Integrative Analysis of Gene Expression and miRNAs Reveal Biological Pathways Associated with Bud Paradormancy and Endodormancy in Grapevine
Abstract
1. Introduction
2. Results
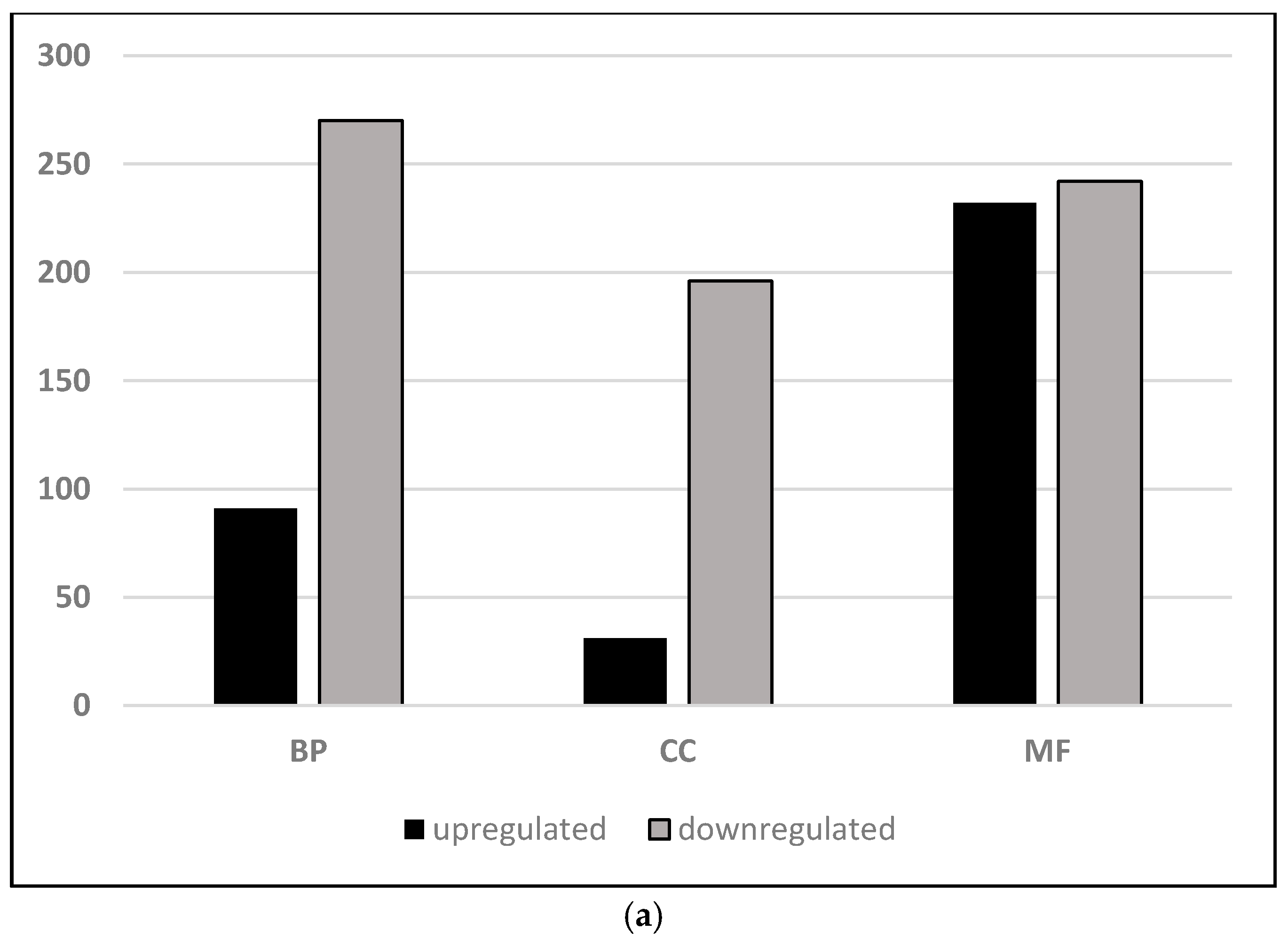
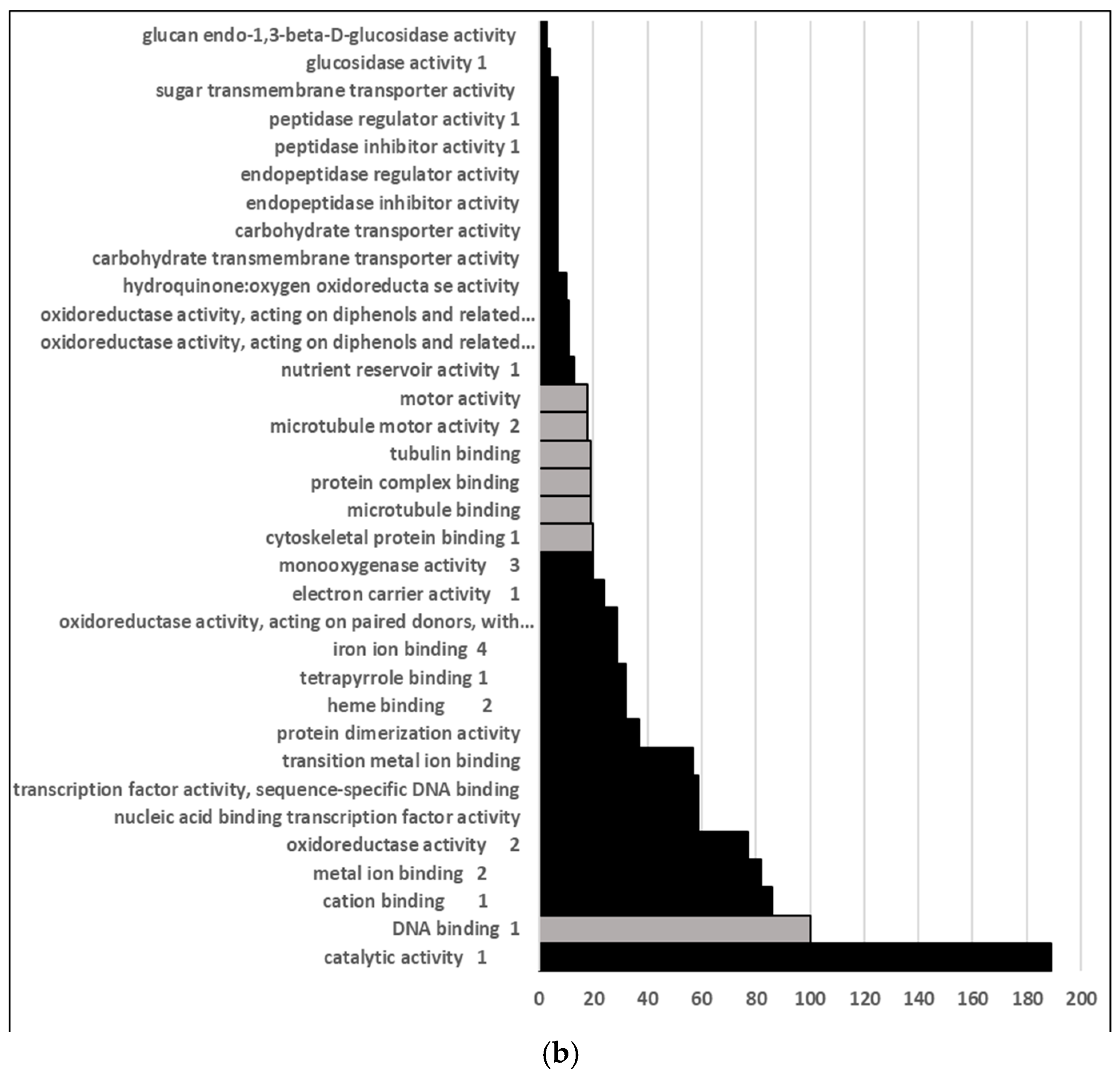
2.1. Endodormancy Is Associated with Greater Down-Regulation of Gene Expression Relative to Paradormancy
2.2. miRNA Abundance Differs in Paradormant (LD) and Endocormant (SD) Buds
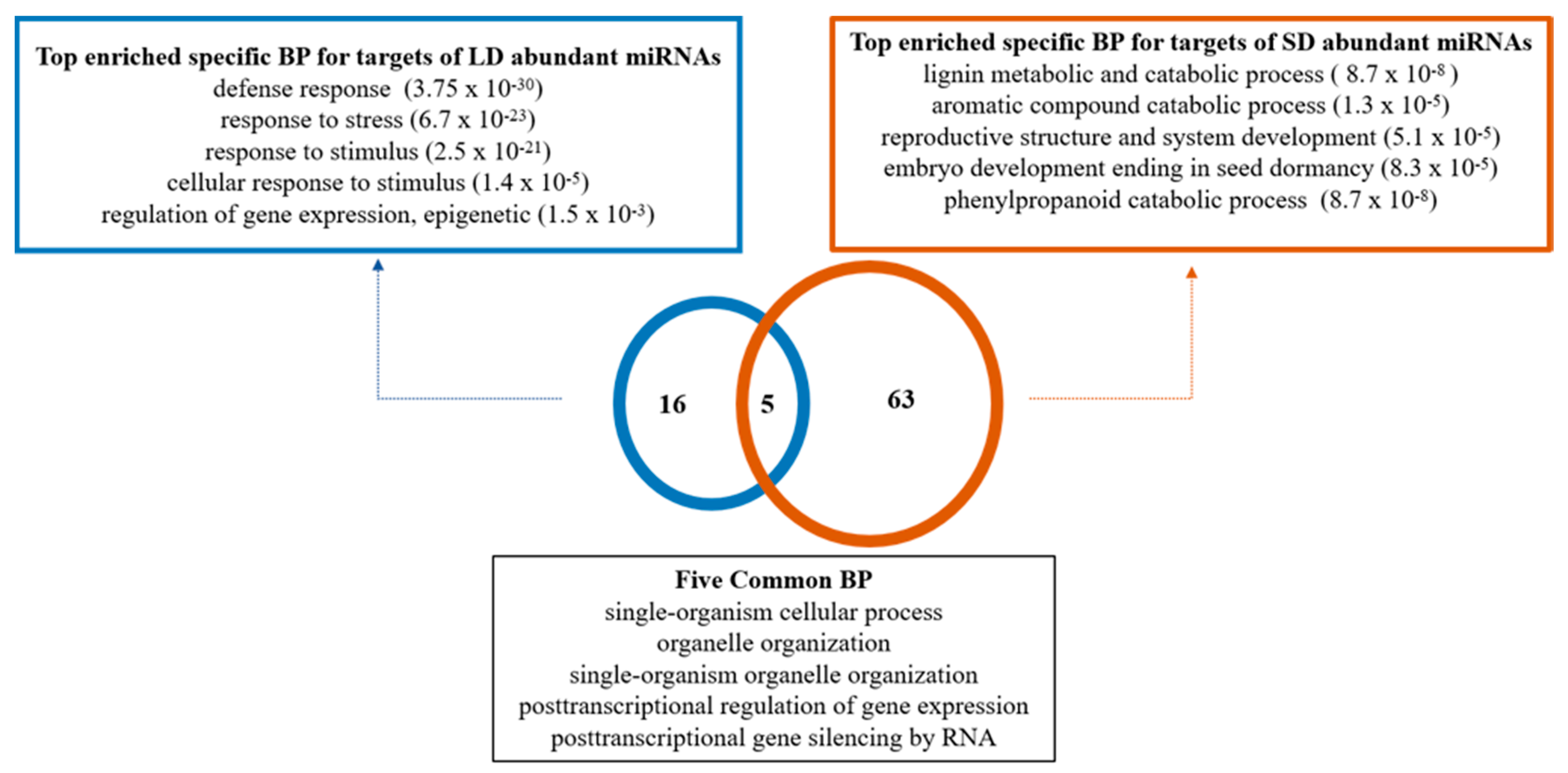
2.3. miRNAs Target Different Genes in Paradormant (LD) and Endodormant (SD) Buds
2.4. Inverse Expression Association of Predicted Target Genes and Abundant miRNAs
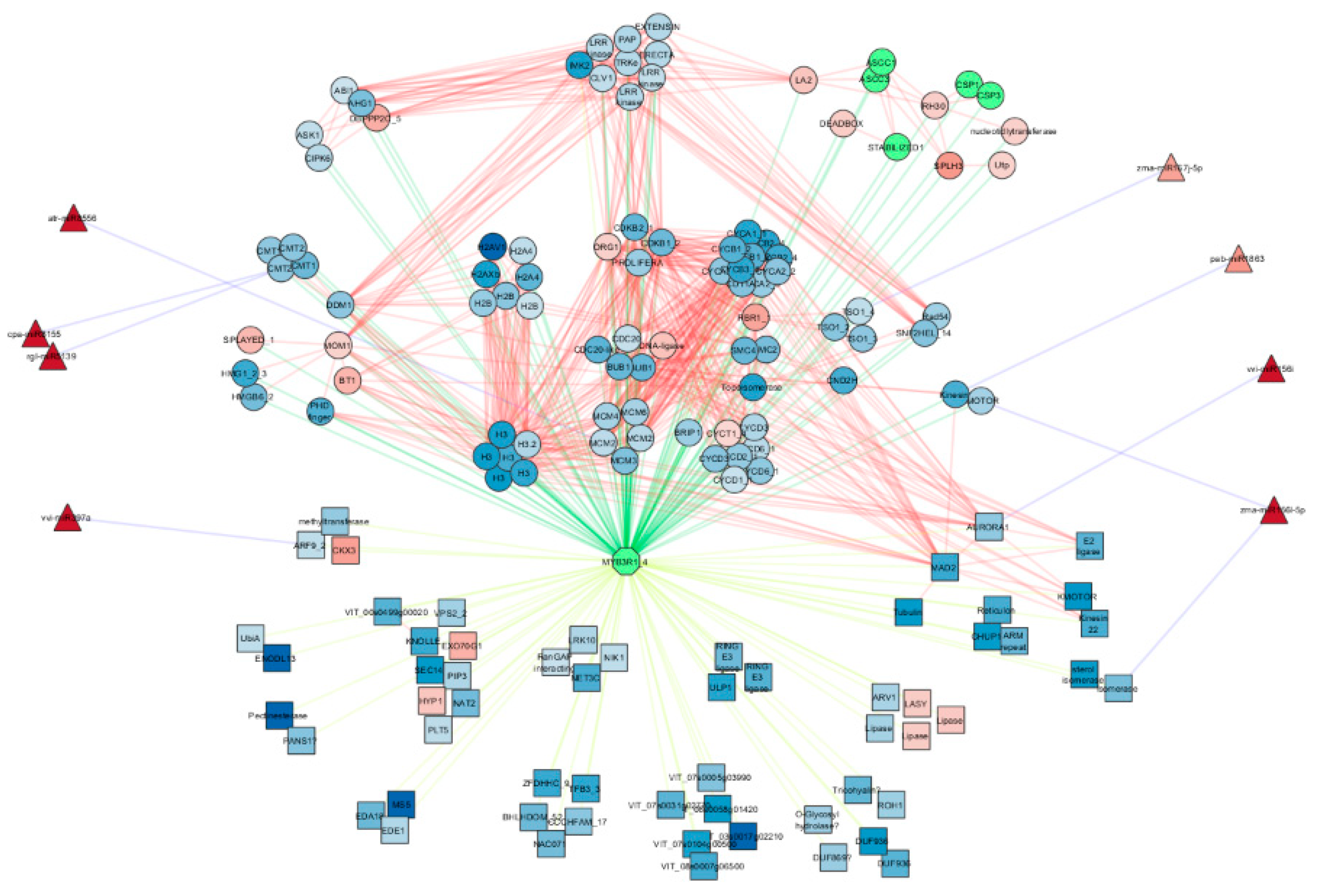
2.5. Predition of Transcription Factor Regulatory Network in LD and SD Abundant miRNAs
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Photoperiod Treatments
4.2. RNA Sequencing and Differential Gene Expression Analysis
4.3. Transcriptome Functional Enrichment Analyses
4.4. VitisNet Gene Set Enrichment Analysis (GSEA)
4.5. Small RNA Library Construction, Sequencing and Processing
4.6. Target Prediction for Abundant miRNAs
4.7. Transcription Factor Motif GSEA and Network
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fennell, A. Freezing Tolerance and Injury in Grapevines. J. Crop. Improv. 2004, 10, 201–235. [Google Scholar] [CrossRef]
- Lang, G.A.; Early, J.D.; Martin, G.C.; Darnell, R.L. Endo-, para-, and ecodormancy: Physiological terminology and classification for dormancy research. HortScience 1987, 22, 371–377. [Google Scholar]
- Mathiason, K.; He, D.; Grimplet, J.; Venkateswari, J.; Galbraith, D.W.; Or, E.; Fennell, A. Transcript profiling in Vitis riparia during chilling requirement fulfillment reveals coordination of gene expression patterns with optimized bud break. Funct. Integr. Genom. 2009, 9, 81–96. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Riquelme, J.; Grimplet, J.; Martínez-Zapater, J.M.; Carmona, M. Transcriptome variation along bud development in grapevine (Vitis vinifera L.). BMC Plant Biol. 2012, 12, 181. [Google Scholar] [CrossRef]
- Fennell, A.Y.; Schlauch, K.A.; Gouthu, S.; Deluc, L.G.; Khadka, V.; Sreekantan, L.; Grimplet, J.; Cramer, G.R.; Mathiason, K.L. Short day transcriptomic programming during induction of dormancy in grapevine. Front. Plant Sci. 2015, 6, 834. [Google Scholar] [CrossRef]
- Ning, D.-L.; Liu, C.-C.; Liu, J.-W.; Shen, Z.; Chen, S.; Liu, F.; Wang, B.-C.; Yang, C.-P. Label-free quantitative proteomics analysis of dormant terminal buds of poplar. Mol. Biol. Rep. 2013, 40, 4529–4542. [Google Scholar] [CrossRef]
- Ruttink, T.; Arend, M.; Morreel, K.; Strome, V.; Rombauts, S.; Fromm, J.; Bhalerao, R.P.; Boerjan, W.; Rohde, A. A Molecular Timetable for Apical Bud Formation and Dormancy Induction in Poplar. Plant Cell 2007, 19, 2370–2390. [Google Scholar] [CrossRef]
- Zhang, Y.X.; Yu, D.; Tian, X.L.; Liu, C.Y.; Gai, S.P.; Zheng, G.S. Differential expression proteins associated with bud dormancy release during chilling treatment of tree peony (Paeonia suffruticosa). Plant Biol. 2015, 17, 114–122. [Google Scholar] [CrossRef]
- Kahlil-Ur-Rehman, M.; Sun, L.; Li, C.-X.; Faheem, M.; Wang, W.; Tao, J.-M. Comparative RNA-seq based transcriptomic analysis of bud dormancy in grape. BMC Plant Biol. 2017, 17, 18. [Google Scholar] [CrossRef]
- Maurya, J.P.; Bhalerao, R.P. Photoperiod- and temperature-mediated control of growth cessation and dormancy in trees: A molecular perspective. Ann. Bot. 2017, 120, 351–360. [Google Scholar] [CrossRef]
- Horvath, D.P.; Chao, W.S.; Suttle, J.C.; Thimmapuram, J.; Anderson, J.V. Transcriptome analysis identifies novel responses and potential regulatory genes involved in seasonal dormancy transitions of leafy spurge (Euphorbia esula L.). BMC Genom. 2008, 9, 536. [Google Scholar] [CrossRef] [PubMed]
- Doğramacı, M.; Foley, M.E.; Chao, W.S.; Christoffers, M.J.; Anderson, J.V. Induction of endodormancy in crown buds of leafy spurge (Euphorbia esula L.) implicates a role for ethylene and cross-talk between photoperiod and temperature. Plant Mol. Biol. 2013, 81, 577–593. [Google Scholar] [CrossRef]
- Horvath, D. Common mechanisms regulate flowering and dormancy. Plant Sci. 2009, 177, 523–531. [Google Scholar] [CrossRef]
- Rohde, A.; Bhalerao, R.P. Plant dormancy in the perennial context. Trends Plant Sci. 2007, 12, 217–223. [Google Scholar] [CrossRef]
- Yamane, H.; Kashiwa, Y.; Ooka, T.; Tao, R.; Yonemori, K. Suppression Subtractive Hybridization and Differential Screening Reveals Endodormancy-associated Expression of an SVP/AGL24-type MADS-box Gene in Lateral Vegetative Buds of Japanese Apricot. J. Am. Soc. Hortic. Sci. 2008, 133, 708–716. [Google Scholar] [CrossRef]
- Heide, O.M.; Prestrud, A.K. Low temperature, but not photoperiod, controls growth cessation and dormancy induction and release in apple and pear. Tree Physiol. 2005, 25, 109–114. [Google Scholar] [CrossRef]
- Rothkegel, K.; Sandoval, P.; Soto, E.; Ulloas, L.; Riveros, A.; Lillo-Carmona, V.; Cáceres-Molina, J.; Almeida, A.M.; Meneses, C. Dormant but active: Chilling accumulation modulates the epigenome and transcriptome of Prunus avium during bud dormancy. Front. Plant Sci. 2020, 11, 1115. [Google Scholar] [CrossRef]
- Barakat, A.; Sriram, A.; Park, J.; Zhebentyayeva, T.; Main, D.; Abbott, A. Genome wide identification of chilling responsive microRNAs in Prunus persica. BMC Genom. 2012, 13, 481. [Google Scholar] [CrossRef] [PubMed]
- Jeyaraj, A.; Chandran, V.; Gajjeraman, P. Differential expression of microRNAs in dormant bud of tea (Camellia sinensis (L.) O. Kuntze]. Plant Cell Rep. 2014, 33, 1053–1069. [Google Scholar] [CrossRef]
- Niu, Q.; Li, J.; Cai, D.; Qin, M.; Jia, H.; Bai, S.; Hussain, S.; Liu, G.; Teng, Y.; Zheng, X. Dormancy-associated MADS-box genes and microRNAs jointly control dormancy transition in pear (Pyrus pyrifolia white pear group) flower bud. J. Exp. Bot. 2016, 67, 239–257. [Google Scholar] [CrossRef]
- Huo, H.; Wei, S.; Bradford, K.J. DELAY OF GERMINATION1 (DOG1) regulates both seed dormancy and flowering time through microRNA pathways. Proc. Natl. Acad. Sci. USA 2016, 113, E2199–E2206. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Saito, T.; Ito, A.; Tuan, P.A.; Xu, Y.; Teng, Y.; Moriguchi, T. Small RNA and PARE sequencing in flower bud reveal the involvement of sRNAs in endodormancy release of Japanese pear (Pyrus pyrifolia ‘Kosui’). BMC Genom. 2016, 17. [Google Scholar] [CrossRef]
- Sun, X.; Fan, G.; Su, L.; Wang, W.; Liang, Z.; Li, S.; Xin, H. Identification of cold-inducible microRNAs in grapevine. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef]
- Pagliarani, C.; Vitali, M.; Ferrero, M.; Vitulo, N.; Incarbone, M.; Lovisolo, C.; Valle, G.; Schubert, A. Accumulation of MicroRNAs Differentially Modulated by Drought Is Affected by Grafting in Grapevine. Plant Physiol. 2017, 173, 2180–2195. [Google Scholar] [CrossRef]
- Vimont, N.; Fouché, M.; Campoy, J.A.; Tong, M.; Arkoun, M.; Yvin, J.-C.; Wigge, P.A.; Dirlewanger, E.; Cortijo, S.; Wenden, B. From bud formation to flowering: Transcriptomic state defines the cherry developmental phases of sweet cherry bud dormancy. BMC Genom. 2019, 20, 974. [Google Scholar] [CrossRef] [PubMed]
- Wake, C.M.F.; Fennell, A. Morphological, physiological and dormancy responses of three Vitis genotypes to short photoperiod. Physiol. Plant 2000, 109, 203–210. [Google Scholar] [CrossRef]
- Garg, R.; Jain, M. RNA-seq for transcriptome analysis in non-model plants. In Legume Genomics, Methods and Protocols; Rose, R.J., Ed.; Humana Press: Totowa, NJ, USA, 2013; pp. 43–58. [Google Scholar]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Crdero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Shangguan, L.; Kibet, K.N.; Wang, X.; Han, J.; Song, C.; Fang, J. Characterization of microRNAs Identified in a Table Grapevine Cultivar with Validation of Computationally Predicted Grapevine miRNAs by miR-RACE. PLoS ONE 2011, 6, e21259. [Google Scholar] [CrossRef]
- Wang, C.; Wang, X.; Kibet, K.N.; Song, C.; Zhang, C.; Li, X.; Han, J.; Fang, J. Deep sequencing of grapevine flower and berry short RNA library for discovery of novel microRNAs and validation of precise sequences of grapevine microRNAs deposited in miRBase. Physiol. Plant 2011, 143, 64–81. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Leng, X.; Zhang, Y.; Kayes, E.; Zhang, Y.; Sun, X.; Fang, J. Transcriptome-wide analysis of dynamic variations in regulation modes of grapevine microRNAs on their target genes during grapevine development. Plant Mol. Biol. 2014, 84, 269–285. [Google Scholar] [CrossRef]
- Kullan, J.B.; Pinto, D.L.P.; Bertolini, E.; Fasoli, M.; Zenoni, S.; Tornielli, G.B.; Pezzotti, M.; Meyers, B.C.; Farina, L.; Pe, M.E.; et al. miRVine, a microRNA expression atlas of grapevine based on small RNA sequencing. BMC Genom. 2015, 16, 393. [Google Scholar]
- Khalil-Ur-Rehman, M.; Wang, W.; Xu, Y.-S.; Haider, M.S.; Li, C.-X.; Tao, J.-M. Comparative Study on Reagents Involved in Grape Bud Break and Their Effects on Different Metabolites and Related Gene Expression during Winter. Front. Plant Sci. 2017, 8, 1340. [Google Scholar] [CrossRef]
- Sakamoto, D.; Moriguchi, T.; Sugiura, T.; Ito, A. Effects of dormancy progression and low-temperature response on changes in the sorbitol concentration in xylem sap of Japanese pear during winter season. Tree Physiol. 2013, 33, 398–408. [Google Scholar]
- Debast, S.; Nunes-Nesi, A.; Hajirezaei, M.R.; Hofmann, J.; Sonnewald, U.; Fernie, A.R.; Bornke, F. Altering trehalose-6-phosphate content in transgenic potato tubers affects tuber growth and alters responsiveness to hormones during sprouting. Plant Physiol. 2011, 156, 1754–1771. [Google Scholar] [CrossRef]
- Sonnewald, S.; Sonnewald, U. Regulation of potato tuber sprouting. Planta 2014, 239, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Fichtner, F.; Barbier, F.F.; Feil, R.; Watanabe, M.; Annunziata, M.G.; Chabikwa, T.G.; Höfgen, R.; Stitt, M.; Beveridge, C.A.; Lunn, J.E. Trehalose 6-phosphate is involved in triggering axillary bud outgrowth in garden pea (Pisum sativum L.). Plant J. 2017, 92, 611–623. [Google Scholar] [CrossRef] [PubMed]
- Sreekantan, L.; Mathiason, K.; Grimplet, J.; Schlauch, K.; Dickerson, J.A.; Fennell, A.Y. Differential floral development and gene expression in grapevines during long and short photoperiods suggests a role for floral genes in dormancy transitioning. Plant Mol. Biol. 2010, 73, 191–205. [Google Scholar] [CrossRef] [PubMed]
- Gou, J.; Fu, C.; Liu, S.; Tang, C.; Debnath, S.; Flanagan, A.; Ge, Y.; Tang, Y.; Qingzhen, J.; Larson, P.R.; et al. The miR156-SPL4 module predominantly regulates aerial axillary bud formation and controls shoot architecture. New Phytol. 2017, 216, 829–840. [Google Scholar] [CrossRef]
- Li, H.; Deng, Y.; Wu, T.; Subramanian, S.; Yu, O. Mis-expression of miR482, miR1512, and miR1515 Increases Soybean Nodulation. Plant Physiol. 2010, 153, 1759–1770. [Google Scholar] [CrossRef]
- Rubio, S.; Dantas, D.; Bressan-Smith, R.; Pérez, F.J. Relationship between Endodormancy and Cold Hardiness in Grapevine Buds. J. Plant Growth Regul. 2016, 35, 266–275. [Google Scholar] [CrossRef]
- Lu, S.; Li, Q.; Wei, H.; Chang, M.-J.; Tunlaya-Anukit, S.; Kim, H.; Liu, T.; Song, T.; Sun, Y.-H.; Yuan, L.; et al. Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc. Natl. Acad. Sci. USA 2013, 110, 10848–10853. [Google Scholar] [CrossRef]
- Swetha, C.; Basu, D.; Pachamuthu, K.; Tirumalai, V.; Nair, A.; Prasad, M.; Shivaprasad, V. Major Domestication-Related phenotypes in Indica Rice are due to loss of miRNA-mediated laccase silencing. Plant Cell 2018, 30, 2649–2662. [Google Scholar] [CrossRef]
- Meitha, K.; Agudelo-Romero, P.; Signorelli, S.; Gibbs, D.J.; Considine, J.A.; Foyer, C.H.; Considine, M.J. Developmental control of hypoxia during bud burst in grapevine. Plant Cell Environ. 2018, 41, 1154–1170. [Google Scholar] [CrossRef] [PubMed]
- Shimizu-Sato, S.; Ike, Y.; Mori, H. PsBR1 encodes a pea retinoblastoma-related protein that is phosphorylated in axillary buds during dormancy-to-growth transition. Plant Mol. Biol. 2008, 66, 125–133. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bourbosse, C.; Vegesna, N.; Law, J.A. SOG1 activator and MYB3R repressors regulate a complex DNA damage network in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, E12453–E122462. [Google Scholar] [CrossRef]
- Chang, S.; Puryear, J.; Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 1993, 11, 113–116. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 862–864. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Sazberg, S.L.; Rin, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Grimplet, J.; Cramer, G.R.; Dickerson, J.A.; Mathiason, K.; Van Hemert, J.; Fennell, A.Y. VitisNet, ‘Omics’ Integration through Grapevine Molecular Networks. PLoS ONE 2009, 4, e8365. [Google Scholar] [CrossRef] [PubMed]
- Reimand, J.; Kull, M.; Peterson, H.; Hansen, J.; Vilo, J. g: Profiler—A web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007, 35, W193–W200. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.T.; Lander, E.S.; et al. Gene set enrichment analysis, A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Accerbi, M.; Schmidt, S.A.; De Paoli, E.; Park, S.; Jeong, D.-H.; Green, P.J. Methods for isolation of total RNA to recover miRNAs and other small RNAs from diverse species. Methods Mol. Biol. 2010, 592, 31–50. [Google Scholar]
- Meyer, B.C. Plant MicroRNAs—Methods and Protocols; Meyers Blake, C., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; Available online: http://www.springer.com/us/book/9781603270045 (accessed on 8 November 2020).
- Lu, C.; Meyers, B.C.; Green, P.J. Construction of small RNA cDNA libraries for deep sequencing. Methods 2007, 43, 110–117. [Google Scholar] [CrossRef]
- Turner, M.; Adhikari, S.; Subramanian, S. Optimizing stem-loop qPCR assays through multiplexed cDNA synthesis of U6 and miRNAs. Plant Signal. Behav. 2013, 8, e24918. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, R.; Bernhart, S.H.; Höner zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofaker, I.L. Vienna RNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. microRNA-directed phasing during grans-Acting siRNA biogenesis in plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef]
- Schwab, R.; Palatnik, J.F.; Riester, M.; Schommer, C.; Schmid, M.; Weigel, D. Specific Effects of MicroRNAs on the Plant Transcriptome. Dev. Cell 2005, 8, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Luo, W.; Friedman, M.; Shedden, K.; Hankenson, K.; Woolf, P. GAGE: Generally applicable gene set enrichment for pathway analysis. BMC Bioinform. 2009, 10, 161. [Google Scholar] [CrossRef] [PubMed]

| miR Family | Fold (LD vs. SD) | FDR Adjusted p-Value | Mean LD Count | Mean SD Count | miRNA Sequences with ≥20 | miR Family Logo 1 |
|---|---|---|---|---|---|---|
| 166 | 2.7 | 9.2 × 10−10 | 604 | 225 | 431 |  |
| 167 | 2.5 | 6.1 × 10−13 | 2933 | 1164 | 170 |  |
| 894 | 4.1 | 4.3 × 10−5 | 237 | 58 | 64 |  |
| 3636 | 4.7 | 4.1 × 10−7 | 929 | 197 | 45 |  |
| 3623 | 3.1 | 7.0 × 10−4 | 629 | 201 | 42 |  |
| 169 | 3.2 | 2.6 × 10−2 | 264 | 82 | 27 |  |
| 390 | 3.0 | 3.4 × 10−2 | 936 | 309 | 20 |  |
| 3640 | 3.6 | 1.4 × 10−2 | 207 | 58 | 19 |  |
| 3637 | 3.0 | 6.6 × 10−3 | 145 | 48 | 16 |  |
| 164 | 2.5 | 1.1 × 10−2 | 25 | 10 | 14 |  |
| 391 | 3.0 | 2.7 × 10−2 | 563 | 185 | 12 |  |
| 2916 | −2.3 | 3.5 × 10−6 | 53 | 121 | 96 |  |
| 3639 | −2.0 | 2.7 × 10−2 | 225 | 445 | 45 |  |
| 156 | −3.6 | 1.8 ×10−2 | 348 | 1254 | 24 |  |
| 397 | −3.4 | 1.3 × 10−2 | 347 | 1168 | 16 |  |
| 408 | −3.8 | 3.6 × 10−2 | 76 | 289 | 15 |  |
| 398 | −3.9 | 1.2 × 10−2 | 141 | 545 | 10 |  |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smita, S.; Robben, M.; Deuja, A.; Accerbi, M.; Green, P.J.; Subramanian, S.; Fennell, A. Integrative Analysis of Gene Expression and miRNAs Reveal Biological Pathways Associated with Bud Paradormancy and Endodormancy in Grapevine. Plants 2021, 10, 669. https://doi.org/10.3390/plants10040669
Smita S, Robben M, Deuja A, Accerbi M, Green PJ, Subramanian S, Fennell A. Integrative Analysis of Gene Expression and miRNAs Reveal Biological Pathways Associated with Bud Paradormancy and Endodormancy in Grapevine. Plants. 2021; 10(4):669. https://doi.org/10.3390/plants10040669
Chicago/Turabian StyleSmita, Shuchi, Michael Robben, Anup Deuja, Monica Accerbi, Pamela J. Green, Senthil Subramanian, and Anne Fennell. 2021. "Integrative Analysis of Gene Expression and miRNAs Reveal Biological Pathways Associated with Bud Paradormancy and Endodormancy in Grapevine" Plants 10, no. 4: 669. https://doi.org/10.3390/plants10040669
APA StyleSmita, S., Robben, M., Deuja, A., Accerbi, M., Green, P. J., Subramanian, S., & Fennell, A. (2021). Integrative Analysis of Gene Expression and miRNAs Reveal Biological Pathways Associated with Bud Paradormancy and Endodormancy in Grapevine. Plants, 10(4), 669. https://doi.org/10.3390/plants10040669






