00063511_51 | SL | 1D | 485.71 | 5.78 | 0.39 | 8.01 |
| Kukri_c62142_683 | SL | 2A | 692.64 | 5.90 | 0.24 | 9.55 |
| BS00064387_51 | AL | 5A | 87.40 | 5.30 | 0.20 | 17.86 |
| wsnp_Ra_c10053_16636851 | AL | 5A | 99.02 | 4.40 | 0.20 | 17.86 |
| wsnp_Ku_c328_679106 | AL | 5A | 104.23 | 5.22 | 0.20 | 17.86 |
| Excalibur_rep_c109881_701 | PV | 7A | 65.97 | 5.29 | 5.40 | 19.35 |
| Trait | h2 | PA | Pacc |
|---|---|---|---|
| VAE | 0.21 | 0.43 | 0.92 |
| PSH | 0.24 | 0.42 | 0.86 |
| AX | 0.28 | 0.45 | 0.85 |
| DFO | 0.2 | 0.38 | 0.85 |
| AX | 0.28 | 0.45 | 0.85 |
| PM | 0.24 | 0.41 | 0.83 |
| OPF | 0.27 | 0.39 | 0.76 |
| AL | 0.3 | 0.4 | 0.73 |
| SL | 0.08 | 0.14 | 0.52 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El Hanafi, S.; Cherkaoui, S.; Kehel, Z.; Al-Abdallat, A.; Tadesse, W. Genome-Wide Association and Prediction of Male and Female Floral Hybrid Potential Traits in Elite Spring Bread Wheat Genotypes. Plants 2021, 10, 895. https://doi.org/10.3390/plants10050895
El Hanafi S, Cherkaoui S, Kehel Z, Al-Abdallat A, Tadesse W. Genome-Wide Association and Prediction of Male and Female Floral Hybrid Potential Traits in Elite Spring Bread Wheat Genotypes. Plants. 2021; 10(5):895. https://doi.org/10.3390/plants10050895
Chicago/Turabian StyleEl Hanafi, Samira, Souad Cherkaoui, Zakaria Kehel, Ayed Al-Abdallat, and Wuletaw Tadesse. 2021. "Genome-Wide Association and Prediction of Male and Female Floral Hybrid Potential Traits in Elite Spring Bread Wheat Genotypes" Plants 10, no. 5: 895. https://doi.org/10.3390/plants10050895
APA StyleEl Hanafi, S., Cherkaoui, S., Kehel, Z., Al-Abdallat, A., & Tadesse, W. (2021). Genome-Wide Association and Prediction of Male and Female Floral Hybrid Potential Traits in Elite Spring Bread Wheat Genotypes. Plants, 10(5), 895. https://doi.org/10.3390/plants10050895







