Molecular Characterization of Potato Virus Y (PVY) Using High-Throughput Sequencing: Constraints on Full Genome Reconstructions Imposed by Mixed Infection Involving Recombinant PVY Strains
Abstract
1. Introduction
2. Results
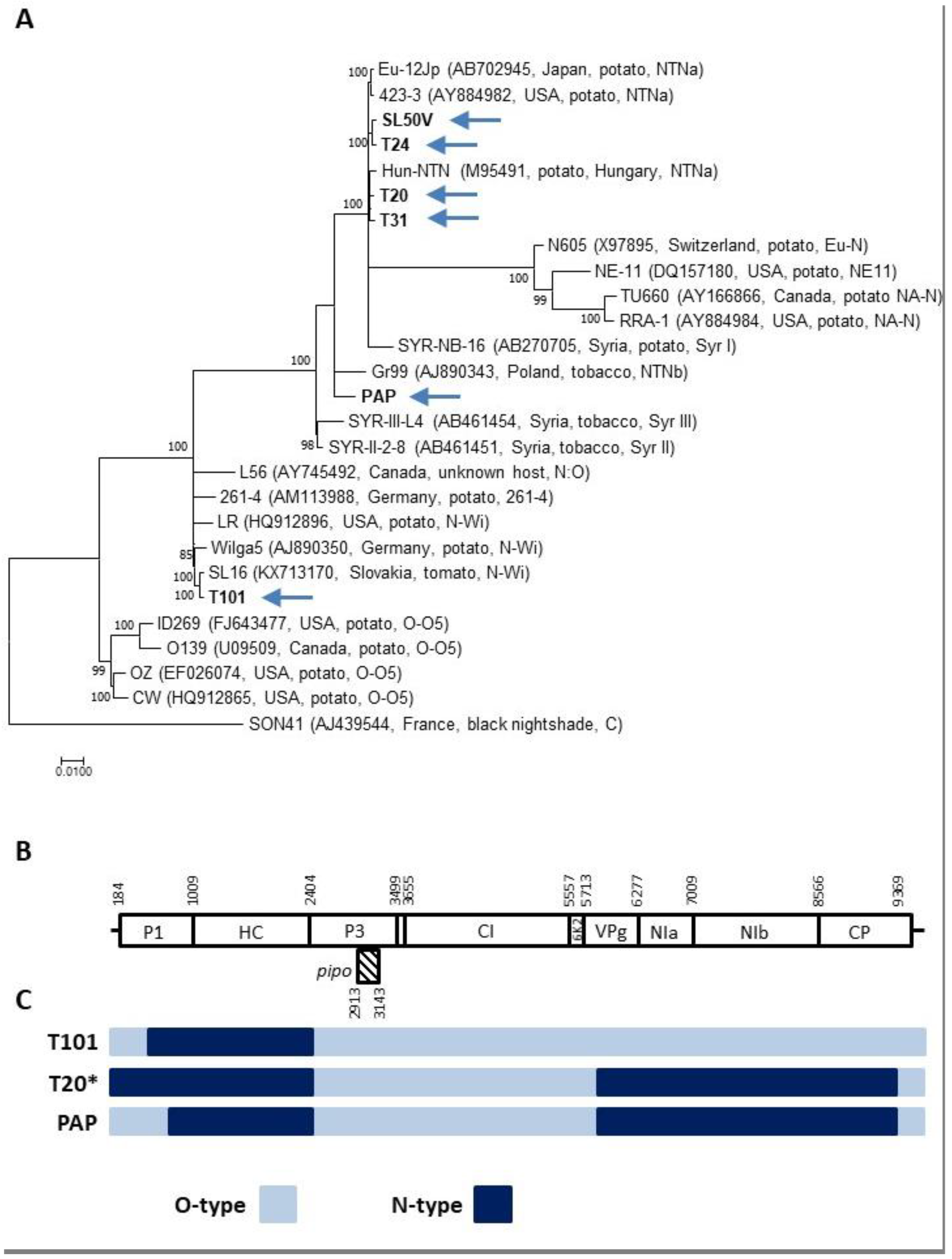
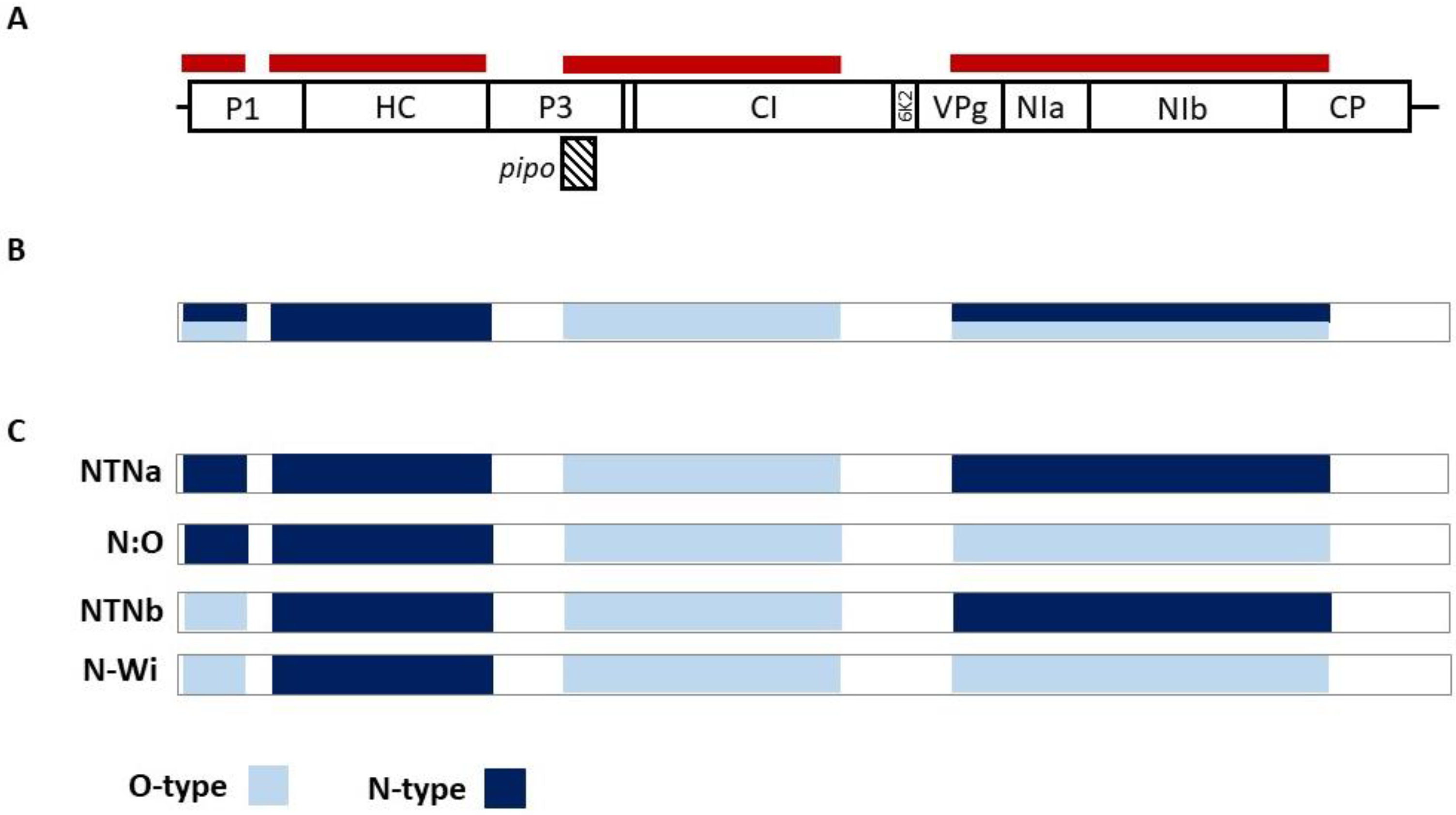
2.1. PVY Isolates Identified in Non-Potato Hosts Belong to Recombinant Strains
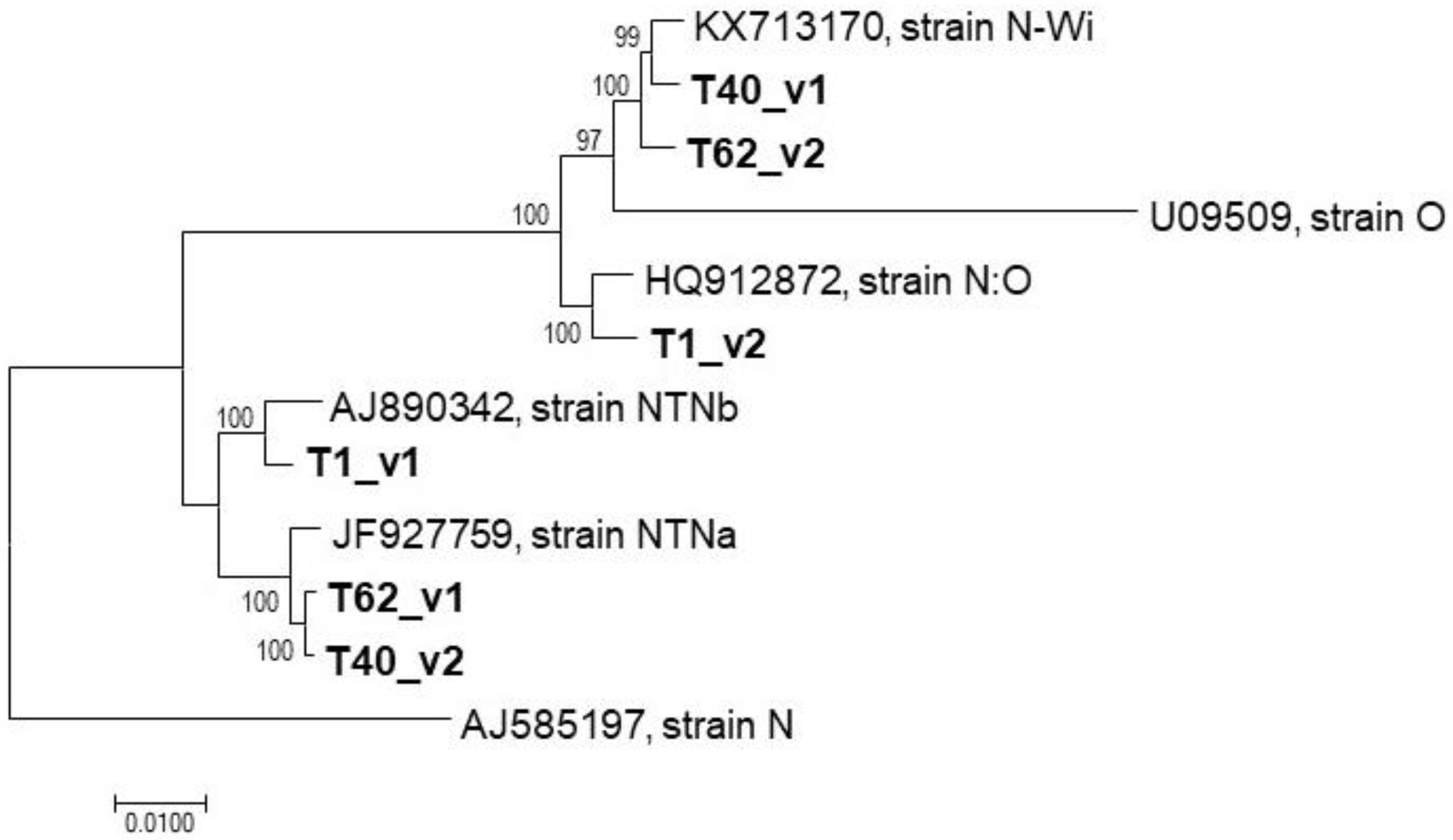
2.2. Mixed Infection Involving Different PVY Isolates
2.3. Attempts to Reconstruct Complete PVY Genomic Sequences from Mixed Infection
3. Discussion
4. Materials and Methods
4.1. Plant Samples
4.2. HTS Analysis
4.3. Strain Specific RT-PCR Assays
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Quenouille, J.; Vassilakos, N.; Moury, B. Potato virus Y: A major crop pathogen that has provided major insights into the evolution of viral pathogenicity. Mol. Plant Pathol. 2013, 14, 439–452. [Google Scholar] [CrossRef] [PubMed]
- Nigam, D.; LaTourrette, K.; Souza, P.F.N.; Garcia-Ruiz, H. Genome-Wide Variation in Potyviruses. Front. Plant Sci. 2019, 10, 1439. [Google Scholar] [CrossRef] [PubMed]
- Lacomme, C.; Jacquot, E. General Characteristics of Potato virus Y (PVY) and Its Impact on Potato Production, An Overview. In Potato virus Y: Biodiversity, Pathogenicity, Epidemiology and Management; Lacomme, C., Glais Lv Bellstedt, D., Dupuis, B., Karasev, A., Jacquot, E., Eds.; Springer: Cham, Switzerland, 2017. [Google Scholar] [CrossRef]
- Hančinský, R.; Mihálik, D.; Mrkvová, M.; Candresse, T.; Glasa, M. Plant Viruses Infecting Solanaceae Family Members in the Cultivated and Wild Environments: A Review. Plants 2020, 9, 667. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, R.; García-Marcos, A.; Barajas, D.; Martiáñez, J.; Tenllado, F. PVX-potyvirus synergistic infections differentially alter microRNA accumulation in Nicotiana benthamiana. Virus Res. 2012, 165, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Funke, C.N.; Nikolaeva, O.V.; Green, K.J.; Tran, L.T.; Chikh-Ali, M.; Quintero-Ferrer, A.; Cating, R.A.; Frost, K.E.; Hamm, P.B.; Olsen, N.; et al. Strain-Specific Resistance to Potato virus Y (PVY) in Potato and Its Effect on the Relative Abundance of PVY Strains in Commercial Potato Fields. Plant Dis. 2017, 101, 20–28. [Google Scholar] [CrossRef]
- Dupuis, B.; Bragard, C.; Schumpp, O. Resistance of Potato Cultivars as a Determinant Factor of Potato virus Y (PVY) Epidemiology. Potato Res. 2019, 62, 123–138. [Google Scholar] [CrossRef]
- Lindner, K.; Trautwein, F.; Kellermann, A.; Bauch, G. Potato virus Y (PVY) in Seed Potato Certification. J. Plant Dis. Prot. 2015, 122, 109–119. [Google Scholar] [CrossRef]
- Rubio, L.; Galipienso, L.; Ferriol, I. Detection of Plant Viruses and Disease Management: Relevance of Genetic Diversity and Evolution. Front. Plant Sci. 2020, 11, 1092. [Google Scholar] [CrossRef]
- Glais, L.; Bellstedt, D.U.; Lacomme, C. Diversity, Characterisation and Classification of PVY. In Potato virus Y: Biodiversity, Pathogenicity, Epidemiology and Management; Lacomme, C., Glais, L., Bellstedt, D., Dupuis, B., Karasev, A., Jacquot, E., Eds.; Springer: Cham, Switzerland, 2017. [Google Scholar] [CrossRef]
- Chikh-Ali, M.; Gray, S.M.; Karasev, A.V. An Improved Multiplex IC-RT-PCR Assay Distinguishes Nine Strains of Potato virus Y. Plant Dis. 2013, 97, 1370–1374. [Google Scholar] [CrossRef]
- Martinelli, F.; Scalenghe, R.; Davino, S.; Panno, S.; Scuderi, G.; Ruisi, P.; Villa, P.; Stroppiana, D.; Boschetti, M.; Goulart, L.R.; et al. Advanced methods of plant disease detection. A review. Agron. Sustain. Dev. 2015, 35, 1–25. [Google Scholar] [CrossRef]
- Syller, J.; Grupa, A. The effects of co-infection by different Potato virus Y (PVY) isolates on virus concentration in solanaceous hosts and efficiency of transmission. Plant Pathol. 2014, 63, 466–475. [Google Scholar] [CrossRef]
- da Silva, W.; Kutnjak, D.; Xu, Y.; Xu, Y.; Giovannoni, J.; Elena, S.F.; Gray, S. Transmission modes affect the population structure of potato virus Y in potato. PLoS Pathog. 2020, 16, e1008608. [Google Scholar] [CrossRef]
- Steinger, T.; Gilliand, H.; Hebeisen, T. Epidemiological analysis of risk factors for the spread of potato viruses in Switzerland. Ann. Appl. Biol. 2014, 164, 200–207. [Google Scholar] [CrossRef]
- Elwan, E.A.; Abdel Aleem, E.E.; Fattouh, F.A.; Green, K.J.; Tran, L.T.; Karasev, A.V. Occurrence of Diverse Recombinant Strains of Potato virus Y Circulating in Potato Fields in Egypt. Plant Dis. 2017, 101, 1463–1469. [Google Scholar] [CrossRef][Green Version]
- Karasev, A.V.; Gray, S.M. Continuous and emerging challenges of Potato virus Y in potato. Ann. Rev. Phytopathol. 2013, 51, 571–586. [Google Scholar] [CrossRef] [PubMed]
- Karasev, A.V.; Hu, X.; Brown, C.J.; Kerlan, C.; Nikolaeva, O.V.; Crosslin, J.M.; Gray, S.M. Genetic diversity of the ordinary strain of Potato virus Y (PVY) and origin of recombinant PVY strains. Phytopathology 2011, 101, 778–785. [Google Scholar] [CrossRef] [PubMed]
- Green, K.J.; Brown, C.J.; Karasev, A.V. Genetic diversity of potato virus Y (PVY): Sequence analyses reveal ten novel PVY recombinant structures. Arch. Virol. 2018, 163, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Green, K.J.; Brown, C.J.; Gray, S.M.; Karasev, A.V. Phylogenetic study of recombinant strains of Potato virus Y. Virology 2017, 507, 40–52. [Google Scholar] [CrossRef]
- Hu, X.; Karasev, A.V.; Brown, C.J.; Lorenzen, J.H. Sequence characteristics of potato virus Y recombinants. J. Gen. Virol. 2009, 90, 3033–3041. [Google Scholar] [CrossRef]
- Chikh-Ali, M.; Maoka, T.; Natsuaki, K.T.; Natsuaki, T. The simultaneous differentiation of Potato virus Y strains including the newly described strain PVYNTN-NW by multiplex PCR assay. J. Virol. Methods 2010, 165, 15–20. [Google Scholar] [CrossRef]
- Pecman, A.; Kutnjak, D.; Gutiérrez-Aguirre, I.; Adams, I.; Fox, A.; Boonham, N.; Ravnikar, M. Next Generation Sequencing for Detection and Discovery of Plant Viruses and Viroids: Comparison of Two Approaches. Front. Microbiol. 2017, 8, 1998. [Google Scholar] [CrossRef]
- Minicka, J.; Zarzyńska-Nowak, A.; Budzyńska, D.; Borodynko-Filas, N.; Hasiów-Jaroszewska, B. High-Throughput Sequencing Facilitates Discovery of New Plant Viruses in Poland. Plants 2020, 9, 820. [Google Scholar] [CrossRef]
- Barba, M.; Czosnek, H.; Hadidi, A. Historical perspective, development and applications of next-generation sequencing in plant virology. Viruses 2014, 6, 106–136. [Google Scholar] [CrossRef]
- Villamil-Garzón, A.; Cuellar, W.J.; Guzmán-Barney, M. Natural co-infection of Solanum tuberosum crops by the Potato yellow vein virus and potyvirus in Colombia. Agron. Colomb. 2014, 32, 213–223. [Google Scholar] [CrossRef]
- Akinyemi, I.A.; Wang, F.; Zhou, B.; Qi, S.; Wu, Q. Ecogenomic survey of plant viruses infecting Tobacco by Next generation sequencing. Virol. J. 2016, 13, 181. [Google Scholar] [CrossRef] [PubMed]
- Tomašechová, J.; Hančinský, R.; Predajňa, L.; Kraic, J.; Mihálik, D.; Šoltys, K.; Vávrová, S.; Böhmer, M.; Sabanadzovic, S.; Glasa, M. High-Throughput Sequencing Reveals Bell Pepper Endornavirus Infection in Pepper (Capsicum annum) in Slovakia and Enables Its Further Molecular Characterization. Plants 2020, 9, 41. [Google Scholar] [CrossRef] [PubMed]
- Alcaide, C.; Rabadán, M.P.; Moreno-Pérez, M.G.; Gómez, P. Implications of mixed viral infections on plant disease ecology and evolution. Adv. Virus Res. 2020, 106, 145–169. [Google Scholar] [CrossRef]
- Moreno, A.B.; López-Moya, J.J. When Viruses Play Team Sports: Mixed Infections in Plants. Phytopathology 2020, 110, 29–48. [Google Scholar] [CrossRef]
- Boonham, N.; Kreuze, J.; Winter, S.; van der Vlugt, R.; Bergervoet, J.; Tomlinson, J.; Mumford, R. Methods in virus diagnostics: From ELISA to next generation sequencing. Virus Res. 2014, 186, 20–31. [Google Scholar] [CrossRef]
- Blawid, R.; Silva, J.; Nagata, T. Discovering and sequencing new plant viral genomes by next-generation sequencing: Description of a practical pipeline. Ann. Appl. Biol. 2017, 170, 301–314. [Google Scholar] [CrossRef]
- Glais, L.; Tribodet, M.; Kerlan, C. Genomic variability in Potato potyvirus Y (PVY): Evidence that PVY(N)W and PVY(NTN) variants are single to multiple recombinants between PVY(O) and PVY(N) isolates. Arch. Virol. 2002, 147, 363–378. [Google Scholar] [CrossRef]
- Sihelská, N.; Predajňa, L.; Nagyová, A.; Šoltys, K.; Budiš, J.; Gubiš, J.; Mrkvová, M.; Kraic, J.; Mihálik, D.; Glasa, M. Detection and molecular characterization of Slovak tomato isolates belonging to two recombinant strains of potato virus Y. Acta Virol. 2016, 60, 347–353. [Google Scholar] [CrossRef]
- Hasiów-Jaroszewska, B.; Stachecka, J.; Minicka, J.; Sowiński, M.; Borodynko, N. Variability of Potato virus Y in Tomato Crops in Poland and Development of a Reverse-Transcription Loop-Mediated Isothermal Amplification Method for Virus Detection. Phytopathology 2015, 105, 1270–1276. [Google Scholar] [CrossRef]
- Nagy, P.D. Recombination in Plant RNA Viruses. In Plant Virus Evolution; Roossinck, M.J., Ed.; Springer: Berlin/Heidelberg, Germany, 2008. [Google Scholar] [CrossRef]
- Sztuba-Solińska, J.; Urbanowicz, A.; Figlerowicz, M.; Bujarski, J.J. RNA-RNA recombination in plant virus replication and evolution. Ann. Rev. Phytopathol. 2011, 49, 415–443. [Google Scholar] [CrossRef]
- MacKenzie, T.D.B.; Lavoie, J.; Nie, X.; Singh, M. Differential Spread of Potato virus Y (PVY) Strains O, N:O and NTN in the Field: Implications for the Rise of Recombinant PVY Strains in New Brunswick, Canada. Am. J. Potato Res. 2018, 95, 301–310. [Google Scholar] [CrossRef]
- MacKenzie, T.D.B.; Nie, X.; Bisht, V.; Singh, M. Proliferation of Recombinant PVY Strains in Two Potato-Producing Regions of Canada, and Symptom Expression in 30 Important Potato Varieties with Different PVY Strains. Plant Dis. 2019, 103, 2221–2230. [Google Scholar] [CrossRef]
- Chikh-Ali, M.; Alruwaili, H.; Vander Pol, D.; Karasev, A.V. Molecular Characterization of Recombinant Strains of Potato virus Y From Saudi Arabia. Plant Dis. 2016, 100, 292–297. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lorenzen, J.H.; Piche, L.M.; Gudmestad, N.C.; Meacham, T.; Shiel, P. A Multiplex PCR Assay to Characterize Potato virus Y Isolates and Identify Strain Mixtures. Plant Dis. 2006, 90, 935–940. [Google Scholar] [CrossRef] [PubMed]
- EFSA PLH Panel (EFSA Panel on Plant Health); Bragard, C.; Dehnen-Schmutz, K.; Gonthier, P.; Jacques, M.A.; Jaques Miret, J.A.; Justesen, A.F.; MacLeod, A.; Magnusson, C.S.; Milonas, P.; et al. Scientific Opinion on the pest categorisation of potato virus Y (non-EU isolates). EFSA J. 2020, 18, 5938. [Google Scholar] [CrossRef]
- Braidwood, L.; Müller, S.Y.; Baulcombe, D. Extensive recombination challenges the utility of Sugarcane mosaic virus phylogeny and strain typing. Sci. Rep. 2019, 9, 20067. [Google Scholar] [CrossRef] [PubMed]
- Visser, J.C.; Bellstedt, D.U.; Pirie, M.D. The Recent Recombinant Evolution of a Major Crop Pathogen, Potato virus Y. PLoS ONE 2012, 7, e50631. [Google Scholar] [CrossRef]
- Della Bartola, M.; Byrne, S.; Mullins, E. Characterization of Potato virus Y Isolates and Assessment of Nanopore Sequencing to Detect and Genotype Potato Viruses. Viruses 2020, 12, 478. [Google Scholar] [CrossRef]
- Ziebell, H.; Carr, J.P. Cross-Protection: A century of mystery. Adv. Virus Res. 2010, 76, 211–264. [Google Scholar] [CrossRef]
- Singh, M.; Singh, R.P. Host dependent cross-protection between PVYN, PVY° and PVA in potato cultivars and Solarium brachycarpum. Can. J. Plant Pathol. 1995, 17, 82–86. [Google Scholar] [CrossRef]
- Grupa, A.; Otulak-Kozieł, K.; Syller, J. Serological, molecular and immunofluorescent evidence for interference competition between isolates of Potato virus Y. Plant Pathol. 2018, 67, 1997–2012. [Google Scholar] [CrossRef]
- Pooggin, M.M. Small RNA-Omics for Plant Virus Identification, Virome Reconstruction, and Antiviral Defense Characterization. Front. Microbiol. 2018, 9, 2779. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
| Sample | Natural Host | Locality | Year of Sampling | Symptom on Leaves | Additional Viruses Identified in the Sample 1 |
|---|---|---|---|---|---|
| T101 | tomato | Pezinok | 2019 | leaf narrowing and deformation | CMV |
| T20 | tomato | Paňa | 2017 | mild leaf distortions | PVM |
| T24 | tomato | Nitra | 2017 | mosaics | CMV |
| T31 | tomato | Švošov | 2017 | symptomless | - |
| PAP | sweet pepper | Čachtice | 2019 | mosaics | RWMV |
| SL50V | tomato | Pezinok | 2018 | curling, mosaics, deformations | CMV |
| PAR-P2 | tomato | Pezinok | 2018 | Mosaics | LRNV |
| T1 | tomato | Paňa | 2017 | deformations, vein clearing | CMV |
| T40 | tomato | Plavecký Mikuláš | 2017 | symptomless | PVM, PVS |
| T62 | tomato | Sološnica | 2017 | curling | CMV, PVM, PVS |
| Sample | Number of Reads | Mean Length (bp) of Reads Mapping PVY | Number of Reads Mapped to the Determined Genome | Mean Sequence Depth | Accession Number | The Closest BLAST Relative (% of nt Identity) | PVY Strain 1 |
|---|---|---|---|---|---|---|---|
| T101 | 3,168,840 | 176.4 | 46,134 | 835.9 | MW595185 | KX713170 (Slovakia, tomato) 99.63% | N-Wi |
| T20 | 3,585,846 | 93.1 | 846,986 | 9196.7 | MW595182 | JF927752 (Poland, tobacco) 99.72% | NTNa |
| T24 | 2,215,328 | 116.7 | 53,849 | 670.5 | MW595183 | KX184818 (Israel, potato) 99.75% | NTNa |
| T31 | 2,666,282 | 129.7 | 58,843 | 793.6 | MW595184 | JF927761 (Poland, tobacco) 99.89% | NTNa |
| PAP | 542,034 | 149.7 | 57,741 | 948.1 | MW595181 | AB185833 (Syria, potato) 98.61% | NTNb |
| SL50V | 2,550,640 | 119.4 | 6075 | 80.5 | MW595187 | MH937417 (Germany, potato) 99.81% | NTNa |
| PAR-P2 | 3,912,270 | 185.8 | 147,409 | 2830.2 | MW595186 | KX184817 (Israel, potato) 99.75% | NTNa |
| nt 37-493 a | nt 704-2407 | nt 3001-4948 | nt 5880-8806 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample | Number of Reads | Mean Length of Reads Mapping PVY (bp) | O-Type Reads (Sequence Depth) | N-Type Reads (Sequence Depth) | O-Type Reads (Sequence Depth) | N-Type Reads (Sequence Depth) | O-Type Reads (Sequence Depth) | N-Type Reads (Sequence Depth) | O–Type Reads (Sequence Depth) | N-Type Reads (Sequence Depth) |
| T1 | 1,855,568 | 115.3 | 961 (250.6x) | 408 (102.4x) | - | 18,488 (1214.5x) | 12,684 (705.5x) | - | 12,276 (478.1x) | 8991 (357.2x) |
| T40 | 2,906,680 | 119.3 | 877 (242.3x) | 2139 (556.2x) | - | 41,975 (2811.3x) | 43,905 (2547.5x) | - | 14,569 (588.5x) | 44,405 (1775.4x) |
| T62 | 2,886,468 | 122.1 | 710 (196.2x) | 1179 (316.3x) | - | 25,723 (1753.9x) | 22,494 (1317.1x) | - | 9351 (381.7x) | 19,524 (795.4x) |
| Primer | Sequence (5′ → 3′) | Orientation | Genome Portion | Specific Target |
|---|---|---|---|---|
| PVY-O-127F | GGAAACCATTTCAACTCAAC | + | UTR-P1 | O |
| PVY-O-469-R | CTGGAAGTGATATTCTTCCC | − | O | |
| PVY-N-125F | GTGTAAGCTATCGTAATTCAG | + | N | |
| PVY-N-487R | AACACTTGACGCAGCCATTTG | − | N | |
| PVY-O-6320F | GCCCAAACAGTTTGTAGGCTG | + | NIa | O |
| PVY-O-6811R | GTAGTTCGTGGTGTGTTTGTTG | − | O | |
| PVY-N-6359F | GGAACGTCTGAAATGTATGGG | + | N | |
| PVY-N-6796R | CACATTATTCGCCAAGCTGTG | − | N |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Glasa, M.; Hančinský, R.; Šoltys, K.; Predajňa, L.; Tomašechová, J.; Hauptvogel, P.; Mrkvová, M.; Mihálik, D.; Candresse, T. Molecular Characterization of Potato Virus Y (PVY) Using High-Throughput Sequencing: Constraints on Full Genome Reconstructions Imposed by Mixed Infection Involving Recombinant PVY Strains. Plants 2021, 10, 753. https://doi.org/10.3390/plants10040753
Glasa M, Hančinský R, Šoltys K, Predajňa L, Tomašechová J, Hauptvogel P, Mrkvová M, Mihálik D, Candresse T. Molecular Characterization of Potato Virus Y (PVY) Using High-Throughput Sequencing: Constraints on Full Genome Reconstructions Imposed by Mixed Infection Involving Recombinant PVY Strains. Plants. 2021; 10(4):753. https://doi.org/10.3390/plants10040753
Chicago/Turabian StyleGlasa, Miroslav, Richard Hančinský, Katarína Šoltys, Lukáš Predajňa, Jana Tomašechová, Pavol Hauptvogel, Michaela Mrkvová, Daniel Mihálik, and Thierry Candresse. 2021. "Molecular Characterization of Potato Virus Y (PVY) Using High-Throughput Sequencing: Constraints on Full Genome Reconstructions Imposed by Mixed Infection Involving Recombinant PVY Strains" Plants 10, no. 4: 753. https://doi.org/10.3390/plants10040753
APA StyleGlasa, M., Hančinský, R., Šoltys, K., Predajňa, L., Tomašechová, J., Hauptvogel, P., Mrkvová, M., Mihálik, D., & Candresse, T. (2021). Molecular Characterization of Potato Virus Y (PVY) Using High-Throughput Sequencing: Constraints on Full Genome Reconstructions Imposed by Mixed Infection Involving Recombinant PVY Strains. Plants, 10(4), 753. https://doi.org/10.3390/plants10040753






