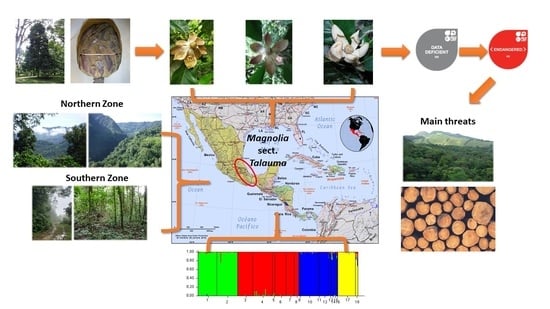
Disentangling Species Delineation and Guiding Conservation of Endangered Magnolias in Veracruz, Mexico
Abstract
1. Introduction
2. Results
2.1. Genetic Analysis and Characterisation
2.1.1. Null Alleles and Linkage Disequilibrium
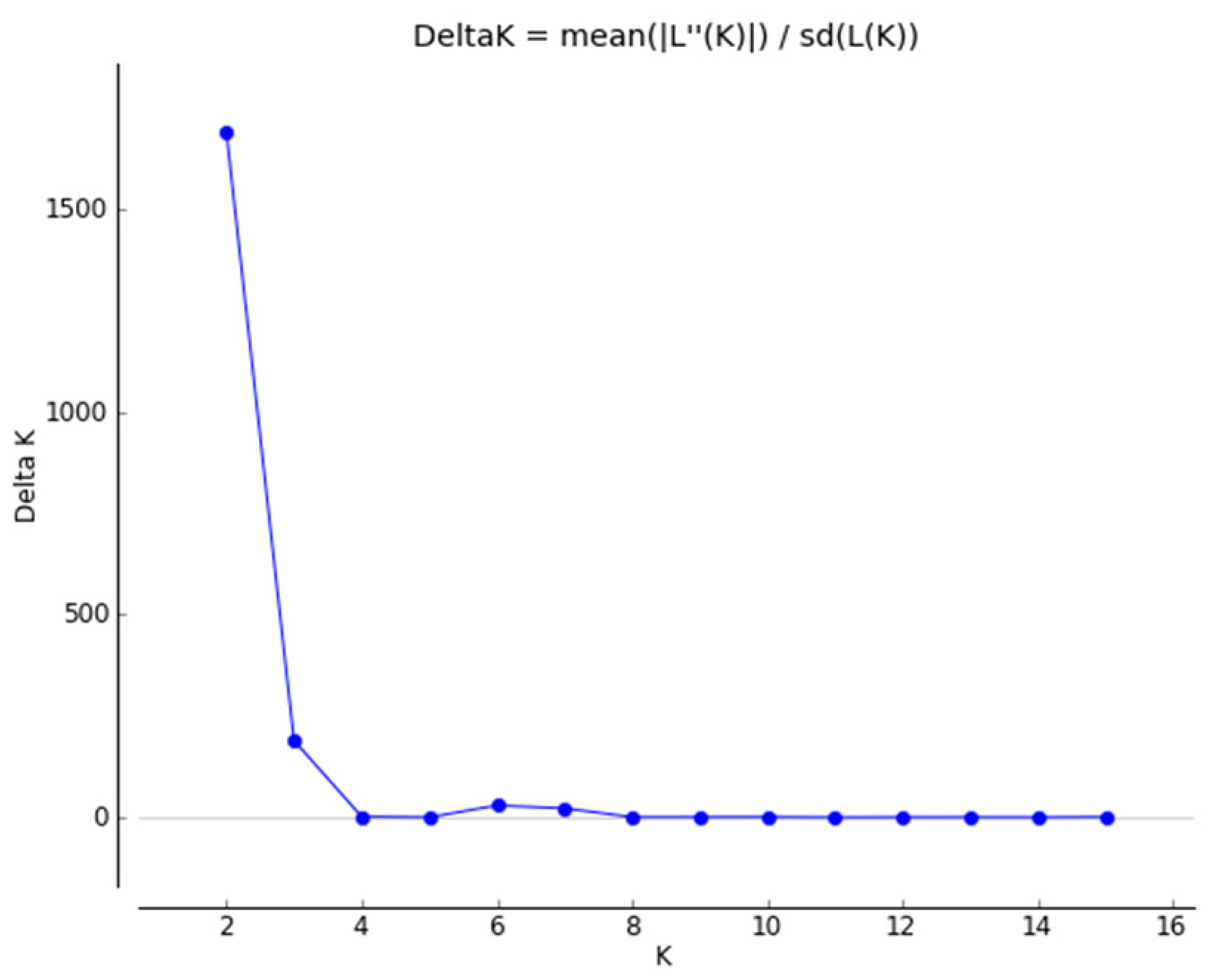
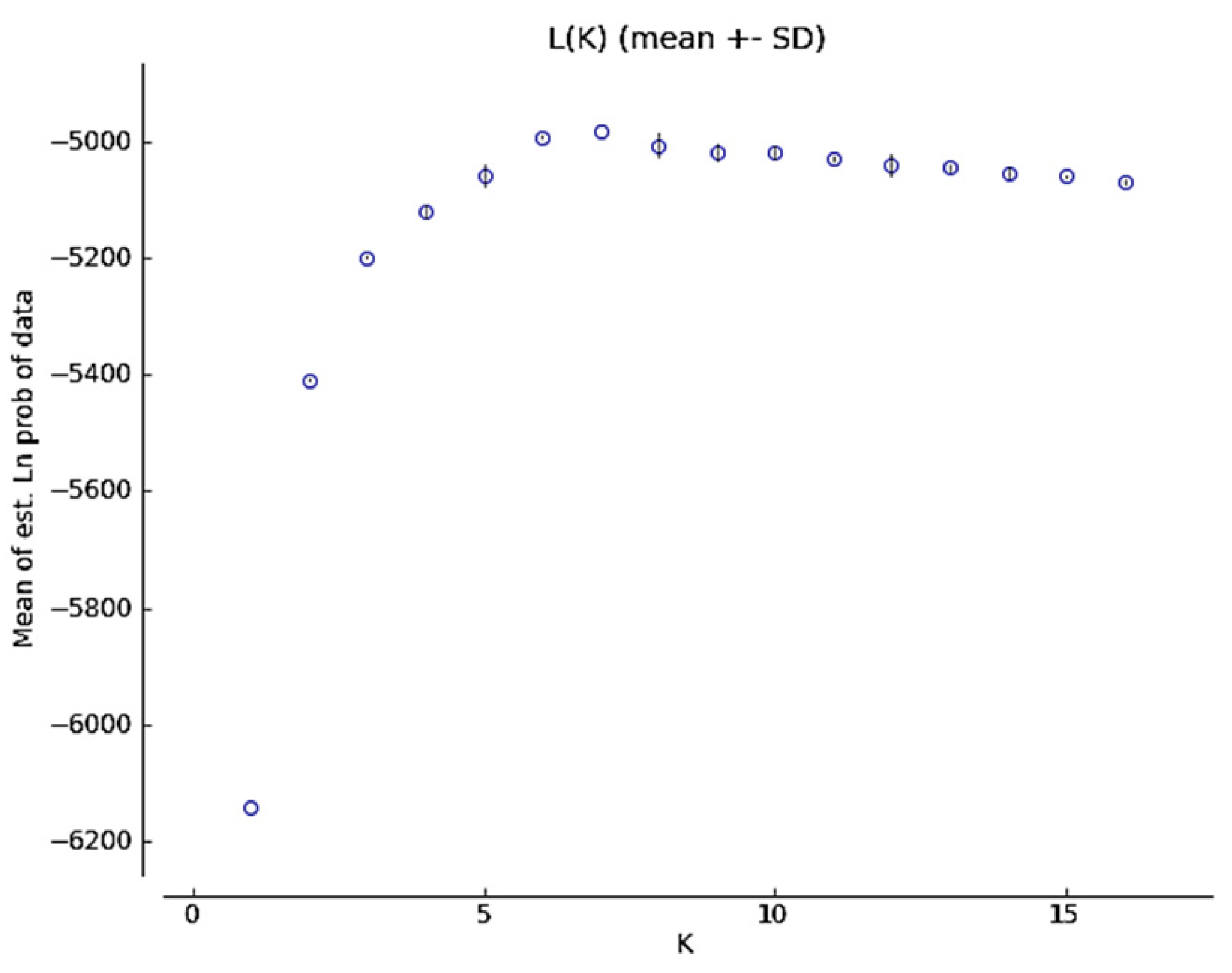
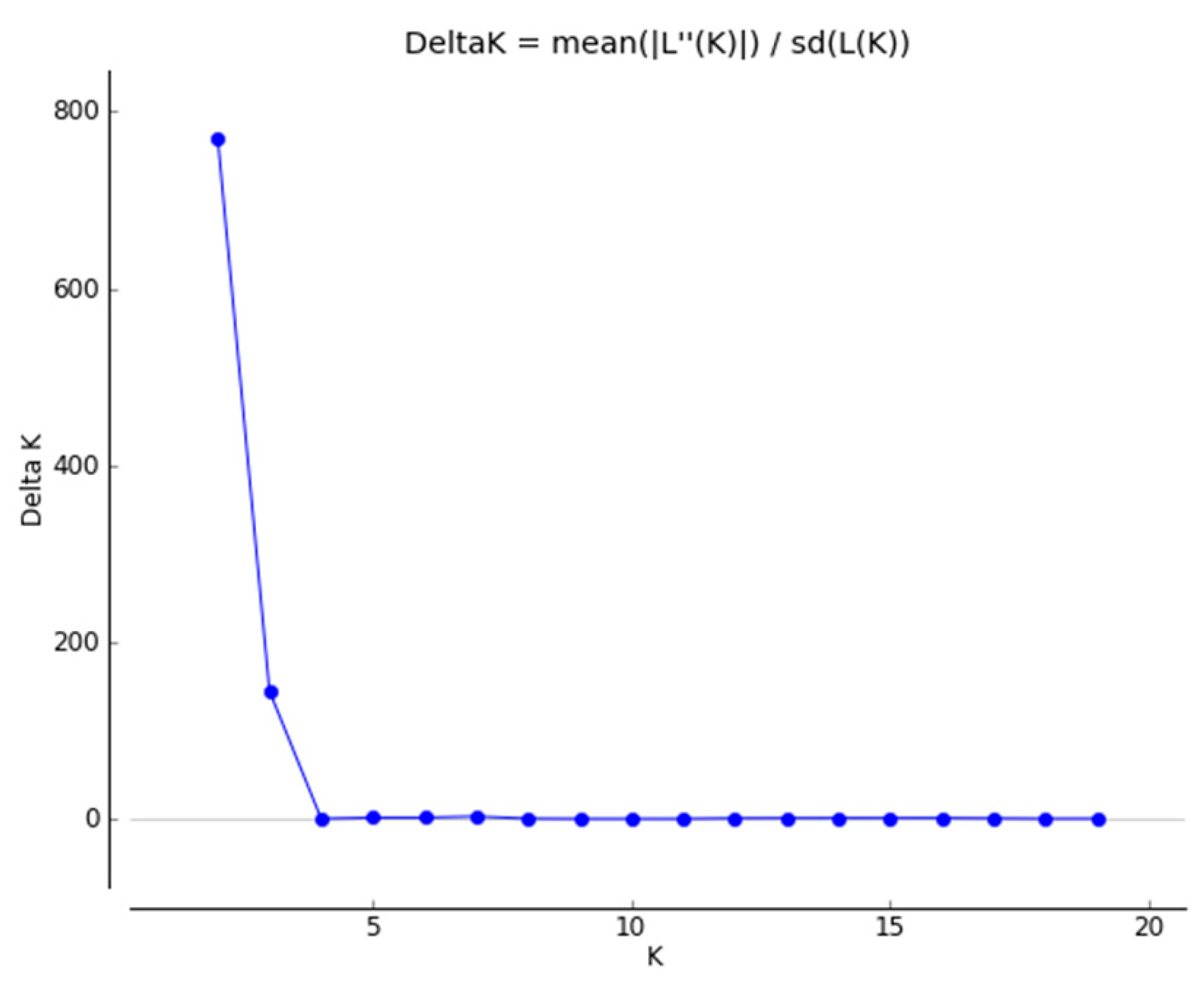
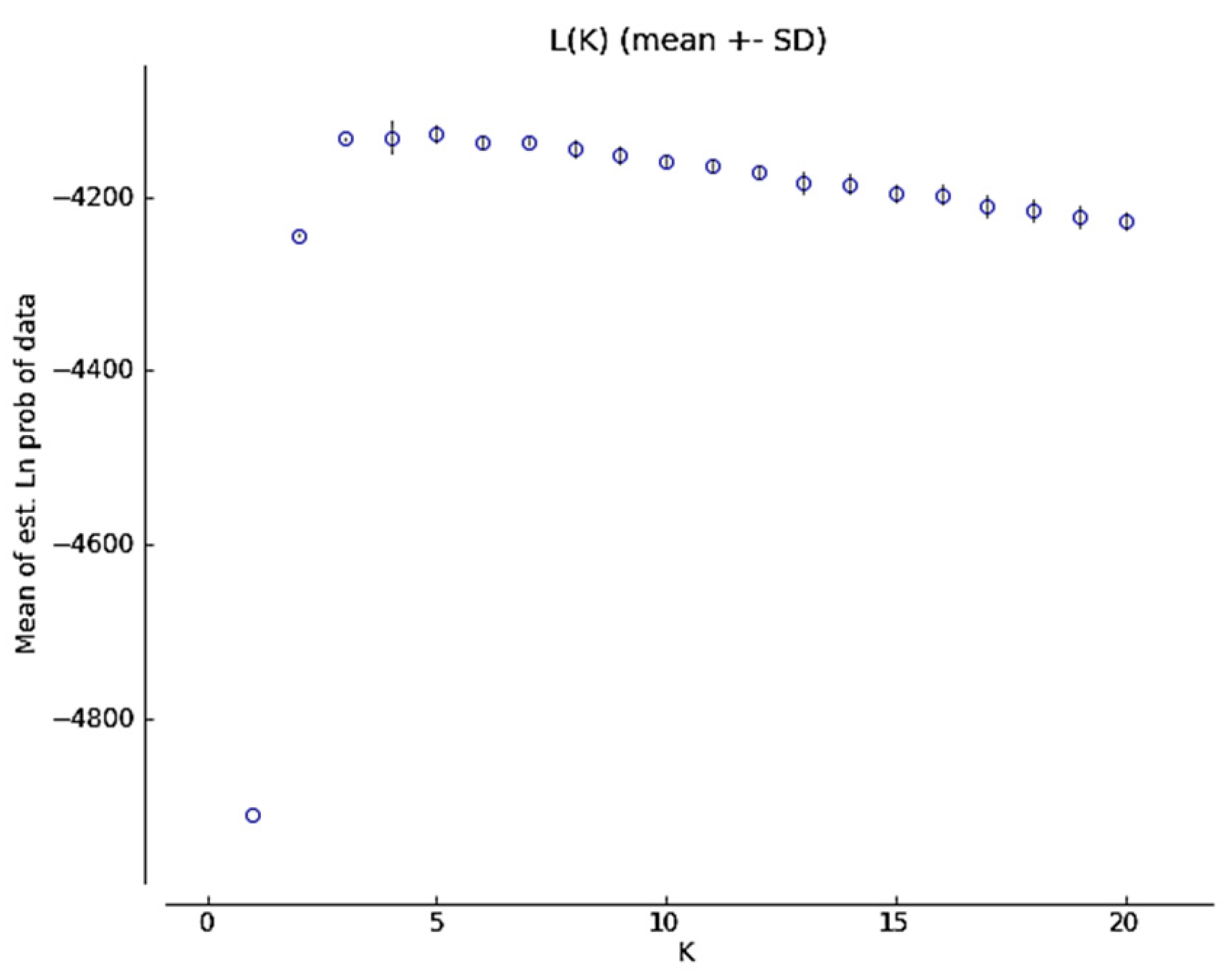
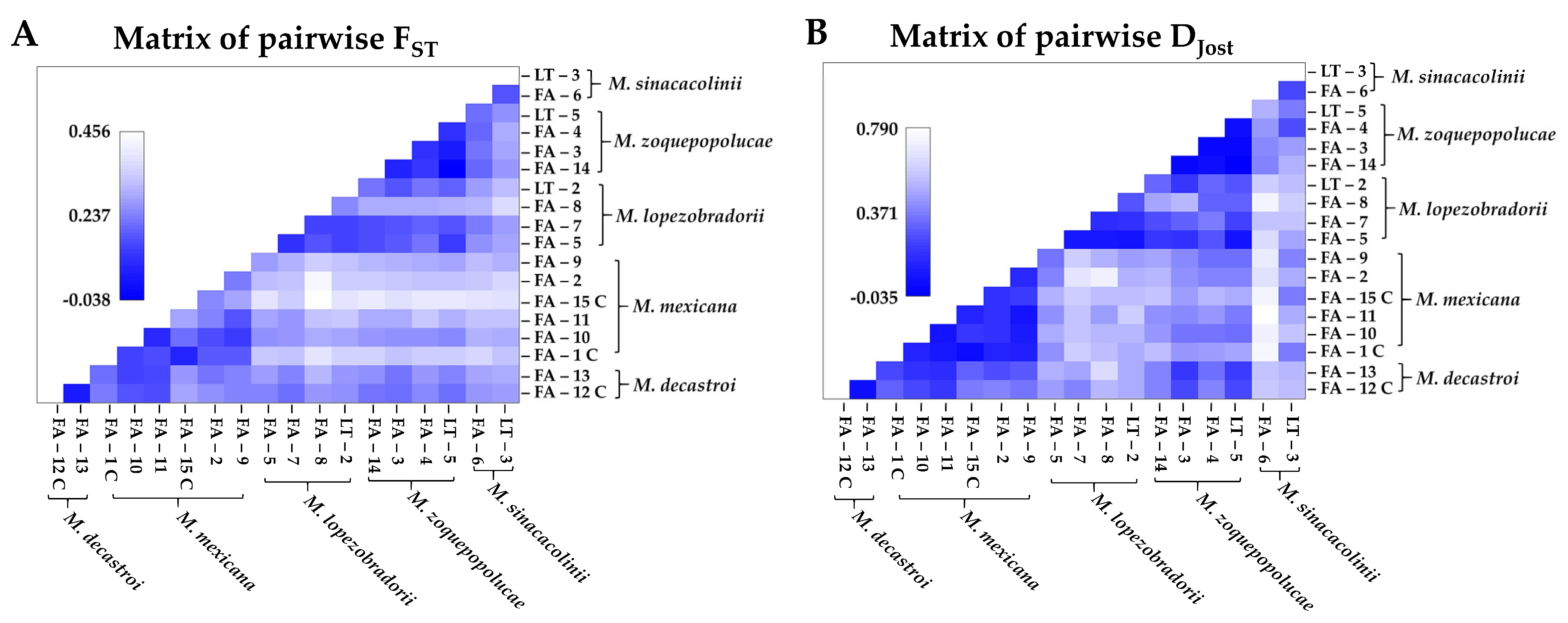
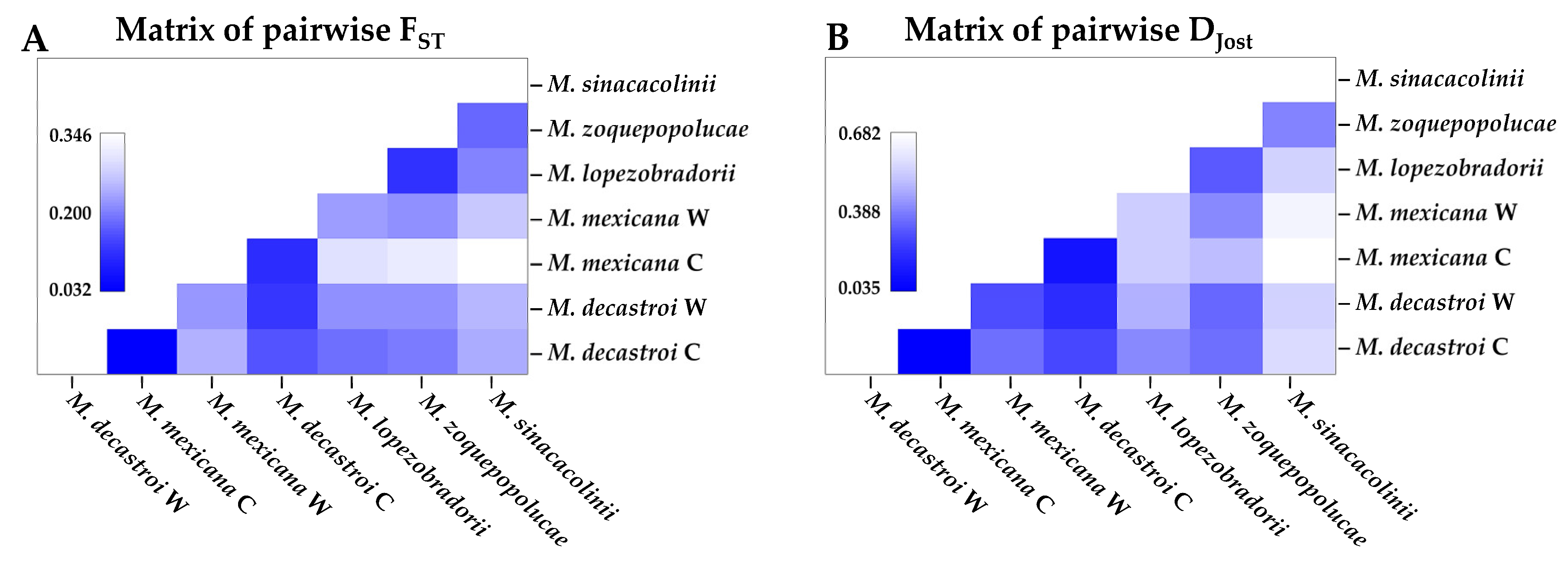
2.1.2. Genetic Structure
2.1.3. Genetic Diversity
2.2. Assessment of Conservation Status
3. Discussion
3.1. Disentangling the Species
3.2. Patterns of Gene Flow Between, and Inbreeding within the Wild Sample Localities
3.3. Defining the Conservation Units of the Magnolias of Veracruz
3.4. Priorities for Magnolia Conservation in Veracruz
3.5. Ex Situ Collections Versus In Situ Populations
3.6. Implications for Conservation
4. Materials and Methods
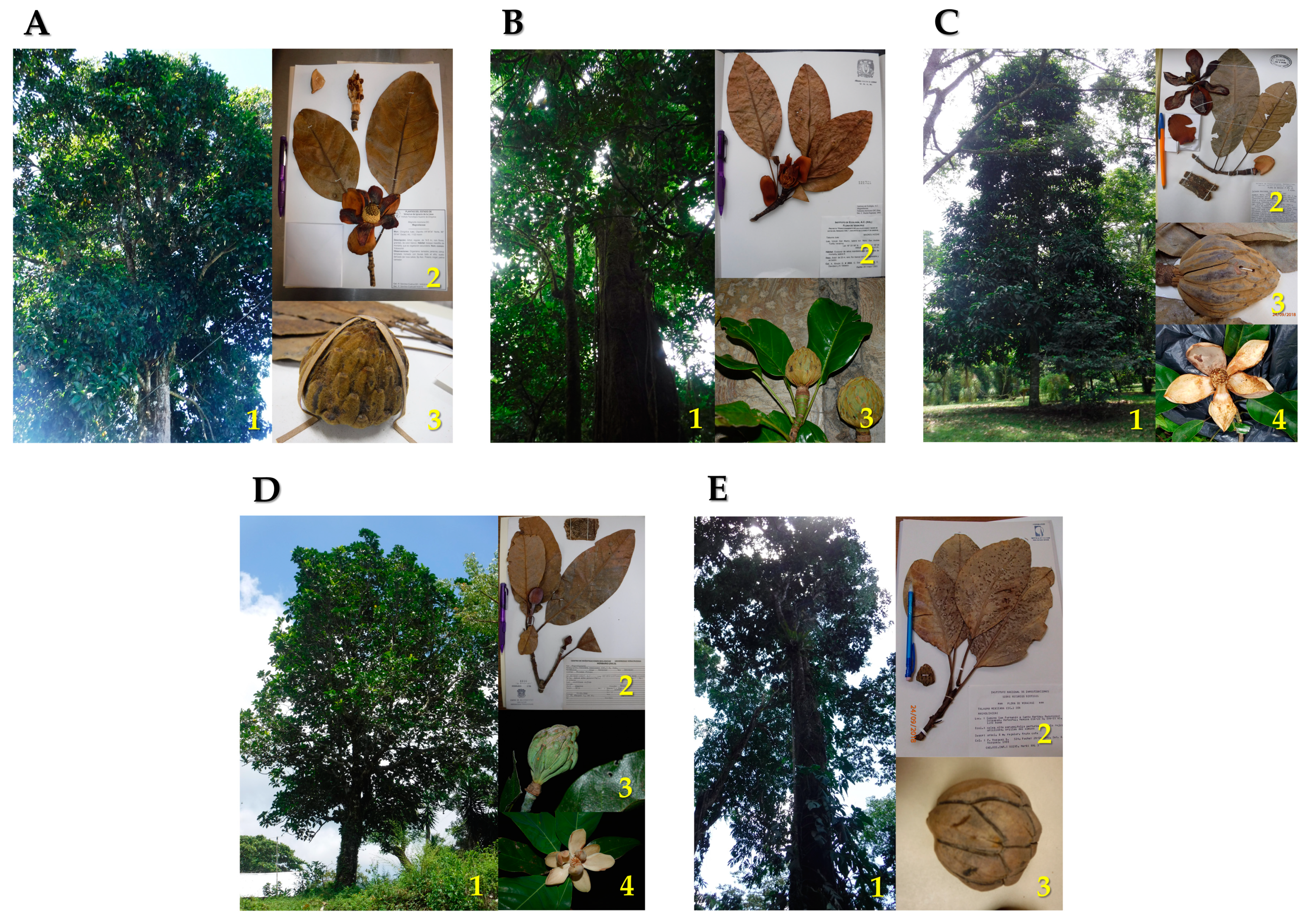
4.1. Study Species, Study Zones, and Sampling
4.2. DNA Extraction and PCR
4.3. Genetic Analysis and Characterisation
4.3.1. Null Alleles and Linkage Disequilibrium
4.3.2. Genetic Structure
4.3.3. Genetic Diversity
4.4. Assessment of Conservation Status
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Ceballos, G.; Ehrlich, P.R.; Barnosky, A.D.; García, A.; Pringle, R.M.; Palmer, T.M. Accelerated Modern Human-induced Species Losses: Entering the Sixth Mass Extinction. Sci. Adv. 2015, 1, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Barnosky, A.D.; Matzke, N.; Tomiya, S.; Wogan, G.O.U.; Swartz, B.; Quental, T.B.; Marshall, C.; McGuire, J.L.; Lindsey, E.L.; Maguire, K.C.; et al. Has the Earth’s Sixth Mass Extinction Already Arrived? Nature 2011, 471, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Morrone, J.J. Regionalización Biogeográfica y Evolución Biótica de México: Encrucijada de la Biodiversidad del Nuevo Mundo. Rev. Mex. Biodivers. 2019, 90, 1–68. [Google Scholar] [CrossRef]
- Llorente-Bousquets, J.; Ocegueda, S. Estado del Conocimiento de la Biota. In Capital Natural de México: Conocimiento Actual de la Biodiversidad; Comisión Nacional para el Conocimiento y Uso de la Biodiversidad (CONABIO): Tlalpan, Mexico, 2008; Volume I, pp. 283–322. ISBN 978-607-7607-03-8. [Google Scholar]
- Villaseñor, J.L. Checklist of the Native Vascular Plants of Mexico. Rev. Mex. Biodivers. 2016, 87, 559–902. [Google Scholar] [CrossRef]
- Lorea-Hernández, F.G.; Durán Espinosa, C.; Gallardo Hernández, C.; Peredo Nava, M. La Diversidad de las Plantas con Semillas de la flora Veracruzana. In La Biodiversidad en Veracruz: Estudio de Estado; Comisión Nacional para el Conocimiento y Uso de la Biodiversidad (CONABIO): Tlalpan, Mexico, 2012; Volume II, pp. 201–211. [Google Scholar]
- Gómez-Pompa, A.; Krömer, T.; Castro-Cortés, R. Atlas de la Flora de Veracruz: Un Patrimonio Natural en Peligro; Comisión del Estado de Veracruz para la Conmemoración de la Independencia Nacional y de la Revolución Mexicana: Tlalnepantla, México, 2010; ISBN 9786079513177. [Google Scholar]
- Ellis, E.A.; Martínez Bello, M.; Monroy Ibarra, R. Focos Rojos para la Conservación de la Biodiversidad. In La Biodiversidad en Veracruz; Cruz Aragón, A., Ed.; Comisión Nacional para el Conocimiento y Uso de la Biodiversidad (CONABIO): Tlalpan, Mexico, 2011; Volume I, pp. 351–367. [Google Scholar]
- Arroyo-Rodríguez, V.; Mandujano, S.; Benítez-Malvido, J. Diversidad y Estructura de la Vegetación en Fragmentos de Selva de Los Tuxtlas. In Biodiversidad de Veracruz. Estudio de Estado; Cruz Angón, A., Ed.; Comisión Nacional para el Conocimiento y Uso de la Biodiversidad (CONABIO): Tlalpan, Mexico, 2011; pp. 235–242. [Google Scholar]
- Castillo-Hernández, L.A.; Flores-Olvera, H. Floristic Composition of the Cloud Forest of the Bicentenario Reserve, Zongolica, Veracruz, México. Bot. Sci. 2017, 95, 1–25. [Google Scholar] [CrossRef][Green Version]
- González-Soriano, E.; Dirzo, R.; Vogt, R. Historia Natural de Los Tuxtlas; Instituto de Biología, UNAM: Mexico City, Mexico, 1997; ISBN 968-36-5646-3. [Google Scholar]
- Vázquez-Torres, M.; Campos-Jimenez, J. Árboles de la Región de Los Tuxtlas; Gobierno del Estado de Veracruz, Secretaría de Educación del Estado de Veracruz, Comisión del Estado de Veracruz de Ignacio de la Llave para la Conmemoración del Bicentenario de la Independencia Nacional y del Centenario de la Revolución: Xalapa, Veracruz, 2010; ISBN 9786073300070. [Google Scholar]
- Diario Oficial de la Federación. Secretaría de Medio Ambiente, Recursos Naturales y Pesca. 1998. Decreto por el que se Declara Área Natural Protegida con el Carácter de Reserva de la Biósfera de la Región Denominada Los Tuxtlas, Ubicada en los Municipios de Angel R. Cabada, Catemaco, Mecayapan, Pajapan, San Andrés Tuxtla, Santiago Tuxtla, Soteapan y Tatahuicapan de Juárez, en el Estado de Veracruz, con una Superficie Total de 155 122 hectáreas. Available online: https://www.dof.gob.mx/nota_detalle.php?codigo=4900167&fecha=23/11/1998 (accessed on 11 December 2020).
- Salazar-Arteaga, H. Estructura y Conectividad del Paisaje en la Reserva de la Biósfera Los Tuxtlas. Master’s Thesis, Instituto de Ecología, A.C., Xalapa, Mexico, 2015. [Google Scholar]
- Rivers, M.; Beech, E.; Murphy, L.; Oldfield, S. The Red List of Magnoliaceae: Revised and Extended; Botanic Gardens Conservation International, 2016; ISBN 9781905164646. [Google Scholar]
- Cires, E.; De Smet, Y.; Cuesta, C.; Goetghebeur, P.; Sharrock, S.; Gibbs, D.; Oldfield, S.; Kramer, A.; Samain, M.-S. Gap Analyses to Support ex situ Conservation of Genetic Diversity in Magnolia, a Flagship Group. Biodivers. Conserv. 2013, 22, 567–590. [Google Scholar] [CrossRef]
- Simberloff, D. Flagships, Umbrellas, and Keystones: Is Single-species Management Passe in the Landscape Era? Biol. Conserv. 1998, 83, 247–257. [Google Scholar] [CrossRef]
- Azuma, H.; García-Franco, J.G.; Rico-Gray, V.; Thien, L.B. Molecular Phylogeny of the Magnoliaceae: The Biogeography of Tropical and Temperate Disjunctions. Am. J. Bot. 2001, 88, 2275–2285. [Google Scholar] [CrossRef]
- Wang, Y.-B.; Liu, B.-B.; Nie, Z.-L.; Chen, H.-F.; Chen, F.-J.; Figlar, R.B.; Wen, J. Major Clades and a Revised Classification of Magnolia and Magnoliaceae Based on Whole Plastid Genome Sequences via Genome Skimming. J. Syst. Evol. 2020, 58, 673–695. [Google Scholar] [CrossRef]
- Nie, Z.L.; Wen, J.; Azuma, H.; Qiu, Y.L.; Sun, H.; Meng, Y.; Sun, W.B.; Zimmer, E.A. Phylogenetic and Biogeographic Complexity of Magnoliaceae in the Northern Hemisphere Inferred from Three Nuclear Data Sets. Mol. Phylogenet. Evol. 2008, 48, 1027–1040. [Google Scholar] [CrossRef]
- Rico, Y.; Gutiérrez Becerril, B.A. Species Delimitation and Genetic Structure of Two Endemic Magnolia species (section Magnolia; Magnoliaceae) in Mexico. Genetica 2019, 147, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Hernández, M.; Palmarola, A.; Veltjen, E.; Asselman, P.; Testé, E.; Larridon, I.; Samain, M.S.; González-Torres, L.R. Population Structure and Genetic Diversity of Magnolia cubensis subsp. acunae (Magnoliaceae): Effects of Habitat Fragmentation and Implications for Conservation. Oryx 2020. [Google Scholar] [CrossRef]
- Castillo, R.E.; Encarnación, Y.; Peguero, B.; Clase, T.; Gratzfeld, J. Plan de Acción de Conservación integrada de las Magnolias (Magnoliaceae) Amenazadas de República Dominicana—Magnolia domingensis, M. hamorii, y M. pallescens; Fundación PROGRESSIO y Jardín Botánico Nacional Dr. Rafael M. Moscoso: Santo Domingo, Dominican Republic, 2018. [Google Scholar]
- BGCI Global Survey of ex situ Magnoliaceae Collections; Botanic Gardens Conservation International: London, UK, 2008.
- Figlar, R.B.; Nooteboom, H.P. Notes on Magnoliaceae IV. Blumea J. Plant Taxon. Plant Geogr. 2004, 49, 87–100. [Google Scholar] [CrossRef]
- Vázquez-García, J.A.; De Castro-Arce, E.; Múñiz-Castro, M.Á.; Cházaro-Basáñez, M.d.J. Magnolia zoquepopolucae (subsection Talauma, Magnoliaceae), a New Species from Sierra de Santa Marta, Veracruz, Mexico. Phytotaxa 2012, 57, 51–55. [Google Scholar] [CrossRef]
- Vázquez-García, J.A.; Múñiz-Castro, M.Á.; Arroyo, F.; Pérez-Castañeda, Á.J.; Serna, M.; Cuevas Guzmán, R.; Domínguez-Yescas, R.; De Castro-Arce, E.; Gurrola-Díaz, C.M. Novelties in Neotropical Magnolia and an Addendum Proposal to the IUCN Red List of Magnoliaceae. In Recursos forestales en el Occidente de México: Diversidad, Manejo, Producción, Aprovechamiento y Conservación; Salcedo-Pérez, E., Hernández-Álvarez, E., Vázquez-García, J.A., Escoto-García, T., Días-Echavarría, N., Eds.; Universidad de Guadalajara: Guadalajara, Mexico, 2013; pp. 458–496. ISBN 9786078072569. [Google Scholar]
- Vázquez-García, J.A.; Múñiz-Castro, M.Á.; De Castro-Arce, E.; Murguía Araiza, R.; Nuño Rubio, A.T.; Cházaro-Basáñez, M.d.J. Twenty New Neotropical Tree Species of Magnolia (Magnoliaceae). In Recursos Forestales en el Occidente de México: Diversidad, Manejo, Producción, Aprovechamiento y Conservación; Salcedo-Pérez, E., Hernández-Álvarez, E., Vázquez-García, J.A., Escoto-García, T., Días-Echavarría, N., Eds.; Universidad de Guadalajara: Guadalajara, México, 2012; pp. 91–130. [Google Scholar]
- Pennington, T.D.; Sarukhán, J. Arboles Tropicales de México. Manual para la Identificación de las Principales Especies; Fondo de Cultura Económica: Mexico City, Mexico, 2005; ISBN 9789703216437. [Google Scholar]
- Vázquez-García, J.A.; Tribouillier-Navas, E.; Archila, F.; Véliz-Pérez, M.E. A Conspectus of Magnolia (Magnoliaceae) in Guatemala: Novelties and Conservation. Phytotaxa 2019, 427, 221–238. [Google Scholar] [CrossRef]
- IUCN Red List of Threatened Species. Version 2020-3. Available online: https://www.iucnredlist.org/ (accessed on 8 February 2021).
- Vázquez-García, J.A.; Gómez-Domínguez, H.; López-Cruz, A. Magnolia perezfarrerae, a New Species and a Key to Mexican Species of Magnolia (section Talauma, subsection Talauma, Magnoliaceae). Bot. Sci. 2013, 91, 417–425. [Google Scholar] [CrossRef]
- Domínguez-Yescas, R.; Vázquez-García, J.A. Flower of the heart, Magnolia yajlachhi (subsect. Talauma, Magnoliaceae), a New Species of Ceremonial, Medicinal, Conservation and Nurse Tree Relevance in the Zapotec Culture, Sierra Norte de Oaxaca, Mexico. Phytotaxa 2019, 393, 21–34. [Google Scholar] [CrossRef]
- McVaugh, R. Botanical Results of the Sessé & Mociño Expedition. VII. A Guide to Relevant Scientific Names of Plants; Hunt Institute for Botanical Documentation; Carnegie Mellon University: Pittsburgh, PA, USA, 2000. [Google Scholar]
- Sutherland, C.N. Material tipo de la colección de Sessé y Mociño en el Real Jardín Botánico de Madrid. An. del Jardín Botánico Madrid 1997, 55, 375–418. [Google Scholar]
- Germán-Ramírez, M.T. Iconografía Inédita de la Flora Mexicana, Obra de Sessé y Mociño, en el Acervo Histórico del Herbario Nacional de México, MEXU. Bot. Sci. 1994, 97, 67. [Google Scholar] [CrossRef]
- McVaugh, R. El Itinerario y las Colectas de Sessé y Mociño en México. Bot. Sci. 1969, 30, 137–142. [Google Scholar] [CrossRef][Green Version]
- Hernández-Cerda, M.E. Magnoliaceae. Flora de Veracruz 1988, 14, 1–38. [Google Scholar]
- Duminil, J.; Di Michele, M. Plant Species Delimitation: A Comparison of Morphological and Molecular Markers. Plant Biosyst. 2009, 143, 528–542. [Google Scholar] [CrossRef]
- González, D. El Uso de Secuencias Génicas para Estudios Taxonómicos. Bot. Sci. 1997, 157, 137. [Google Scholar] [CrossRef]
- Rentaría-Alcántara, M. Breve Revisión de los Marcadores Moleculares. In Ecología molecular; Eguiarte, L.E., Souza, V., Aguirre, X., Eds.; Secretaría de Medio Ambiente y Recursos Naturales de los Estados Unidos Mexicanos (SEMARNAT): Mexico City, Mexico, 2007; pp. 541–566. [Google Scholar]
- Veltjen, E.; Asselman, P.; Hernández Rodríguez, M.; Palmarola Bejerano, A.; Testé Lozano, E.; González Torres, L.R.; Goetghebeur, P.; Larridon, I.; Samain, M.-S. Genetic Patterns in Neotropical Magnolias (Magnoliaceae) Using de novo Developed Microsatellite Markers. Heredity (Edinb.) 2019, 122, 485–500. [Google Scholar] [CrossRef]
- Medrano-Hernández, J.M.; Rodríguez de la O, J.L.; Reyes-Trejo, B.; Peña-Ortega, M.G. Molecular Characterization Using ISSR Primers of Magnolia mexicana DC. from Two Regions in Zongolica, Veracruz, Mexico. Rev. Chapingo Ser. Ciencias For. y del Ambient. 2017, 23, 427–436. [Google Scholar] [CrossRef]
- Arteaga-Rios, L.D.; Mejía-Carraza, J.; Piña-Escutia, J.L.; González-Díaz, J.G.; Rivera-Colín, A. Comparación Molecular y Morfológica entre Ejemplares de Magnolia mexicana “Yoloxóchitl” del Estado de México y Veracruz. Polibotánica 2020, 48, 107–124. [Google Scholar] [CrossRef]
- Li, J.; Conran, J.G. Phylogenetic Relationships in Magnoliaceae subfam. Magnolioideae: A Morphological Cladistic Analysis. Plant Syst. Evol. 2003, 242, 33–47. [Google Scholar] [CrossRef]
- Kim, S.; Suh, Y. Phylogeny of Magnoliaceae Based on Ten Chloroplast DNA Regions. J. Plant Biol. 2013, 56, 290–305. [Google Scholar] [CrossRef]
- Aldaba Núñez, F.A. Sistemática, Diversidad Genética y Conservación de Magnolia en Veracruz, México. Master’s Thesis, Instituto de Ecología, A.C., México, Xalapa, Mexico, 2020. [Google Scholar]
- Waples, R.S. Separating the Wheat from the Chaff: Patterns of Genetic Differentiation in High Gene Flow Species. J. Hered. 1998, 89, 438–450. [Google Scholar] [CrossRef]
- Hamrick, J.L.; Godt, M.J.W.; Sherman-Broyles, S.L. Factors Influencing Levels of Genetic Diversity in Woody Plant Species. New For. 1992, 6, 95–124. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the Analysis of Population Structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef] [PubMed]
- Jost, L. GST and its Relatives do not Measure Differentiation. Mol. Ecol. 2008, 17, 4015–4026. [Google Scholar] [CrossRef]
- Aldaba Núñez, F.A.; Fuentes, A.C.D.; Martínez Salas, E.; Samain, M.-S. Magnolia sinacacolinii; The IUCN Red List of Threatened Species 2020: Gland, Switzerland, 2020; e.T67513605A176997455. [Google Scholar] [CrossRef]
- Aldaba Núñez, F.A.; Fuentes, A.C.D.; Martínez Salas, E.; Samain, M.-S. Magnolia zoquepopolucae; The IUCN Red List of Threatened Species 2020: Gland, Switzerland, 2020; e.T67513642A176997582. [Google Scholar] [CrossRef]
- de Queiroz, K. The General Lineage Concept of species, Species Criteria, and the Process of Speciation and Terminological Recommendations. Endless Forms Species Speciat. 1998, 57–75. [Google Scholar]
- Lacy, R.C. Loss of Genetic Diversity from Managed Populations: Interacting Effects of Drift, Mutation, Immigration, Selection, and Population Subdivision. Conserv. Biol. 1987, 1, 143–158. [Google Scholar] [CrossRef]
- Wang, J. On the Measurements of Genetic Differentiation among Populations. Genet. Res. (Camb.) 2012, 94, 275–289. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software STRUCTURE: A Simulation Study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Chávez-Cortázar, A.; Oyama, K.; Zavala-Ochoa, M.; Mata-Rosas, M.; Veltjen, E.; Samain, M.-S.; Quesada, A.M. Conservation Genetics of Relict Tropical Species of Magnolia (section Macrophylla). Conserv. Genet. 2021, 22, 259–273. [Google Scholar] [CrossRef]
- Hartl, D.; Clark, A. Principles of Population Genetics; Sinauer Associates, Inc.: Sunderland, MA, USA, 1997. [Google Scholar]
- Aparicio, A.; Hampe, A.; Fernández-Carrillo, L.; Albaladejo, R.G. Fragmentation and Comparative Genetic Structure of Four Mediterranean Woody Species: Complex interactions between life history traits and the landscape context. Divers. Distrib. 2012, 18, 226–235. [Google Scholar] [CrossRef]
- Petit, R.J.; Hampe, A. Some Evolutionary Consequences of Being a Tree. Annu. Rev. Ecol. Evol. Syst. 2006, 37, 187–214. [Google Scholar] [CrossRef]
- Tamaki, I.; Setsuko, S.; Tomaru, N. Estimation of Outcrossing Rates at Hierarchical Levels of Fruits, Individuals, Populations and Species in Magnolia stellata. Heredity (Edinb.) 2009, 102, 381–388. [Google Scholar] [CrossRef]
- Antonovics, J. The Effects of a Heterogeneous Environment on the Genetics of Natural Populations: The Realization That Environments Differ Has Had a Profound Effect on Our Views of the Origin and Role of Genetic Variability in Populations. Am. Sci. 1971, 59, 593–599. [Google Scholar] [PubMed]
- Hamrick, J.L.; Linhart, Y.B.; Mitton, J.B. Relationships between Life History Characteristics and Electrophoretically Detectable Genetic Variation in Plants. Annu. Rev. Ecol. Syst. 1979, 10, 173–200. [Google Scholar] [CrossRef]
- Hamrick, J.L.; Murawski, D.A. The Breeding Structure of Tropical Tree Populations. Plant Species Biol. 1990, 5, 157–165. [Google Scholar] [CrossRef]
- Loveless, M.D. Isozyme Variation in Tropical Trees: Patterns of Genetic Organization. New For. 1992, 6, 67–94. [Google Scholar] [CrossRef]
- López-Barrera, G. Diversidad y Estructura Genética de Brosimum alicastrum. Master’s Thesis, Universidad Nacional Autónoma de México, Mexico City, Mexico, 2018. [Google Scholar]
- Coates, D.J.; Byrne, M.; Moritz, C. Genetic Diversity and Conservation Units: Dealing with the Species-population Continuum in the Age of Genomics. Front. Ecol. Evol. 2018, 6, 1–13. [Google Scholar] [CrossRef]
- Taylor, B.L.; Dizon, A.E. First Policy then Science: Why a Management Unit Based Solely on Genetic Criteria Cannot Work. Mol. Ecol. 1999, 8, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Veltjen, E. The Caribbean Magnolia Species (Magnoliaceae): Assessment of the Genetic Diversity and the Underlying Evolutionary History. Ph.D. Thesis, Ghent University, Ghent, Belgium, 2020. [Google Scholar]
- IUCN. Documentation Standards and Consistency Checks for IUCN Red List Assessments and Species Accounts. Version 2; IUCN: Cambridge, UK, 2013. [Google Scholar]
- Reyes-Chilpa, R.; Guzmán-Gutiérrez, S.L.; Campos-Lara, M.; Bejar, E.; Osuna-Fernández, H.R.; Hernández-Pasteur, G. On the First Book of Medicinal Plants Written in the American Continent: The Libellus Medicinalibus Indorum Herbis from Mexico, 1552. A review. Bol. Latinoam. y del Caribe Plantas Med. y Aromat. 2020, 20, 1–27. [Google Scholar] [CrossRef]
- CONABIO. Estrategia Mexicana para la Conservación Vegetal 2012-2030; Comisión Nacional para el Conocimiento y Uso de la Biodiversidad (CONABIO): Mexico City, Mexico, 2012; ISBN 9786077607687. [Google Scholar]
- CONABIO-CONANP-SEMARNAT. Estrategia Mexicana para la Conservación Vegetal: Objetivos y Metas; CONABIO-CONANP-SEMARNAT: Mexico City, Mexico, 2008; ISBN 9786077607687. [Google Scholar]
- Hernández, M.; Testé, E.; Palmarola, A.; Albelo, N.; Moscoso, J.L.; Valle, O.; González-Torres, L.R. Conservación del «Mantequero» en Guamuhaya, a Diez Años de los Primeros Pasos. Bissea 2020, 14, 2. [Google Scholar]
- Palmarola, A.; Hernández, M. Proyecto de Conservación de Magnolias Cubanas. Bissea 2016, 10, 148. [Google Scholar] [CrossRef]
- Rodríguez-Acosta, M. Estrategia para los Jardines Botánicos Mexicanos; Asociación Mexicana de Jardines Botánicos, A. C.: Mexico City, Mexico, 2000; ISBN 968-7369-04-3. [Google Scholar]
- Vovides, A.P. The Mexican Living Cycad Collection at the Jardín Botánico Francisco Javier Clavijero, Xalapa. In Proceedings of the 4th Global Botanic Gardens Congress, Dublin, Irish Republic, June 2010; pp. 1–9. [Google Scholar]
- Palacios-Ríos, M. Cultivation of Tropical Pteridophytes in the Jardín Botánico “Francisco J. Clavijero” of Xalapa, Veracruz, México. In Fern Horticulture: Past, Present and Future Perspectives; Thomas, B.A., Ide, J., Eds.; Fern Gazette-Intercept: Andover, MA, USA, 1992; pp. 263–266. [Google Scholar]
- Salazar, V.M.; Mata, M. Micropropagación y Conservación de Orquídeas Mexicanas en el Jardín Botánico Clavijero. Lankesteriana 2003, 7, 151–153. [Google Scholar] [CrossRef]
- Edmands, S.; Timmerman, C.C. Modeling Factors Affecting the Severity of Outbreeding Depression. Conserv. Biol. 2003, 17, 883–892. [Google Scholar] [CrossRef]
- Hufford, K.M.; Krauss, S.L.; Veneklaas, E.J. Inbreeding and Outbreeding Depression in Stylidium hispidum: Implications for Mixing Seed Sources for Ecological Restoration. Ecol. Evol. 2012, 2, 2262–2273. [Google Scholar] [CrossRef]
- Peakbagger Sierra Madre Oriental. Available online: https://www.peakbagger.com/range.aspx?rid=172 (accessed on 1 August 2020).
- Peakbagger Sierra Madre del Sur. Available online: https://www.peakbagger.com/range.aspx?rid=175 (accessed on 1 August 2020).
- Stacy, L. Mexico and The United States; Marshall Cavendish: New York, NY, USA, 2002. [Google Scholar]
- Thiers, B. Index Herbariorum: A Global Directory of Public Herbaria and Associated Staff. Available online: http://sweetgum.nybg.org/science/ih/ (accessed on 18 December 2018).
- Larridon, I.; Walter, H.E.; Guerrero, P.C.; Duarte, M.; Cisternas, M.A.; Hernández, C.P.; Bauters, K.; Asselman, P.; Goetghebeur, P.; Samain, M.-S. An Integrative Approach to Understanding the Evolution and Diversity of Copiapoa (Cactaceae), a Threatened Endemic Chilean Genus from the Atacama Desert. Am. J. Bot. 2015, 102, 1506–1520. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Glaubitz, J.C. CONVERT: A User-friendly Program to Reformat Diploid Genotypic Data for Commonly Used Population Genetic Software Packages. Mol. Ecol. Notes 2004, 4, 309–310. [Google Scholar] [CrossRef]
- Coombs, J.A.; Letcher, B.H.; Nislow, K.H. Create: A Software to Create Input Files from Diploid Genotypic Data for 52 Genetic Software Programs. Mol. Ecol. Resour. 2008, 8, 578–580. [Google Scholar] [CrossRef]
- Lischer, H.E.L.; Excoffier, L. PGDSpider: An Automated Data Conversion Tool for Connecting Population Genetics and Genomics Programs. Bioinformatics 2012, 28, 298–299. [Google Scholar] [CrossRef] [PubMed]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. MICRO-CHECKER: Software for Identifying and Correcting Genotyping Errors in Microsatellite Data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Kalinowski, S.T.; Taper, M.L. Maximum Likelihood Estimation of the Frequency of Null Alleles at Microsatellite Loci. Conserv. Genet. 2006, 7, 991–995. [Google Scholar] [CrossRef]
- Raymond, M.; Rousset, F. GENEPOP (Version 1.2): Population Genetics Software for Exact Tests and Ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar] [CrossRef]
- Rousset, F. GENEPOP’007: A Complete Re-implementation of the GENEPOP Software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Rice, W.R. Analyzing Tables of Statistical Tests. Evolution (N. Y.) 1989, 43, 223–225. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of Sample Group Information. Mol. Ecol. Resour. 2009, 9, 1322–1332. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; von Holdt, B.M. STRUCTURE HARVESTER: A Website and Program for Visualizing STRUCTURE Output and Implementing the Evanno Method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Rosenberg, N.A. DISTRUCT: A Program for the Graphical Display of Population Structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- R Core Development Team. R: A Language and Environment for statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2008; Volume 2, ISBN 3-900051-07-0. [Google Scholar]
- Jombart, T.; Devillard, S.; Dufour, A.B.; Pontier, D. Revealing Cryptic Spatial Patterns in Genetic Variability by a New Multivariate Method. Heredity (Edinb.) 2008, 101, 92–103. [Google Scholar] [CrossRef]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant Analysis of Principal Components: A New Method for the Analysis of Genetically Structured Populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef]
- Miller, J.M.; Cullingham, C.I.; Peery, R.M. The influence of a priori grouping on inference of Genetic Clusters: Simulation Study and literature Review of the DAPC Method. Heredity (Edinb.) 2020, 125, 269–280. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin Suite ver 3.5: A New Series of Programs to perform Population Genetics Analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Keenan, K.; Mcginnity, P.; Cross, T.F.; Crozier, W.W.; Prodöhl, P.A. DiveRsity: An R Package for the Estimation and Exploration of Population Genetics Parameters and Their Associated Errors. Methods Ecol. Evol. 2013, 4, 782–788. [Google Scholar] [CrossRef]
- Goudet, J. FSTAT (Version 1.2): A Computer Program to Calculate F-Statistics. J. Hered. 1995, 86, 485–486. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenALEx 6.5: Genetic Analysis in Excel. Population Genetic Software for teaching and Research -An Update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Bachman, S.; Moat, J.; Hill, A.W.; de laTorre, J.; Scott, B. Supporting Red List Threat Assessments with GeoCAT: Geospatial Conservation Assessment Tool. Zookeys 2011, 150, 117–126. [Google Scholar] [CrossRef] [PubMed]
- IUCN Threats Classification Scheme (Version 3.2). Available online: https://www.iucnredlist.org/resources/threat-classification-scheme (accessed on 2 June 2020).



| Species | Locality | N | SSR Locus | ||||
|---|---|---|---|---|---|---|---|
| MA39_009 | MA39_224 | MA39_287 | MA42_421 | MA42_471 | |||
| M. decastroi | FA-12 (C) | 30 | No | YES (0.122) | No | No | YES (0.061) |
| FA-13 | 33 | No | YES (0.256) | No | No | No | |
| M. lopezobradorii | FA-5 | 3 | NA | NA | NA | NA | NA |
| FA-7 | 27 | No | YES (0.249) | No | No | No | |
| FA-8 | 3 | NA | NA | NA | NA | NA | |
| LT-2 | 14 | No | YES (0.219) | No | No | No | |
| M. mexicana | FA-1 (C) | 24 | No | YES (0.202) | No | No | No |
| FA-2 | 13 | No | YES (0.187) | No | No | No | |
| FA-9 | 7 | No | No (0.330) | No | No | No | |
| FA-10 | 31 | No | YES (0.369) | No | No | YES (0.011) | |
| FA-11 | 4 | No | No (0.202) | No | No | No | |
| FA-15 (C) | 18 | No | YES (0.396) | No | No | No | |
| M. sinacacolinii | FA-6 | 28 | YES (0.263) | YES (0.339) | YES (0.348) | YES (0.339) | No |
| LT-3 | 4 | NA | NA (0.278) | NA | NA | NA | |
| M. zoquepopolucae | FA-3 | 5 | No | YES (0.286) | No | No | No |
| FA-4 | 3 | NA | NA | NA | NA | NA | |
| FA-14 | 4 | No | YES (0.387) | No | No | No | |
| LT-5 | 3 | NA | NA | NA | NA | NA | |
| Species | M. decastroi | M. mexicana | M. lopezobradorii | M. zoquepopolucae | M. sinacacolinii | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Localities | FA-12 | FA-13 | FA-1 | FA-10 | FA-11 | FA-15 | FA-2 | FA-9 | FA-5 | FA-7 | FA-8 | LT-2 | FA-14 | FA-3 | FA-4 | LT-5 | FA-6 | LT-3 | |
| M. decastroi | FA-12 (30) (C) | – | 0.03 | 0.30 | 0.23 | 0.19 | 0.36 | 0.38 | 0.35 | 0.45 | 0.39 | 0.52 | 0.49 | 0.39 | 0.23 | 0.39 | 0.25 | 0.57 | 0.53 |
| FA-13 (33) | 0.03 | – | 0.23 | 0.17 | 0.15 | 0.31 | 0.25 | 0.29 | 0.51 | 0.46 | 0.63 | 0.49 | 0.38 | 0.19 | 0.34 | 0.21 | 0.56 | 0.52 | |
| M. mexicana | FA-1 (24) (C) | 0.21 | 0.18 | – | 0.12 | 0.08 | 0.02 | 0.12 | 0.11 | 0.43 | 0.58 | 0.53 | 0.49 | 0.53 | 0.43 | 0.45 | 0.47 | 0.75 | 0.37 |
| FA-10 (31) | 0.14 | 0.11 | 0.11 | – | 0.06 | 0.18 | 0.16 | 0.08 | 0.48 | 0.56 | 0.51 | 0.52 | 0.44 | 0.35 | 0.35 | 0.34 | 0.72 | 0.56 | |
| FA-11 (4) | 0.14 | 0.13 | 0.13 | 0.06 | – | 0.13 | 0.17 | 0.06 | 0.42 | 0.55 | 0.45 | 0.59 | 0.41 | 0.41 | 0.44 | 0.36 | 0.79 | 0.49 | |
| FA-15 (18) (C) | 0.26 | 0.24 | 0.06 | 0.19 | 0.26 | – | 0.17 | 0.20 | 0.50 | 0.59 | 0.53 | 0.54 | 0.54 | 0.45 | 0.52 | 0.48 | 0.73 | 0.36 | |
| FA-2 (13) | 0.23 | 0.20 | 0.15 | 0.13 | 0.22 | 0.22 | – | 0.14 | 0.39 | 0.66 | 0.72 | 0.50 | 0.51 | 0.42 | 0.39 | 0.39 | 0.65 | 0.48 | |
| FA-9 (7) | 0.22 | 0.22 | 0.16 | 0.11 | 0.14 | 0.26 | 0.21 | – | 0.35 | 0.58 | 0.50 | 0.46 | 0.46 | 0.40 | 0.33 | 0.33 | 0.70 | 0.39 | |
| M. lopezobradorii | FA-5 (3) | 0.21 | 0.25 | 0.32 | 0.24 | 0.27 | 0.38 | 0.30 | 0.26 | – | 0.07 | 0.06 | 0.06 | 0.19 | 0.17 | 0.28 | 0.06 | 0.63 | 0.47 |
| FA-7 (27) | 0.18 | 0.22 | 0.31 | 0.24 | 0.25 | 0.33 | 0.31 | 0.28 | 0.08 | – | 0.15 | 0.17 | 0.26 | 0.30 | 0.38 | 0.22 | 0.55 | 0.56 | |
| FA-8 (3) | 0.24 | 0.29 | 0.39 | 0.27 | 0.31 | 0.46 | 0.43 | 0.33 | 0.14 | 0.12 | – | 0.27 | 0.46 | 0.52 | 0.30 | 0.31 | 0.73 | 0.59 | |
| LT-2 (14) | 0.23 | 0.25 | 0.34 | 0.27 | 0.32 | 0.38 | 0.33 | 0.31 | 0.12 | 0.11 | 0.22 | – | 0.33 | 0.18 | 0.31 | 0.27 | 0.59 | 0.54 | |
| M. zoquepopolucae | FA-14 (4) | 0.20 | 0.23 | 0.34 | 0.24 | 0.28 | 0.40 | 0.34 | 0.29 | 0.14 | 0.13 | 0.28 | 0.19 | – | 0.00 | 0.03 | -0.03 | 0.39 | 0.49 |
| FA-3 (5) | 0.18 | 0.20 | 0.32 | 0.23 | 0.28 | 0.37 | 0.32 | 0.28 | 0.16 | 0.15 | 0.28 | 0.14 | 0.06 | – | 0.00 | 0.01 | 0.39 | 0.44 | |
| FA-4 (3) | 0.23 | 0.25 | 0.34 | 0.23 | 0.32 | 0.39 | 0.31 | 0.27 | 0.20 | 0.16 | 0.28 | 0.20 | 0.09 | 0.08 | – | 0.04 | 0.43 | 0.25 | |
| LT-5 (3) | 0.18 | 0.21 | 0.33 | 0.22 | 0.28 | 0.39 | 0.31 | 0.26 | 0.10 | 0.14 | 0.28 | 0.17 | -0.04 | 0.04 | 0.09 | – | 0.50 | 0.37 | |
| M. sinacacolinii | FA-6 (28) | 0.25 | 0.26 | 0.35 | 0.28 | 0.32 | 0.38 | 0.32 | 0.31 | 0.24 | 0.21 | 0.29 | 0.25 | 0.17 | 0.20 | 0.17 | 0.19 | – | 0.23 |
| LT-3 (4) | 0.25 | 0.28 | 0.31 | 0.27 | 0.31 | 0.38 | 0.35 | 0.28 | 0.26 | 0.25 | 0.36 | 0.30 | 0.25 | 0.26 | 0.27 | 0.24 | 0.15 | – | |
| Species | M. decastroi (C) | M. decastroi (W) | M. mexicana (C) | M. mexicana (W) | M. lopezobradorii | M. zoquepopolucae | M. sinacacolinii |
|---|---|---|---|---|---|---|---|
| M. decastroi (C) | – | 0.04 | 0.32 | 0.25 | 0.38 | 0.33 | 0.55 |
| M. decastroi (W) | 0.03 | – | 0.26 | 0.19 | 0.45 | 0.32 | 0.54 |
| M. mexicana (C) | 0.23 | 0.21 | – | 0.1 | 0.52 | 0.49 | 0.68 |
| M. mexicana (W) | 0.15 | 0.12 | 0.1 | – | 0.51 | 0.38 | 0.64 |
| M. lopezobradorii | 0.18 | 0.20 | 0.29 | 0.2 | – | 0.28 | 0.54 |
| M. zoquepopolucae | 0.19 | 0.21 | 0.31 | 0.21 | 0.11 | – | 0.36 |
| M. sinacacolinii | 0.23 | 0.25 | 0.35 | 0.26 | 0.19 | 0.17 | – |
| Species | Locality | N | Adults | C | NG | P | A | AR(12) | AR(24) | AR (14) | AP | HO | HE | FIS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M. decastroi | TOTAL | 63 | 10 | 30 | 63 | 100% | 5.7 | NA | NA | 4.85 | 8 | 0.67 | 0.67 | 0.01 |
| FA-12 | 30 | 0 | 30 | 30 | 100% | 5.62 | 4.81 | 5.16 | NA | 2 | 0.66 | 0.67 | 0.03 * | |
| FA-13 | 33 | 10 | 0 | 33 | 100% | 5.08 | 4.54 | 4.78 | NA | 1 | 0.68 | 0.64 | −0.04 | |
| M. lopezobradorii | TOTAL | 47 | 19 | 0 | 47 | 100% | 7.54 | NA | NA | 5.95 | 12 | 0.65 | 0.71 | 0.09 |
| FA-5 | 3 | 2 | 0 | 3 | 100% | 3.08 | NA | NA | NA | 0 | 0.62 | 0.5 | 0.08 | |
| FA-7 | 27 | 3 | 0 | 27 | 100% | 6.7 | 5.7 | 6.34 | NA | 4 | 0.70 | 0.69 | 0.00 | |
| FA-8 | 3 | 3 | 0 | 3 | 92.31% | 2.85 | NA | NA | NA | 2 | 0.67 | 0.49 | −0.17 | |
| LT-2 | 14 | 11 | 0 | 14 | 100% | 4.15 | 4.09 | NA | NA | 0 | 0.56 | 0.59 | 0.09 * | |
| M. mexicana | TOTAL | 97 | 42 | 27 | 97 | 100% | 5.62 | NA | NA | 4.57 | 5 | 0.52 | 0.60 | 0.14 |
| FA-2 | 13 | 5 | 0 | 13 | 100% | 3.46 | 3.46 | NA | NA | 2 | 0.47 | 0.46 | 0.01 | |
| FA-9 | 7 | 6 | 0 | 7 | 92.31% | 2.92 | NA | NA | NA | 0 | 0.50 | 0.49 | 0.06 | |
| FA-10 | 31 | 10 | 0 | 31 | 100% | 4.46 | 4.13 | 4.32 | NA | 1 | 0.61 | 0.62 | 0.04 * | |
| FA-11 | 4 | 3 | 0 | 4 | 100% | 3.00 | NA | NA | NA | 0 | 0.58 | 0.50 | −0.01 | |
| FA-1 | 24 | 10 | 8 | 24 | 92.31% | 3.54 | 3.3 | 3.46 | NA | 0 | 0.47 | 0.49 | 0.06 | |
| FA-15 | 18 | 8 | 18 | 18 | 84.62% | 2.85 | 2.69 | NA | NA | 0 | 0.45 | 0.41 | −0.09 | |
| M. sinacacolinii | TOTAL | 32 | 16 | 0 | 30.77 | 92.31% | 8.69 | NA | NA | 7.08 | 41 | 0.54 | 0.68 | 0.21 |
| FA-6 | 28 | 12 | 0 | 27 | 92.31% | 7.7 | 6.5 | 7.23 | NA | 33 | 0.5 | 0.6 | 0.19 * | |
| LT-3 | 4 | 4 | 0 | 3.77 | 76.92% | 3.15 | NA | NA | NA | 4 | 0.53 | 0.50 | 0.10 * | |
| M. zoquepopolucae | TOTAL | 15 | 9 | 0 | 14.69 | 100% | 6.46 | NA | 8.00 | 6.39 | 8 | 0.68 | 0.6 | −0.01 |
| FA-3 | 5 | 2 | 0 | 5 | 92.31% | 3.85 | NA | NA | NA | 1 | 0.69 | 0.58 | −0.08 | |
| FA-4 | 3 | 2 | 0 | 3 | 84.62% | 3.23 | NA | NA | NA | 2 | 0.72 | 0.54 | −0.14 | |
| FA-14 | 4 | 3 | 0 | 4 | 92.31% | 3.7 | NA | NA | NA | 0 | 0.65 | 0.58 | 0.01 | |
| LT-5 | 3 | 2 | 0 | 2.692 | 84.62% | 2.92 | NA | NA | NA | 1 | 0.67 | 0.54 | 0.01 |
| Species | Locality | State | Municipality | N | Voucher |
|---|---|---|---|---|---|
| M. decastroi | FA-12 | Veracruz | Zongolica | 30 | NA 1 |
| FA-13 | 33 | Aldaba 224 | |||
| M. lopezobradorii | FA-5 | Veracruz | San Andrés Tuxtla | 3 | Aldaba 241 |
| FA-7 | 27 | Aldaba 242 | |||
| FA-8 | 3 | Aldaba 245 | |||
| LT-2 | 21 | Samain & Martínez 2016–03 | |||
| M. mexicana | FA-2 | Puebla | Cuetzalan del Progreso | 13 | Aldaba 215 |
| FA-10 | Xicotepec | 31 | Aldaba 219 | ||
| FA-11 | Hueytamalco | 4 | Aldaba 202 | ||
| FA-1 | Veracruz | Xalapa | 24 | Aldaba 210 | |
| FA-9 | Yecuatla | 7 | Aldaba 218 | ||
| FA-15 | Coatepec | 18 | Aldaba 227 | ||
| M. sinacacolinii | FA-6 | Veracruz | Catemaco | 29 | Aldaba 235 |
| LT-3 | San Andrés Tuxtla | 4 | Samain & Martínez 2016-07 | ||
| M. zoquepopolucae | FA-3 | Veracruz | Soteapan | 5 | Aldaba 239 |
| FA-4 | 3 | Aldaba 240 | |||
| FA-14 | 4 | Aldaba 247 | |||
| LT-5 | 3 | Samain & Martínez 2016–12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aldaba Núñez, F.A.; Veltjen, E.; Martínez Salas, E.M.; Samain, M.-S. Disentangling Species Delineation and Guiding Conservation of Endangered Magnolias in Veracruz, Mexico. Plants 2021, 10, 673. https://doi.org/10.3390/plants10040673
Aldaba Núñez FA, Veltjen E, Martínez Salas EM, Samain M-S. Disentangling Species Delineation and Guiding Conservation of Endangered Magnolias in Veracruz, Mexico. Plants. 2021; 10(4):673. https://doi.org/10.3390/plants10040673
Chicago/Turabian StyleAldaba Núñez, Fabián Augusto, Emily Veltjen, Esteban Manuel Martínez Salas, and Marie-Stéphanie Samain. 2021. "Disentangling Species Delineation and Guiding Conservation of Endangered Magnolias in Veracruz, Mexico" Plants 10, no. 4: 673. https://doi.org/10.3390/plants10040673
APA StyleAldaba Núñez, F. A., Veltjen, E., Martínez Salas, E. M., & Samain, M.-S. (2021). Disentangling Species Delineation and Guiding Conservation of Endangered Magnolias in Veracruz, Mexico. Plants, 10(4), 673. https://doi.org/10.3390/plants10040673