Analysis of Cadmium Root Retention for Two Contrasting Rice Accessions Suggests an Important Role for OsHMA2
Abstract
:1. Introduction
2. Results
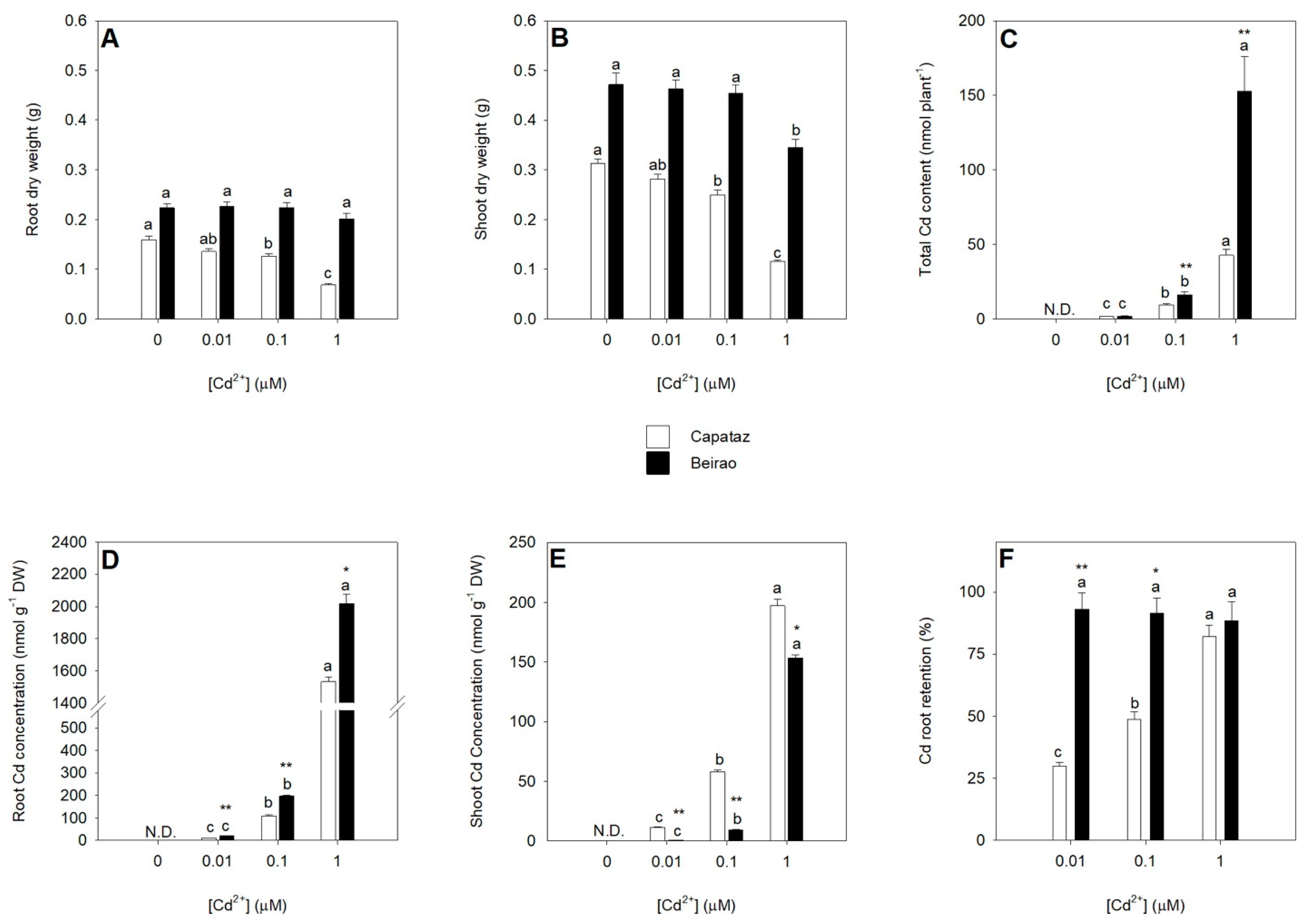
2.1. Plant Growth under Cd Stress and Cd Partitioning between Roots and Shoots
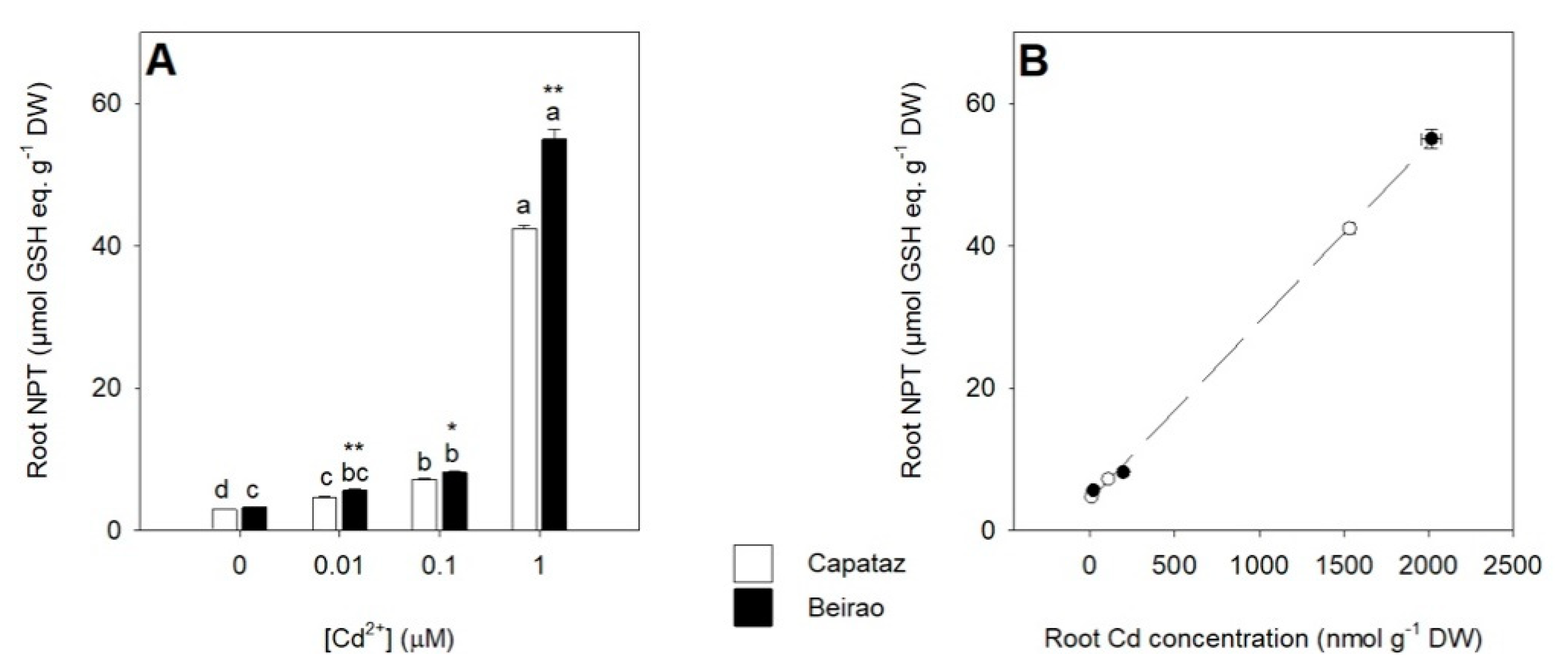
2.2. Effects of Cd Exposure on Thiol Accumulation
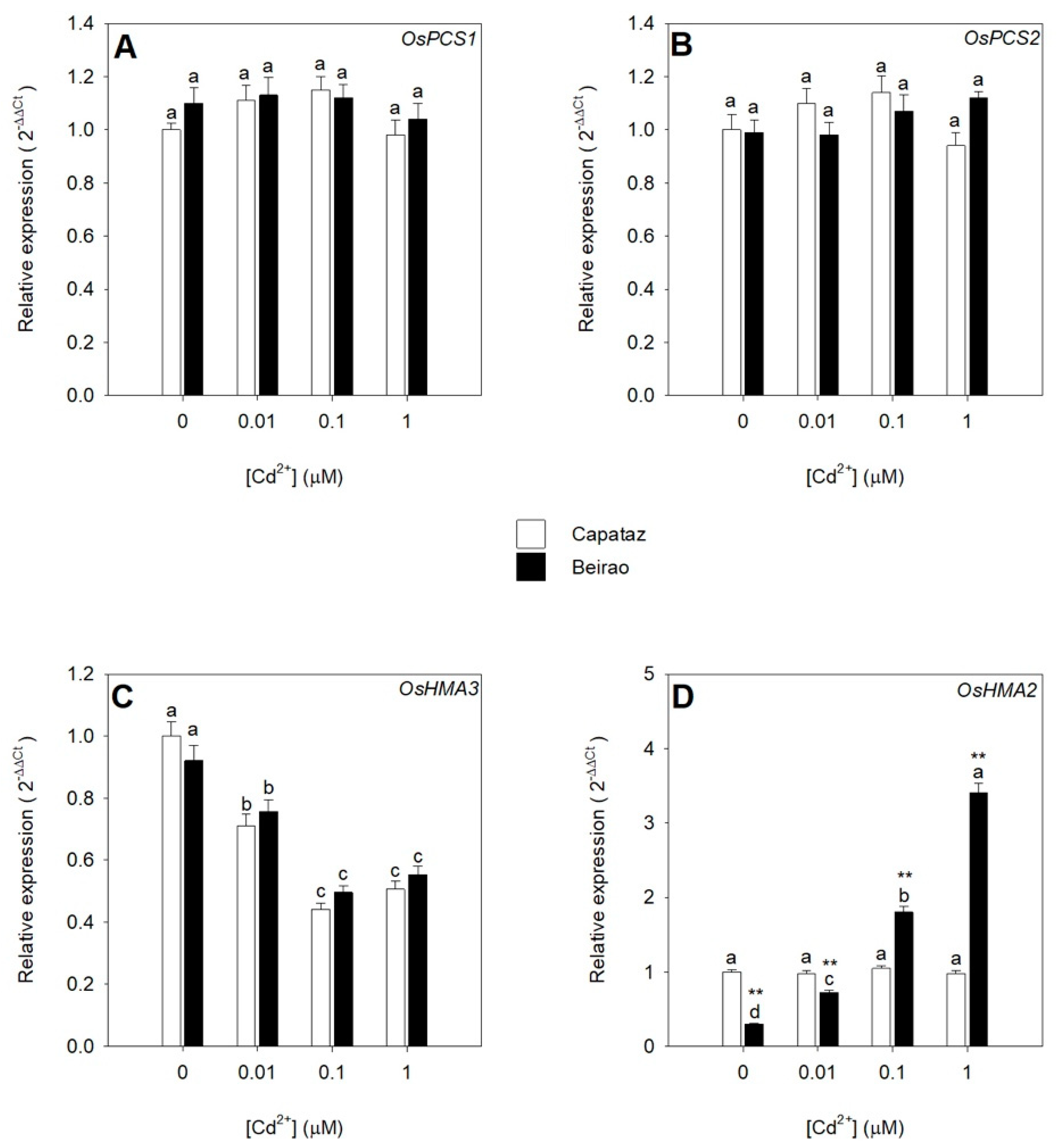
2.3. Effects of Cd Exposure on the Relative Expression of Genes Involved in Cd Chelation, Compartmentalization, and Translocation
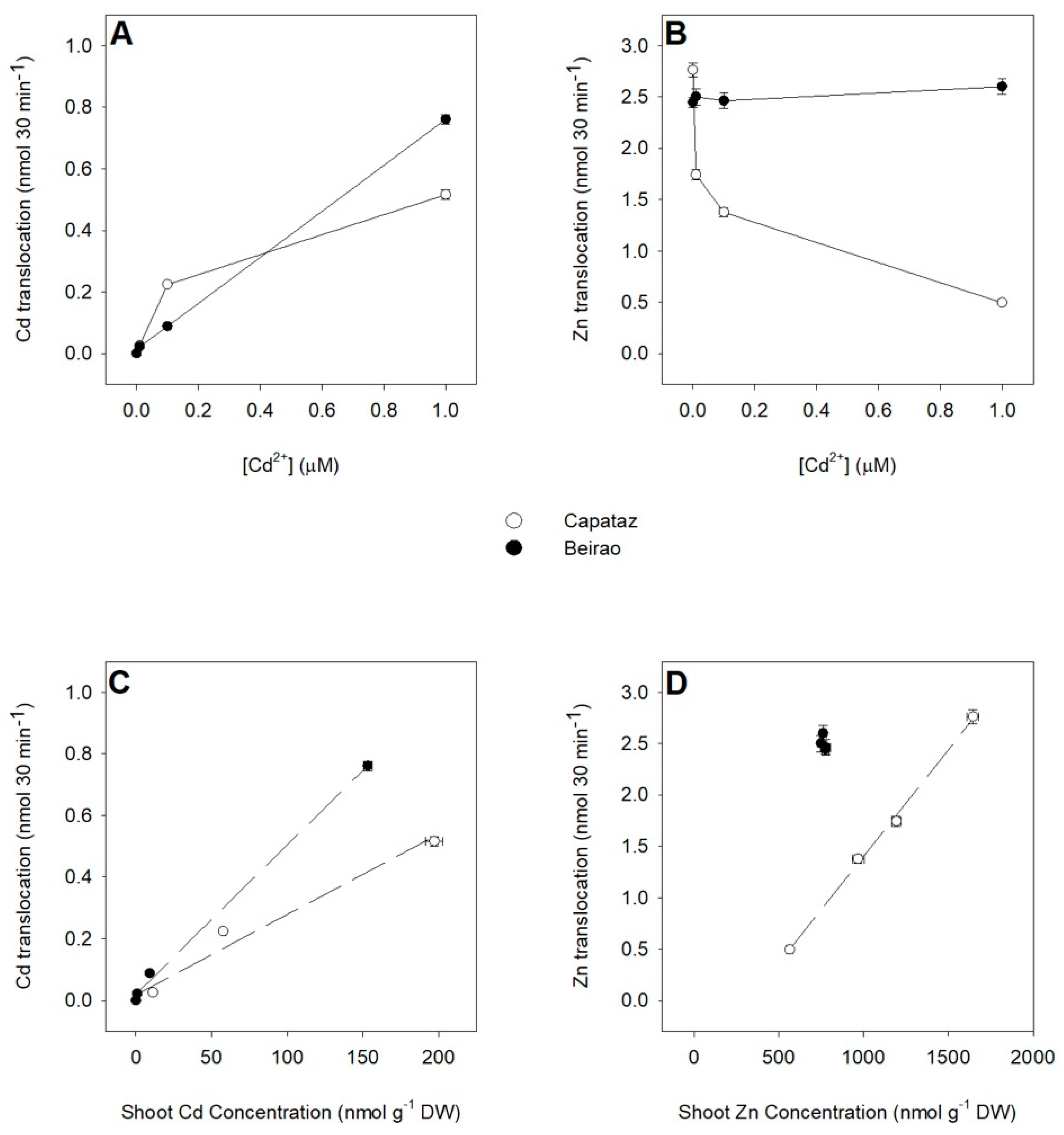
2.4. Effects of Cd on Zn Partitioning between Roots and Shoots
2.5. Analysis of Root-to-Shoot Cd/Zn Translocation
3. Discussion
4. Materials and Methods
4.1. Plant Material, Growth Conditions, and Sampling
4.2. Determination of Cd, Zn and Total Non-Protein Thiols (NPTs)
4.3. Analysis of Root-to-Shoot Cd Translocation
4.4. RNA Extraction and qRT-PCR Analysis
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alloway, B.J.; Steinnes, E. Anthropogenic addition of cadmium to soils. In Cadmium in Soil and Plants; McLaughlin, M.J., Singh, B.R., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1999; pp. 97–123. [Google Scholar]
- McLaughlin, M.; Parker, D.R.; Clarke, J.M. Metals and micronutrients—Food safety issues. Field Crops Res. 1999, 60, 143–163. [Google Scholar] [CrossRef]
- Nordberg, G.F. Historical perspectives on cadmium toxicology. Toxicol. Appl. Pharm. 2009, 238, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Clemens, S.; Aarts, M.G.; Thomine, S.; Verbruggen, N. Plant science: The key to preventing slow cadmium poisoning. Trends Plant Sci. 2013, 18, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Wagner, G.J. Accumulation of cadmium in crop plants and its consequences to human health. Adv. Agron. 1993, 51, 173–212. [Google Scholar] [CrossRef]
- EFSA. Cadmium in food—Scientific opinion of the panel on contaminants in the food chain. EFSA J. 2009, 980, 1–139. [Google Scholar] [CrossRef]
- FAO/WHO. Evaluation of Certain Food Additives and Contaminants. 2001. Available online: https://www.who.int/ipcs/publications/jecfa/reports/trs940.pdf (accessed on 6 April 2021).
- Kobayashi, J. Pollution by cadmium and the itai-itai disease in Japan. In Toxicity of Heavy Metals in the Environment; Oehme, F.W., Ed.; Marcel Dekker Inc.: New York, NY, USA, 1978; pp. 199–260. [Google Scholar]
- Cai, S.; Yue, L.; Hu, Z.; Zhong, X.; Ye, Z.; Xu, H.; Liu, Y.R.; Ji, R.D.; Zhang, W.H.; Zhang, F. Cadmium exposure and health effects among residents in an irrigation area with ore dressing wastewater. Sci. Total Environ. 1990, 90, 67–73. [Google Scholar] [CrossRef]
- Kobayashi, E.; Okubo, Y.; Suwazono, Y.; Kido, T.; Nogawa, K. Dose–response relationship between total cadmium intake calculated from cadmium concentration in rice collected from each household of farmers and renal dysfunction in inhabitants of the Jinzu River basin, Japan. J. Appl. Toxicol. 2002, 22, 431–436. [Google Scholar] [CrossRef]
- CODEX STAN 193–1995. General Standard for Contaminants and Toxins in Foods and Feed. 2019. Available online: http://www.fao.org/fao-who-codexalimentarius/sh-proxy/en/?lnk=1&url=https%253A%252F%252Fworkspace.fao.org%252Fsites%252Fcodex%252FStandards%252FCXS%2B193-1995%252FCXS_193e.pdf (accessed on 6 April 2021).
- Murakami, M.; Ae, N.; Ishikawa, S. Phytoextraction of cadmium by rice (Oryza sativa L.), soybean (Glycine max (L.) Merr.), and maize (Zea mays L.). Environ. Pollut. 2007, 145, 96–103. [Google Scholar] [CrossRef]
- Makino, T.; Takano, H.; Kamiya, T.; Itou, T.; Sekiya, N.; Inahara, M.; Sakurai, Y. Restoration of cadmium-contaminated paddy soils by washing with ferric chloride: Cd extraction mechanism and bench-scale verification. Chemosphere 2008, 70, 1035–1043. [Google Scholar] [CrossRef]
- Grant, C.A.; Clarke, J.M.; Duguid, S.; Chaney, R.L. Selection and breeding of plant cultivars to minimize cadmium accumulation. Sci. Total Environ. 2008, 390, 301–310. [Google Scholar] [CrossRef]
- Uraguchi, S.; Mori, S.; Kuramata, M.; Kawasaki, A.; Arao, T.; Ishikawa, S. Root-to-shoot Cd translocation via the xylem is the major process determining shoot and grain cadmium accumulation in rice. J. Exp. Bot. 2009, 60, 2677–2688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueno, D.; Yamaji, N.; Kono, I.; Huang, C.F.; Ando, T.; Yano, M.; Ma, J.F. Gene limiting cadmium accumulation in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 16500–16505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.L.; Schulz, R.; Marschner, H. Genotypic differences in uptake and distribution of cadmium and nickel in plants. Angew. Bot. 1995, 69, 42–48. [Google Scholar] [CrossRef]
- Grant, C.A.; Buckley, W.T.; Bailey, L.D.; Selles, F. Cadmium accumulation in crops. Can. J. Plant Sci. 1998, 78, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Cakmak, I.; Welch, R.M.; Hart, J.; Norvell, W.A.; Oztürk, L.; Kochian, L.V. Uptake and retranslocation of leaf-applied cadmium (109Cd) in diploid, tetraploid and hexaploid wheats. J. Exp. Bot. 2000, 51, 221–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, J.M.; Norvell, W.A.; Clarke, F.R.; Buckley, W.T. Concentration of cadmium and other elements in the grain of near-isogenic durum lines. Can. J. Plant Sci. 2002, 82, 27–33. [Google Scholar] [CrossRef]
- Dunbar, K.R.; McLaughlin, M.J.; Reid, R.J. The uptake and partitioning of cadmium in two cultivars of potato (Solanum tuberosum L.). J. Exp. Bot. 2003, 54, 349–354. [Google Scholar] [CrossRef]
- Lozano-Rodríguez, E.; Hernández, L.E.; Bonay, P.; Carpena-Ruiz, R.O. Distribution of Cd in shoot and root tissues of maize and pea plants: Physiological disturbances. J. Exp. Bot. 1997, 48, 123–128. [Google Scholar] [CrossRef] [Green Version]
- Puig, S.; Peñarrubia, L. Placing metal micronutrients in context: Transport and distribution in plants. Curr. Opin. Plant Biol. 2009, 12, 299–306. [Google Scholar] [CrossRef]
- Verbruggen, N.; Hermans, C.; Schat, H. Mechanisms to cope with arsenic or cadmium excess in plants. Curr. Opin. Plant Biol. 2009, 12, 364–372. [Google Scholar] [CrossRef]
- Nocito, F.F.; Lancilli, C.; Dendena, B.; Lucchini, G.; Sacchi, G.A. Cadmium retention in rice roots is influenced by cadmium availability, chelation and translocation. Plant Cell Environ. 2011, 34, 994–1008. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Cózatl, D.G.; Jobe, T.O.; Hauser, F.; Schroeder, J.I. Long-distance transport, vacuolar sequestration, tolerance, and transcriptional responses induced by cadmium and arsenic. Curr. Opin. Plant Biol. 2011, 14, 554–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fontanili, L.; Lancilli, C.; Suzui, N.; Dendena, B.; Yin, Y.-G.; Ferri, A.; Ishii, S.; Kawachi, N.; Lucchini, G.; Fujimaki, S.; et al. Kinetic analysis of zinc/cadmium reciprocal competitions suggests a possible Zn-insensitive pathway for root-to-shoot cadmium translocation in rice. Rice 2016, 9, 16. [Google Scholar] [CrossRef] [Green Version]
- Clemens, S. Toxic metal accumulation, responses to exposure and mechanisms of tolerance in plants. Biochimie 2006, 88, 1707–1719. [Google Scholar] [CrossRef] [PubMed]
- Zenk, M.H. Heavy metal detoxification in higher plants—A review. Gene 1996, 179, 21–30. [Google Scholar] [CrossRef]
- Cobbett, C.S. Phytochelatins and their roles in heavy metal detoxification. Plant Physiol. 2000, 123, 825–832. [Google Scholar] [CrossRef] [Green Version]
- Cobbett, C.; Goldsbrough, P. Phytochelatins and metallothioneins: Roles in heavy metal detoxification and homeostasis. Annu. Rev. Plant Biol. 2002, 53, 159–182. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, J.; Ali, A.A.; Baig, M.A.; Iqbal, M.; Haq, I.; Qureshi, M.I. Role of phytochelatins in cadmium stress tolerance in plants. In Cadmium Toxicity and Tolerance in Plants; Hasanuzzaman, M., Prasad, M.N.V., Fujita, M., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 185–212. [Google Scholar]
- Takahashi, R.; Ishimaru, Y.; Shimo, H.; Ogo, Y.; Senoura, T.; Nishizawa, N.K.; Nakanishi, H. The OsHMA2 transporter is involved in root-to-shoot translocation of Zn and Cd in rice. Plant Cell Environ. 2012, 35, 1948–1957. [Google Scholar] [CrossRef]
- Satoh-Nagasawa, N.; Mori, M.; Nakazawa, N.; Kawamoto, T.; Nagato, Y.; Sakurai, K.; Takahashi, H.; Watanabe, A.; Akagi, H. Mutations in rice (Oryza sativa) heavy metal ATPase 2 (OsHMA2) restrict the translocation of zinc and cadmium. Plant Cell Physiol. 2012, 53, 213–224. [Google Scholar] [CrossRef]
- Yamaji, N.; Xia, J.; Mitani-Ueno, N.; Yokosho, K.; Ma, J.F. Preferential delivery of zinc to developing tissues in rice is mediated by P-type heavy metal ATPase OsHMA2. Plant Physiol. 2013, 162, 927–939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verkleij, J.A.C.; Koevoets, P.; Van’t Riet, J.; Bank, R.; Nijdam, Y.; Ernst, W.H.O. Poly(y-glutamylcysteinyl)glycines or phytochelatins and their role in cadmium tolerance of Silene vulgaris. Plant Cell Environ. 1990, 13, 913–921. [Google Scholar] [CrossRef]
- Wong, C.K.E.; Cobbett, C.S. HMA P-type ATPases are the major mechanism for root-to-shoot Cd translocation in Arabidopsis thaliana. New Phytol. 2009, 181, 71–78. [Google Scholar] [CrossRef]
- Sghayar, S.; Ferri, A.; Lancilli, C.; Lucchini, G.; Abruzzese, A.; Porrini, M.; Ghnaya, T.; Nocito, F.F.; Abdelly, C.; Sacchi, G.A. Analysis of cadmium translocation, partitioning and tolerance in six barley (Hordeum vulgare L.) cultivars as a function of thiol metabolism. Biol. Fertil. Soils 2015, 51, 311–320. [Google Scholar] [CrossRef]
- Nocito, F.F.; Lancilli, C.; Giacomini, B.; Sacchi, G.A. Sulfur metabolism and cadmium stress in higher plants. Plant Stress 2007, 1, 142–156. [Google Scholar]
- Miydate, H.; Adachi, S.; Hiraizumi, A.; Tezuka, K.; Nakazawa, N.; Kawamoto, T.; Katou, K.; Kodama, I.; Sakurai, K.; Takahashi, H.; et al. OsHMA3, a P1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytol. 2011, 189, 190–199. [Google Scholar] [CrossRef]
- Rauser, W.E.; Meuwly, P. Retention of cadmium in roots of maize seedlings. Role of complexation by phytochelatins and related thiol peptides. Plant Physiol. 1995, 109, 195–202. [Google Scholar] [CrossRef] [Green Version]
- Clemens, S.; Ma, F.J. Toxic heavy metal and metalloid accumulation in crop plants and foods. Annu. Rev. Plant Biol. 2016, 67, 489–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, J.; Wang, P.; Wang, P.; Yang, M.; Lian, X.; Tang, Z.; Huang, C.F.; Salt, D.E.; Zhao, F.J. A loss-of-function allele of OsHMA3 associated with high cadmium accumulation in shoots and grain of Japonica rice cultivars. Plant Cell Environ. 2016, 39, 1941–1954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, C.; Yang, M.; Li, Y.; Tian, J.; Zhang, Y.; Liang, L.; Liu, Z.; Chen, K.; Li, Y.; Lv, K.; et al. Comprehensive analysis of variation of cadmium accumulation in rice and detection of a new weak allele of OsHMA3. J. Exp. Bot. 2019, 70, 6389–6400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sui, F.; Zhao, D.; Zhu, H.; Gong, Y.; Tang, Z.; Huang, X.-Y.; Zhang, G.; Zhao, F.-J. Map-based cloning of a new total loss-of-function allele of OsHMA3 causing high cadmium accumulation in rice grain. J. Exp. Bot. 2019, 70, 2857–2871. [Google Scholar] [CrossRef]
- Biscarini, F.; Cozzi, P.; Casella, L.; Riccardi, P.; Vattari, A.; Orasen, G.; Perrini, R.; Tacconi, G.; Tondelli, A.; Biselli, C.; et al. Genome-wide association study for traits related to plant and grain morphology, and root architecture in temperate rice accessions. PLoS ONE 2016, 11, e0155425. [Google Scholar] [CrossRef] [PubMed]
- Volante, A.; Desiderio, F.; Tondelli, A.; Perrini, R.; Orasen, G.; Biselli, C.; Riccardi, P.; Vattari, A.; Cavalluzzo, D.; Urso, S.; et al. Genome-wide analysis of japonica rice performance under limited water and permanent flooding conditions. Front. Plant Sci. 2017, 8, 1862. [Google Scholar] [CrossRef] [PubMed]
- Rauser, W.E. Compartmental efflux analysis and removal of extracellular cadmium from roots. Plant Physiol. 1987, 85, 62–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livaka, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using Real-Time Quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maghrebi, M.; Baldoni, E.; Lucchini, G.; Vigani, G.; Valè, G.; Sacchi, G.A.; Nocito, F.F. Analysis of Cadmium Root Retention for Two Contrasting Rice Accessions Suggests an Important Role for OsHMA2. Plants 2021, 10, 806. https://doi.org/10.3390/plants10040806
Maghrebi M, Baldoni E, Lucchini G, Vigani G, Valè G, Sacchi GA, Nocito FF. Analysis of Cadmium Root Retention for Two Contrasting Rice Accessions Suggests an Important Role for OsHMA2. Plants. 2021; 10(4):806. https://doi.org/10.3390/plants10040806
Chicago/Turabian StyleMaghrebi, Moez, Elena Baldoni, Giorgio Lucchini, Gianpiero Vigani, Giampiero Valè, Gian Attilio Sacchi, and Fabio Francesco Nocito. 2021. "Analysis of Cadmium Root Retention for Two Contrasting Rice Accessions Suggests an Important Role for OsHMA2" Plants 10, no. 4: 806. https://doi.org/10.3390/plants10040806
APA StyleMaghrebi, M., Baldoni, E., Lucchini, G., Vigani, G., Valè, G., Sacchi, G. A., & Nocito, F. F. (2021). Analysis of Cadmium Root Retention for Two Contrasting Rice Accessions Suggests an Important Role for OsHMA2. Plants, 10(4), 806. https://doi.org/10.3390/plants10040806







