Genome-Wide Analysis and Expression Profiles of the Dof Family in Cleistogenes songorica under Temperature, Salt and ABA Treatment
Abstract
1. Introduction
2. Results
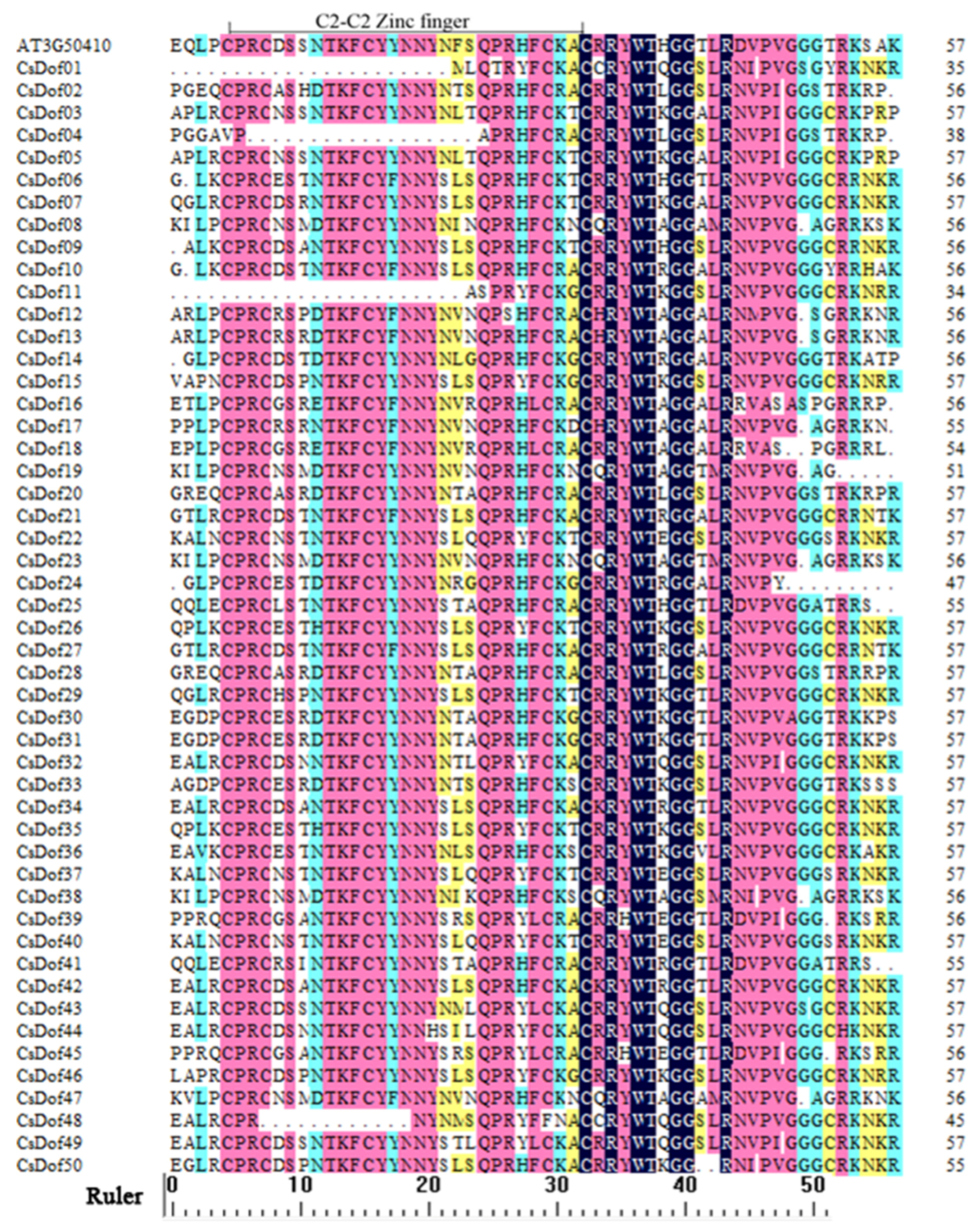
2.1. Identification and Structure Analysis of Dof Genes
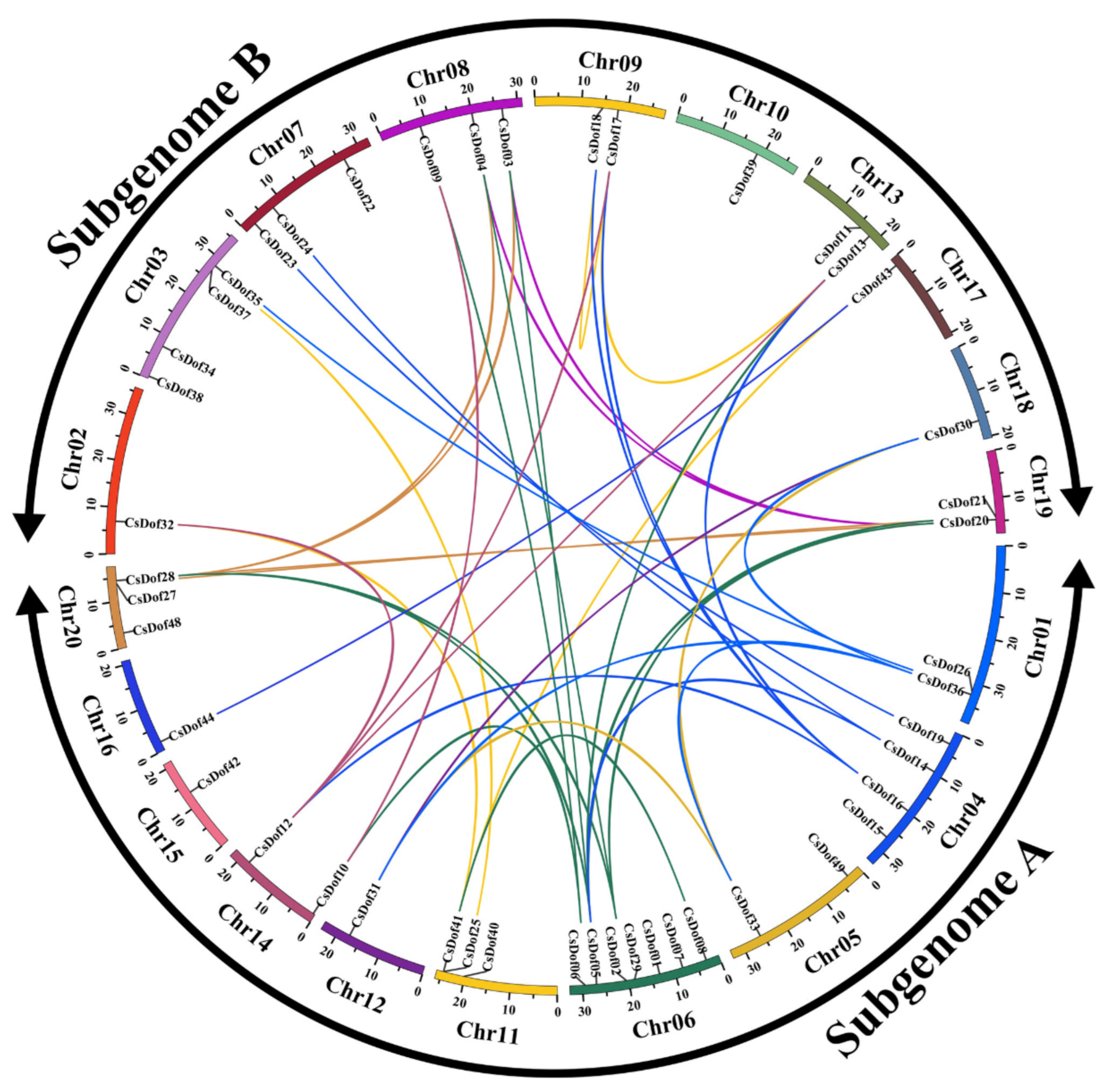
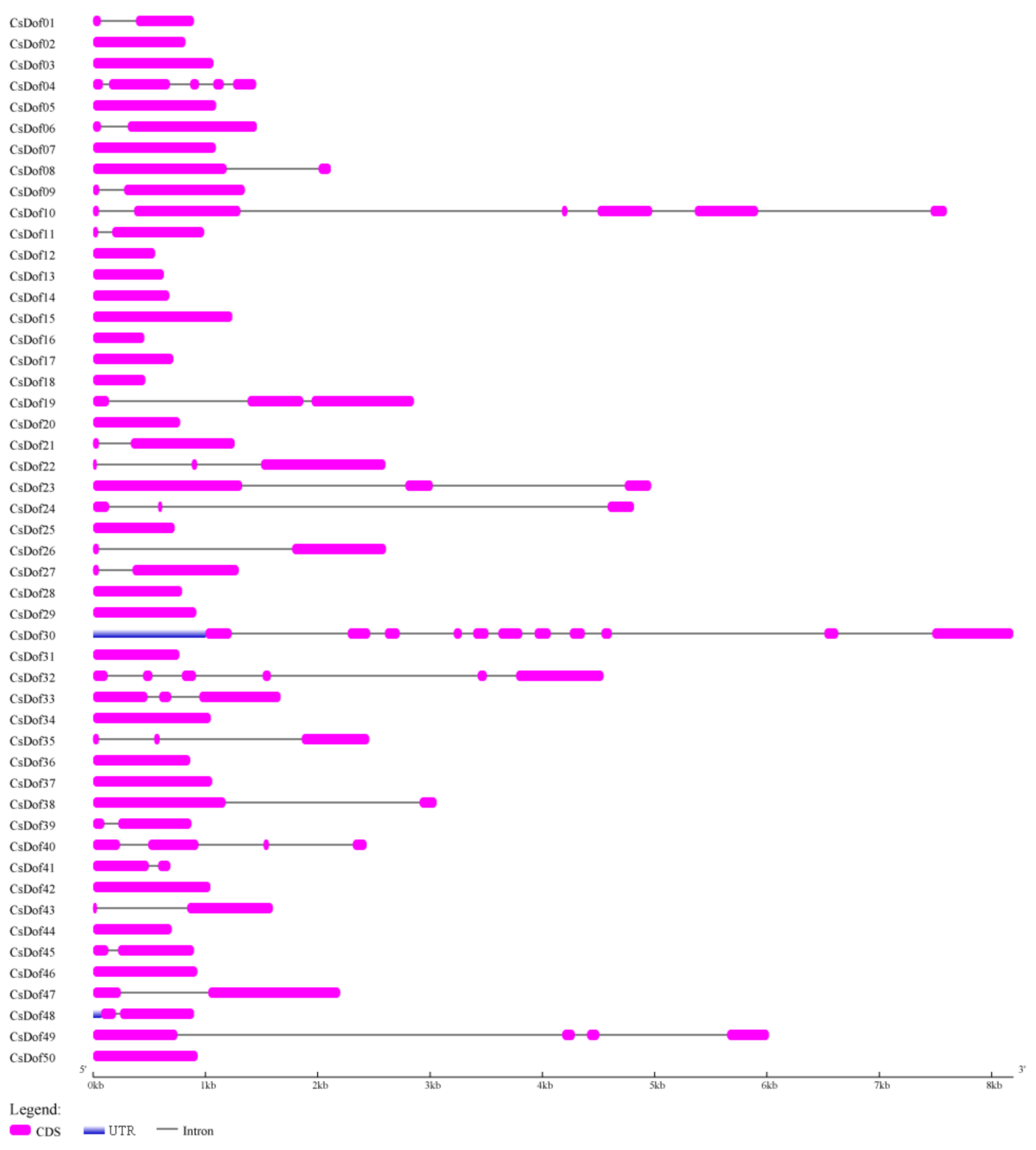
2.2. Chromosomal Localization, Gene Duplication, and Gene Structure of CsDof Gene
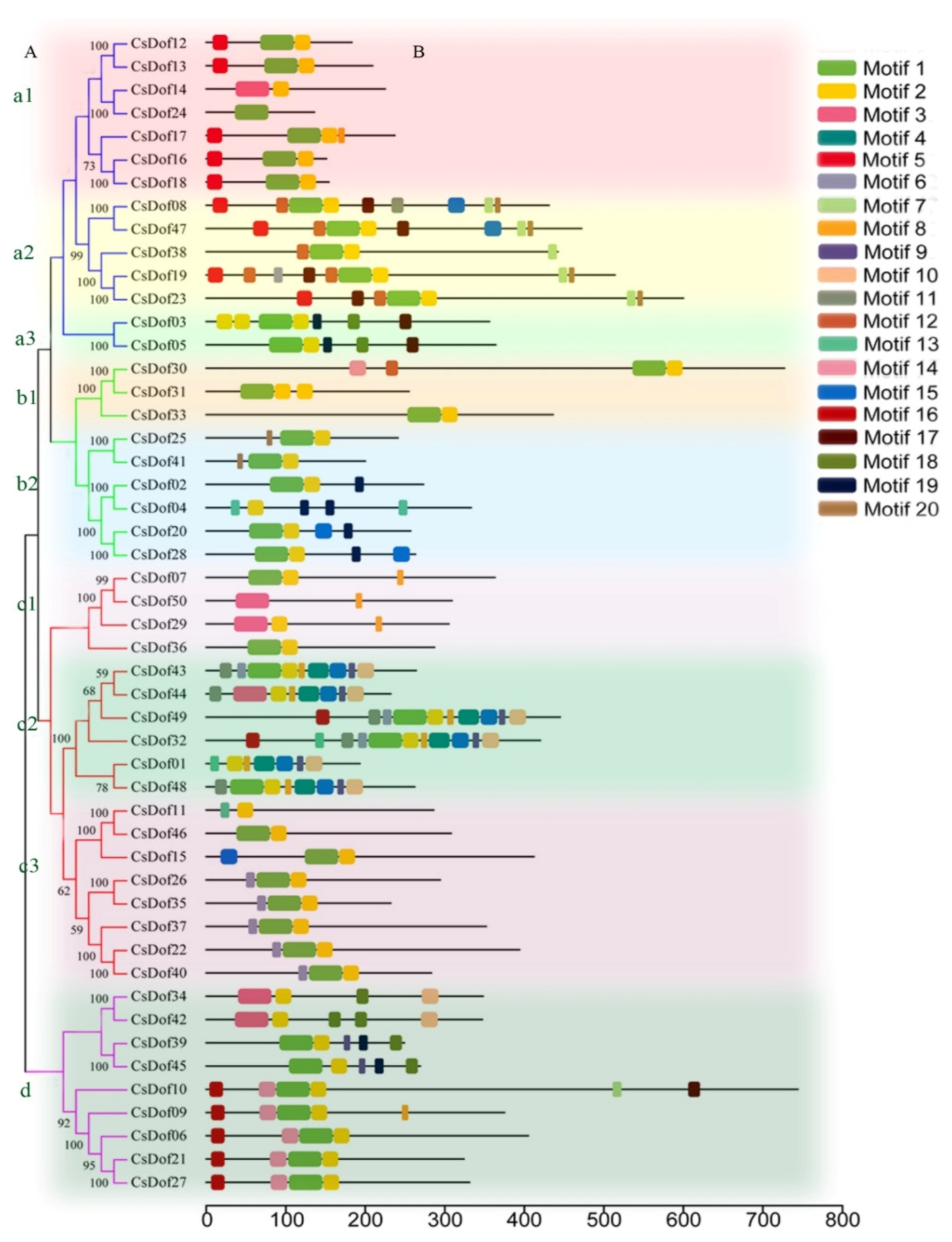
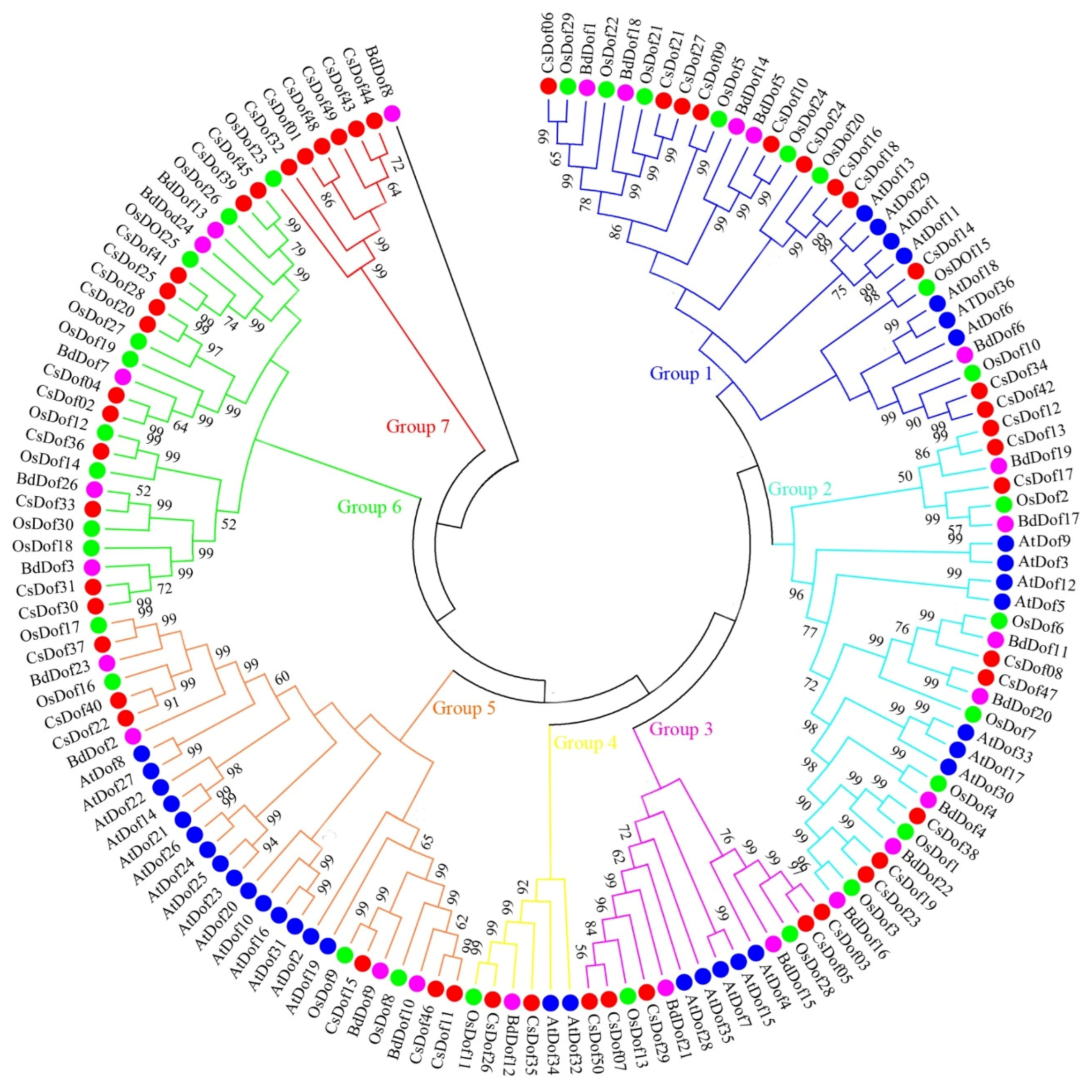
2.3. Conserved Motif, Phylogenetic Analysis, and Classification of the Dof Transcription Factor Family
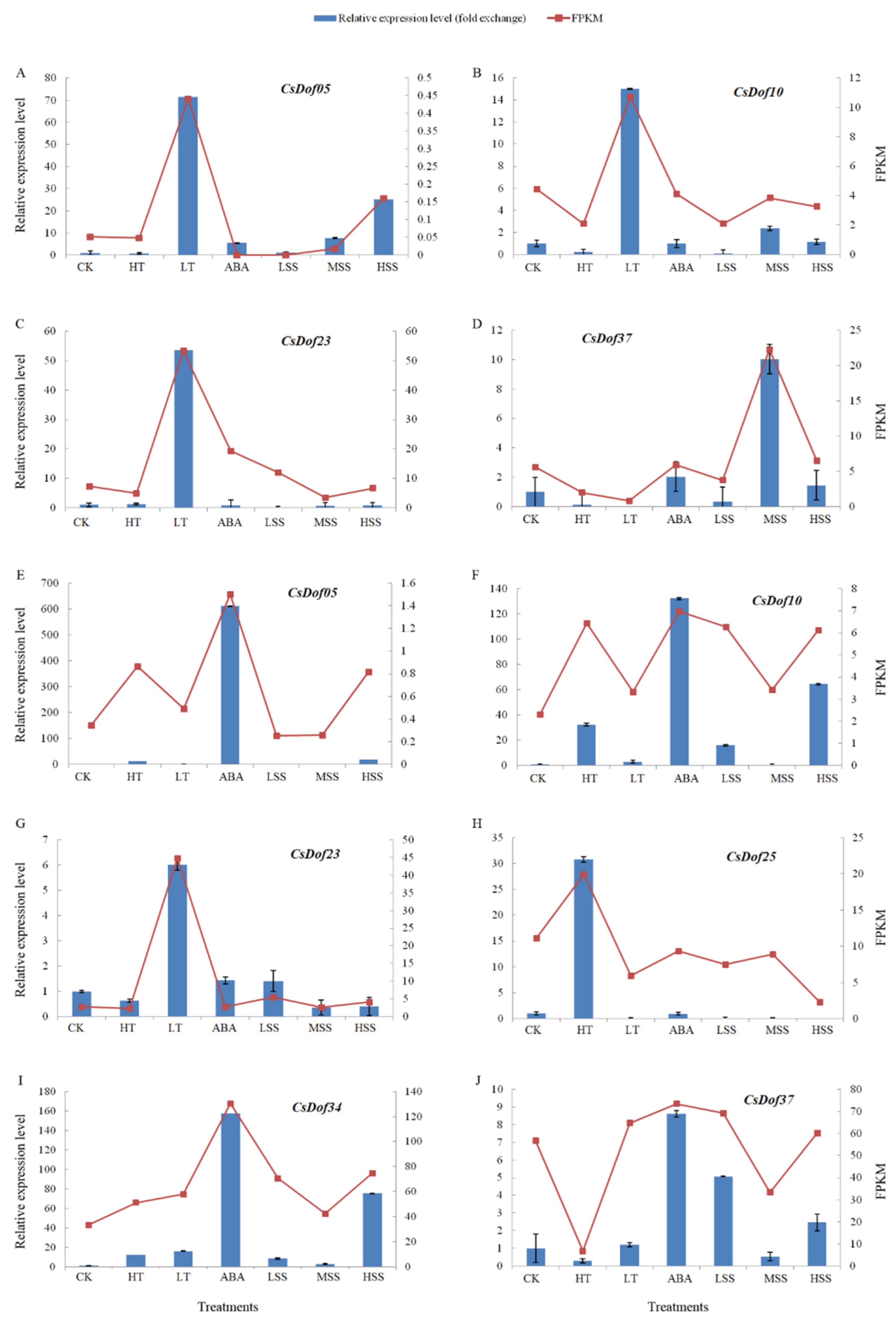
2.4. Expression Profiles of Dof Genes in C. songorica
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Stress Treatments
4.2. Identification and Structural Analysis of Dof Genes in C. songorica
4.3. Chromosomal Location and Gene Structure Analysis
4.4. Conserved Motif and Phylogenetic Analysis
4.5. Gene Duplication and Syntenic Analysis
4.6. RNA-Seq and qPCR Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Qu, L.-J.; Zhu, Y.-X. Transcription factor families in Arabidopsis: Major progress and outstanding issues for future research-Commentary. Curr. Opin. Plant Biol. 2006, 9, 544–549. [Google Scholar] [CrossRef]
- Yu, Q.; Li, C.; Zhang, J.; Tian, Y.; Wang, H.; Zhang, Y.; Zhang, Z.; Xiang, Q.; Han, X.; Zhang, L. Genome-wide identification and expression analysis of the Dof gene family under drought stress in tea (Camellia sinensis). PeerJ 2020, 8, e926. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Wu, F.; Ma, T.; Zong, X.; Ma, Q.; Li, J.; Zhao, Y.; Wang, Y.; Zhang, J. Comprehensive analysis of bZIP transcription factors uncovers their roles during dimorphic floret differentiation and stress response in Cleistogenes songorica. BMC Genom. 2019, 20, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Wasserman, W.W.; Sandelin, A. Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 2004, 5, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, S. Dof DNA-binding domains of plant transcription factors contribute to multiple protein-protein interactions. Eur. J. Biochem. 1997, 250, 403–410. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Ohtsuki, N.; Tsujimoto, Y.; Fujimura, T.; Shinshi, H. Identification of genes of the plant-specific transcription-factor families cooperatively regulated by ethylene and jasmonate in Arabidopsis thaliana. J. Plant Res. 2006, 119, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, S. The Dof family of plant transcription factors. Trends Plant Sci. 2002, 7, 555–560. [Google Scholar] [CrossRef]
- Wei, Q.; Wang, W.; Hu, T.; Hu, H.; Bao, C. Genome-wide identification and characterization of Dof transcription factors in eggplant (Solanum melongena L.). PeerJ 2018, 6, e4481. [Google Scholar] [CrossRef]
- Wen, C.L.; Cheng, Q.; Zhao, L.; Mao, A.; Yang, J.; Yu, S.; Weng, Y.; Xu, Y. Identification and characterisation of Dof transcription factors in the cucumber genome. Sci. Rep. 2016, 6, 23072. [Google Scholar] [CrossRef]
- Chen, M.; Liu, X.; Huan, L.; Sun, M.; Liu, L.; Chen, X.; Gao, D.; Li, L. Genome-wide analysis of Dof family genes and their expression during bud dormancy in peach (Prunus persica). Sci. Hortic. 2017, 214, 18–26. [Google Scholar] [CrossRef]
- Zou, Z.; Zhang, X. Genome-wide identification and comparative evolutionary analysis of the Dof transcription factor family in physic nut and castor bean. PeerJ 2019, 7, e6354. [Google Scholar] [CrossRef]
- Yanagisawa, S. Dof Domain Proteins: Plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol. 2004, 45, 386–391. [Google Scholar] [CrossRef]
- Lijavetzky, D.; Carbonero, P.; Vicente-Carbajosa, J. Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol. Biol. 2003, 3, 17. [Google Scholar] [CrossRef]
- Sara, H.A.; Virginia, G.C.; Pilar, C.; Cristina, B.S. The family of DOF transcription factors in Brachypodium distachyon: Phylogenetic comparison with rice and barley DOFs and expression profiling. BMC Plant Biol. 2012, 12, 202. [Google Scholar]
- Min, G.E.; Yuan-Da, L.; Tan, L.I.; Zhang, T.F.; Zhang, X.L.; Zhao, H. Genome-wide identification and analysis of Dof transcription factor family in maize. Sci. Agric. Sin. 2014, 47, 4563–4572. [Google Scholar]
- Zou, Z.; Yang, J.; Zhang, X. Insights into genes encoding respiratory burst oxidase homologs (RBOHs) in rubber tree (Hevea brasiliensis Muell. Arg.). Ind. Crops Prod. 2019, 128, 126–139. [Google Scholar] [CrossRef]
- Wu, Z.; Cheng, J.; Cui, J.; Xu, X.; Liang, G.; Luo, X.; Chen, X.; Tang, X.; Hu, K.; Cheng, Q. Genome-wide identification and expression profile of Dof transcription factor gene family in pepper (Capsicum annuum. L.). Front. Plant Sci. 2016, 7, 574. [Google Scholar] [CrossRef]
- Dong, C.; Hu, H.; Xie, J. Genome-wide analysis of the DNA-binding with one zinc finger (Dof) transcription factor family in bananas. Genome 2016, 59, 1085. [Google Scholar] [CrossRef]
- Shu, Y.J.; Song, L.L.; Zhang, J.; Liu, Y.; Guo, C.H. Genome-wide identification and characterization of the Dof gene family in Medicago truncatula. Genet. Mol. Res. 2015, 14, 10645. [Google Scholar] [CrossRef]
- Shaw, L.; Mcintyre, C.; Gresshoff, P.; Xue, G. Members of the Dof transcription factor family in Triticum aestivum are associated with light-mediated gene regulation. Funct. Integr. Genom. 2009, 9, 485–498. [Google Scholar] [CrossRef]
- Guo, Y.; Qiu, L.J. Genome-wide analysis of the Dof transcription factor gene family reveals soybean-specific duplicable and functional characteristics. PLoS ONE 2013, 8, e76809. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, J.; Park, S.W. Genome-wide analysis and expression profiling of DNA-binding with one zinc finger (Dof) transcription factor family in potato. Plant Physiol. Biochem. 2015, 94, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Yue, J.J.; Wang, X.J.; Xu, L.; Li, L.B.; Gu, X.P. Genome-wide identification and characterization of the Dof gene family in moso bamboo (Phyllostachys heterocycla var. pubescens). Genes Genom. 2016, 38, 733–745. [Google Scholar] [CrossRef]
- Yanagisawa, S. Dof1 and Dof2 transcription factors are associated with expression of multiple genes involved in carbon metabolism in maize. Plant J. 2000, 21, 281–288. [Google Scholar] [CrossRef]
- Fornara, F.; Panigrahi, K.C.; Gissot, L.; Sauerbrunn, N.; Rühl, M.; Jarillo, J.A.; Coupland, G. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 2009, 17, 75–86. [Google Scholar] [CrossRef]
- Kumar, R.; Taware, R.; Gaur, V.S.; Guru, S.; Kumar, A. Influence of nitrogen on the expression of TaDof1 transcription factor in wheat and its relationship with photo synthetic and ammonium assimilating efficiency. Mol. Biol. Rep. 2009, 36, 2209–2220. [Google Scholar] [CrossRef]
- Le Hir, R.; Bellini, C. The plant-specific Dof transcription factors family: New players involved in vascular system development and functioning in Arabidopsis. Front. Plant Sci. 2013, 4, 164. [Google Scholar] [CrossRef]
- Kumar, A.; Kanwal, P.; Gupta, A.K.; Singh, B.; Gaur, V.S. A full-length Dof1 transcription factor of finger millet and its response to a circadian cycle. Plant Mol. Biol. Report. 2014, 32, 419–427. [Google Scholar] [CrossRef]
- Zou, H.-F.; Zhang, Y.-Q.; Wei, W.; Chen, H.-W.; Song, Q.-X.; Liu, Y.-F.; Zhao, M.-Y.; Wang, F.; Zhang, B.-C.; Lin, Q. The transcription factor AtDOF4. 2 regulates shoot branching and seed coat formation in Arabidopsis. Biochem. J. 2013, 449, 373–388. [Google Scholar] [CrossRef]
- Corrales, A.-R.; Nebauer, S.G.; Carrillo, L.; Fernández-Nohales, P.; Marqués, J.; Renau-Morata, B.; Granell, A.; Pollmann, S.; Vicente-Carbajosa, J.; Molina, R.-V. Characterization of tomato Cycling Dof Factors reveals conserved and new functions in the control of flowering time and abiotic stress responses. J. Exp. Bot. 2014, 65, 995–1012. [Google Scholar] [CrossRef]
- Ma, J.; Li, M.-Y.; Wang, F.; Tang, J.; Xiong, A.-S. Genome-wide analysis of Dof family transcription factors and their responses to abiotic stresses in Chinese cabbage. BMC Genom. 2015, 16, 1–15. [Google Scholar] [CrossRef]
- Imaizumi, T.; Schultz, T.F.; Harmon, F.G.; Ho, L.A.; Kay, S.A. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 2005, 309, 293–297. [Google Scholar] [CrossRef]
- Skirycz, A.; Radziejwoski, A.; Busch, W.; Hannah, M.A.; Czeszejko, J.; Kwaśniewski, M.; Zanor, M.; Lohmann, J.U.; De Veylder, L.; Witt, I.; et al. The DOF transcription factor OBP1 is involved in cell cycle regulation in Arabidopsis thaliana. Plant J. 2010, 56, 779–792. [Google Scholar] [CrossRef]
- Ward, J.M.; Cufr, C.A.; Denzel, M.A.; Neff, M.M. The Dof transcription factor OBP3 modulates phytochrome and cryptochrome signaling in Arabidopsis. Plant Cell 2005, 17, 475–485. [Google Scholar] [CrossRef]
- Yang, Q.; Chen, Q.; Zhu, Y.; Li, T. Identification of MdDof genes in apple and analysis of their response to biotic or abiotic stress. Funct. Plant Biol. 2017, 45, 528–541. [Google Scholar] [CrossRef]
- Yan, Q.; Wu, F.; Yan, Z.; Li, J.; Ma, T.; Zhang, Y.; Zhao, Y.; Wang, Y.; Zhang, J. Differential co-expression networks of long non-coding RNAs and mRNAs in Cleistogenes songorica under water stress and during recovery. BMC Plant Biol. 2019, 19, 1–19. [Google Scholar] [CrossRef]
- Yu, X.; Wang, Y.; Zeng, Y.; Su, D. Effects of temperature and osmotic potential on seed germination of Cleistogenes songorica and Plantago lessingii. Acta Ecol. Sin. 2004, 24, 883–887. [Google Scholar]
- Kong, L.; Zhang, J.; Liu, Z.; Wang, Y. Cloning of a S-adenosyl methionine synthetase gene from Cleistogenes songorica and its expression under drought stress. Acta Prataculturae Sin. 2013, 22, 268–275. [Google Scholar]
- Zhang, J.; Kong, L.; Liu, Z.; Jahufer, Z.; Duan, Z.; Huo, Y.; Di, H.; Wang, Y. Stress-induced expression in Arabidopsis with a Dehydrin LEA protein from Cleistogenes songorica, a xerophytic desert grass. Plant Omics 2015, 8, 485. [Google Scholar]
- Zhang, J.; Duan, Z.; Jahufer, Z.; An, S.; Wang, Y. Stress-inducible expression of a Cleistogenes songorica ALDH gene enhanced drought tolerance in transgenic. Plant Omics 2014, 7, 438. [Google Scholar]
- Zhang, J.; Wu, F.; Yan, Q.; John, U.; Cao, M.; Xu, P.; Zhang, Z.; Ma, T.; Zong, X.; Li, J.; et al. The genome of Cleistogenes songorica provides a blueprint for functional dissection of dimorphic flower differentiation and drought adaptability. Plant Biotechnol. J. 2021, 19, 532–547. [Google Scholar] [CrossRef]
- Yang, X.; Tuskan, G.A.; Cheng, M.Z. Divergence of the Dof gene families in poplar, Arabidopsis, and rice suggests Multiple modes of gene evolution after duplication. Plant Physiol. 2006, 142, 820–830. [Google Scholar] [CrossRef]
- Morenorisueno, M.Á.; Martínez, M.; Vicentecarbajosa, J.; Carbonero, P. The family of DOF transcription factors: From green unicellular algae to vascular plants. Mol. Genet. Genom. 2007, 277, 379–390. [Google Scholar] [CrossRef]
- Kushwaha, H.; Gupta, S.; Singh, V.K.; Rastogi, S.; Yadav, D. Genome wide identification of Dof transcription factor gene family in sorghum and its comparative phylogenetic analysis with rice and Arabidopsis. Mol. Biol. Rep. 2011, 16, 372–384. [Google Scholar] [CrossRef] [PubMed]
- Zong, X.; Yan, Q.; Wu, F.; Ma, Q.; Zhang, J. Genome-wide analysis of the role of NAC family in flower development and abiotic stress responses in Cleistogenes songorica. Genes 2020, 11, 927. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Huang, W.; Liu, Z.W.; Wang, Y.X.; Zhuang, J. Transcriptome-based analysis of Dof family transcription factors and their responses to abiotic stress in tea plant (Camellia sinensis). Int. J. Genom. 2016, 2016, 1–15. [Google Scholar] [CrossRef]
- Liu, J.; Cheng, Z.; Xie, L.; Li, X.; Gao, J. Multifaceted role of PheDof12-1 in the regulation of flowering time and abiotic stress responses in moso bamboo (Phyllostachys edulis). Int. J. Mol. Sci. 2019, 20, 424. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Shi, F.; Liu, X.; Huang, G.; Zhou, M. Phylogenetic analysis and evolution of aromatic amino acid hydroxylase. FEBS Lett. 2010, 584, 4775–4782. [Google Scholar] [CrossRef]
- Fang, Z.; Jiang, W.; He, Y.; Ma, D.; Liu, Y.; Wang, S.; Zhang, Y.; Yin, J. Genome-wide identification, structure characterization, and expression profiling of Dof transcription factor gene family in wheat (Triticum aestivum L.). Agronomy 2020, 10, 294. [Google Scholar] [CrossRef]
- Noguero, M.; Atif, R.M.; Ochatt, S.; Thompson, R.D. The role of the DNA-binding One Zinc Finger (DOF) transcription factor family in plants. Plant Sci. 2013, 209, 32–45. [Google Scholar] [CrossRef]
- Gupta, S.; Malviya, N.; Kushwaha, H.; Nasim, J.; Bisht, N.C.; Singh, V.; Yadav, D. Insights into structural and functional diversity of Dof (DNA binding with one finger) transcription factor. Planta 2015, 241, 549–562. [Google Scholar] [CrossRef]
- Zhou, Y.; Cheng, Y.; Wan, C.; Li, J.; Yang, Y.; Chen, J. Genome-wide characterization and expression analysis of the Dof gene family related to abiotic stress in watermelon. PeerJ 2020, 8, e8358. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, N.; Deng, X.; Liu, D.; Li, M.; Cui, D.; Hu, Y.; Yan, Y. Genome-wide analysis of wheat DNA-binding with one finger (Dof) transcription factor genes: Evolutionary characteristics and diverse abiotic stress responses. BMC Genom. 2020, 21, 1–18. [Google Scholar] [CrossRef]
- Cai, X.; Zhang, Y.; Zhang, C.; Zhang, T.; Hu, T.; Ye, J.; Zhang, J.; Wang, T.; Li, H.; Ye, Z. Genome-wide analysis of plant-specific Dof transcription factor family in Tomato. J. Integr. Plant Biol. 2013, 55, 552–566. [Google Scholar] [CrossRef]
- Yan, Q.; Zong, X.; Wu, F.; Li, J.; Ma, T.; Zhao, Y.; Ma, Q.; Wang, P.; Wang, Y.; Zhang, J. Integrated analysis of co-expression, conserved genes and gene families reveal core regulatory network of heat stress response in Cleistogenes songorica, a xerophyte perennial desert plant. BMC Genom. 2020, 21, 1–18. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.E.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Springer: Berlin/Heidelberg, Germany, 2005; pp. 571–607. [Google Scholar]
- Guo, A.Y.; Zhu, Q.H.; Chen, X.; Luo, J.C. GSDS: A gene structure display server. Hereditas 2007, 29, 1023–1026. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Han, F.; Zhu, B. Evolutionary analysis of three gibberellin oxidase genesin rice, Arabidopsis, and soybean. Gene 2011, 473, 23–35. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Wang, Y.R.; Nan, Z.B. Relative and absolute quantification expression analysis of CsSAMDC gene as a case. China Biotechnol. 2009, 29, 86–91. [Google Scholar]
- Zhu, W.; Lee, Y.S. Dexel-based force-torque rendering and volume updating for 5-DOF haptic product prototyping and virtual sculpting. Comput. Ind. 2004, 55, 125–145. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, P.; Yan, Z.; Zong, X.; Yan, Q.; Zhang, J. Genome-Wide Analysis and Expression Profiles of the Dof Family in Cleistogenes songorica under Temperature, Salt and ABA Treatment. Plants 2021, 10, 850. https://doi.org/10.3390/plants10050850
Wang P, Yan Z, Zong X, Yan Q, Zhang J. Genome-Wide Analysis and Expression Profiles of the Dof Family in Cleistogenes songorica under Temperature, Salt and ABA Treatment. Plants. 2021; 10(5):850. https://doi.org/10.3390/plants10050850
Chicago/Turabian StyleWang, Penglei, Zhuanzhuan Yan, Xifang Zong, Qi Yan, and Jiyu Zhang. 2021. "Genome-Wide Analysis and Expression Profiles of the Dof Family in Cleistogenes songorica under Temperature, Salt and ABA Treatment" Plants 10, no. 5: 850. https://doi.org/10.3390/plants10050850
APA StyleWang, P., Yan, Z., Zong, X., Yan, Q., & Zhang, J. (2021). Genome-Wide Analysis and Expression Profiles of the Dof Family in Cleistogenes songorica under Temperature, Salt and ABA Treatment. Plants, 10(5), 850. https://doi.org/10.3390/plants10050850






