Genome-Wide Analysis of Tubulin Gene Family in Cassava and Expression of Family Member FtsZ2-1 during Various Stress
Abstract
1. Introduction
2. Results
2.1. Identification of the Tubulin Proteins in Cassava
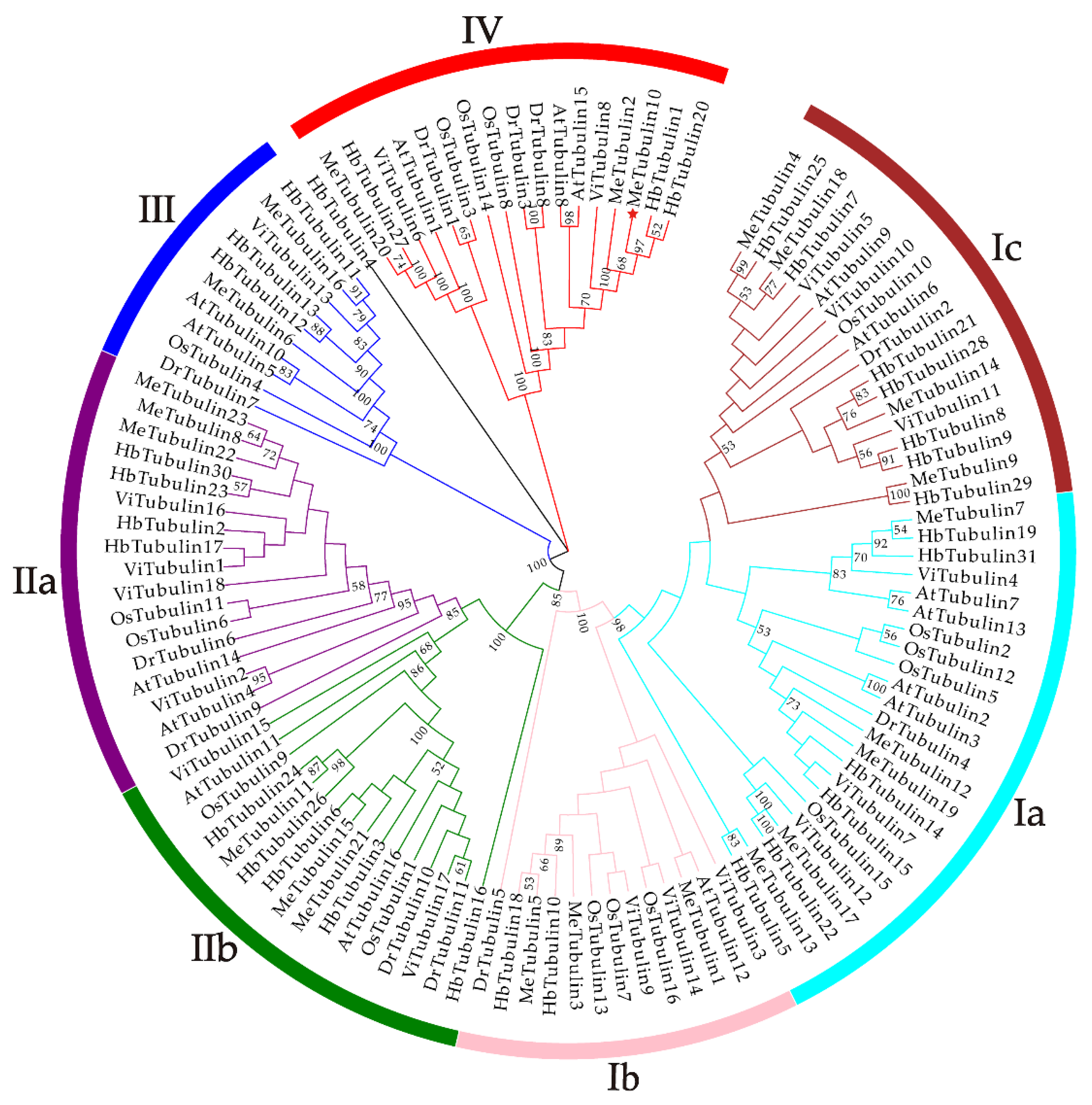
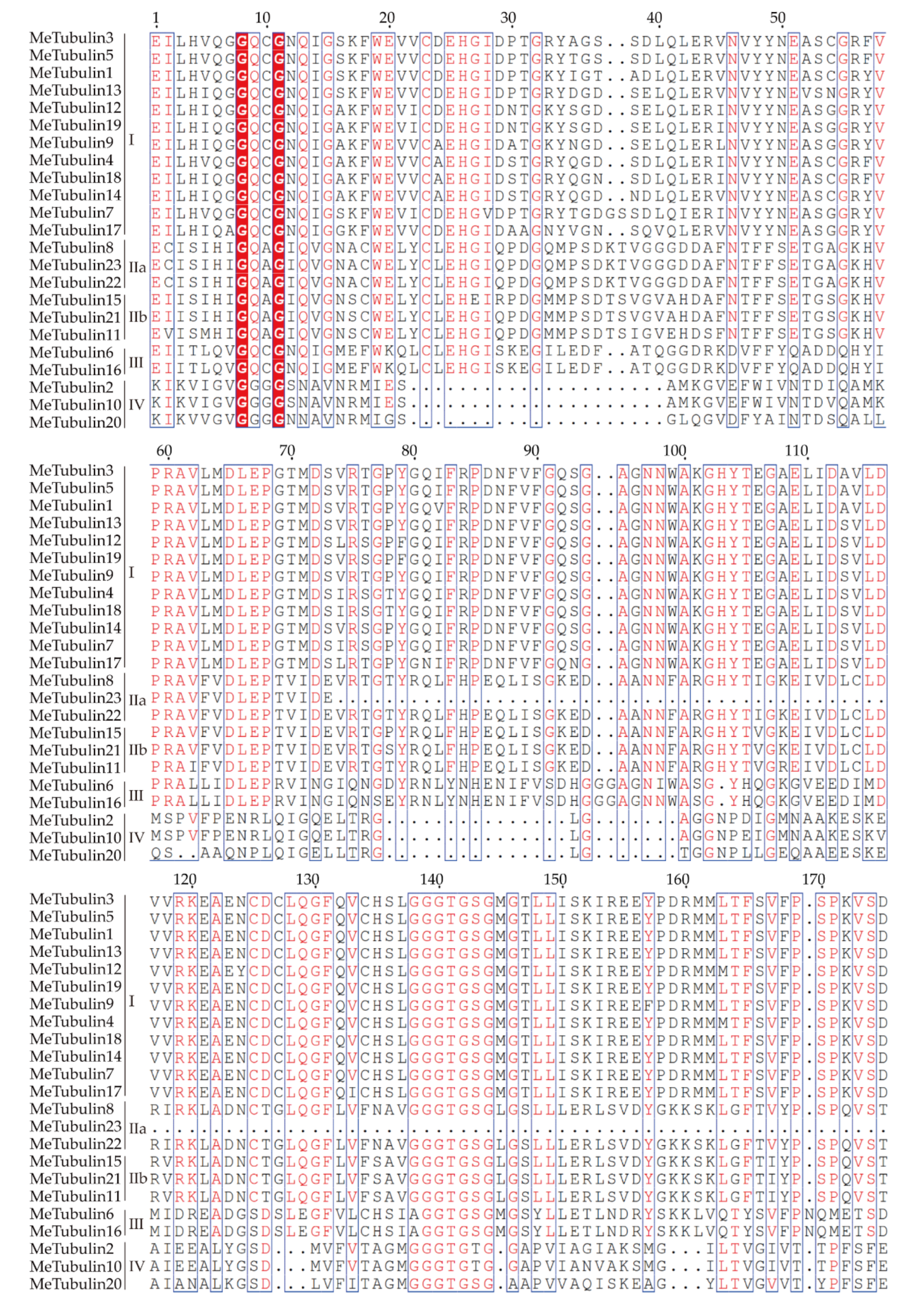
2.2. Multiple Sequence Alignment, Phylogenetic Analysis, and Classification of MeTubulins
2.3. Gene Structure and Motif Composition of MeTubulins
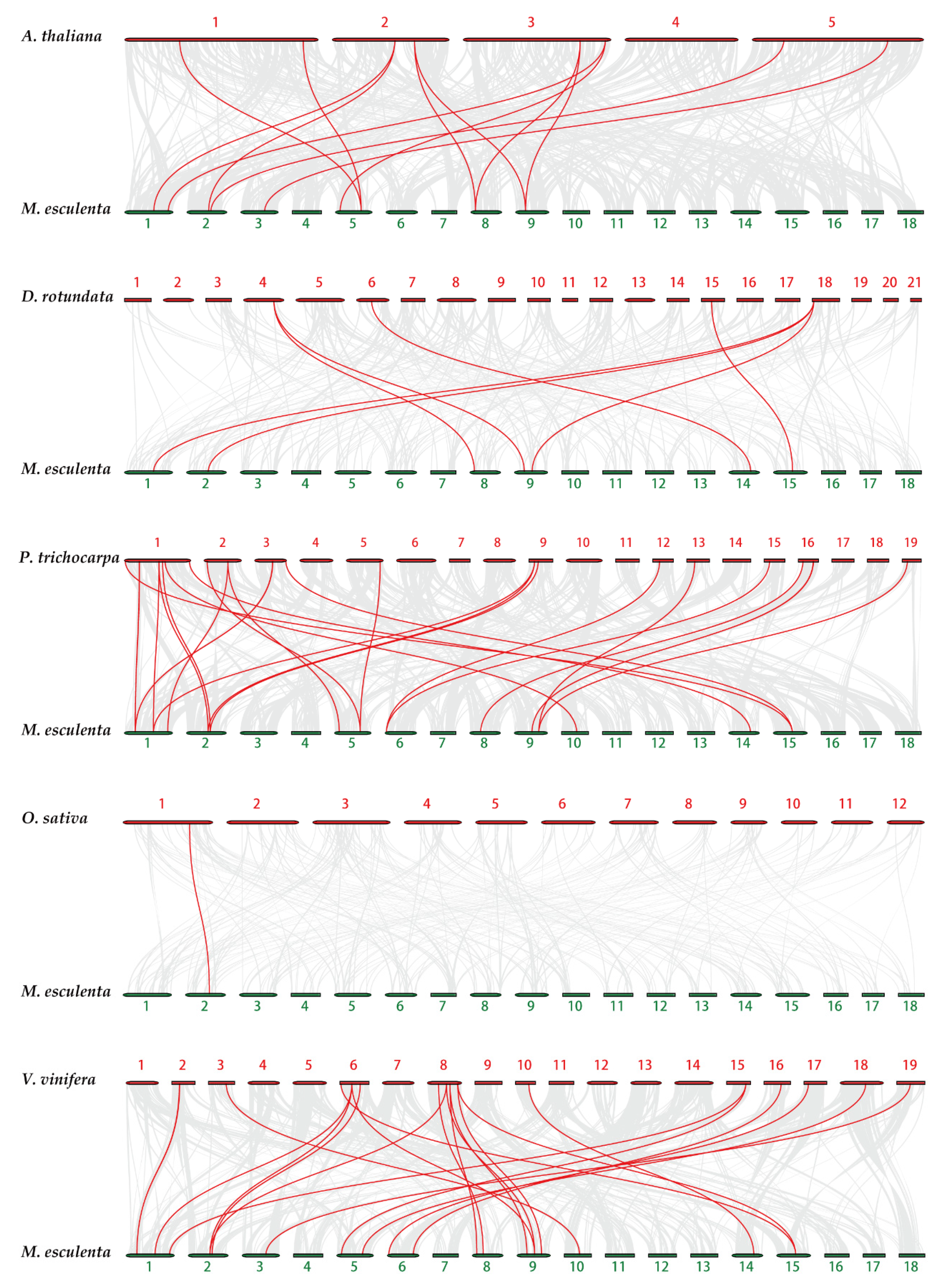
2.4. Chromosome Distribution and Synteny Analysis of MeTubulins Gene
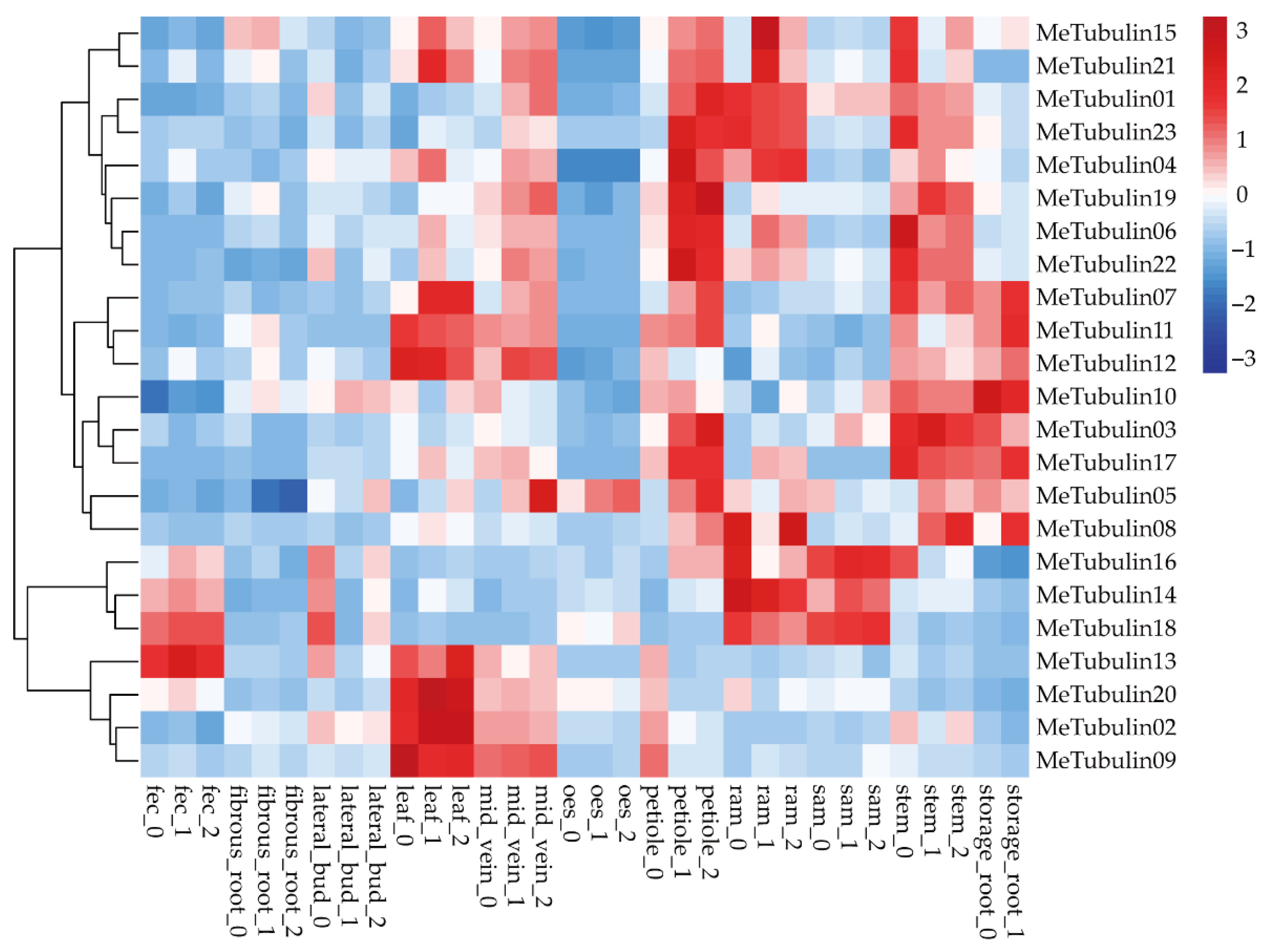
2.5. Expression Profiles of MeTubulins in Different Tissue Types
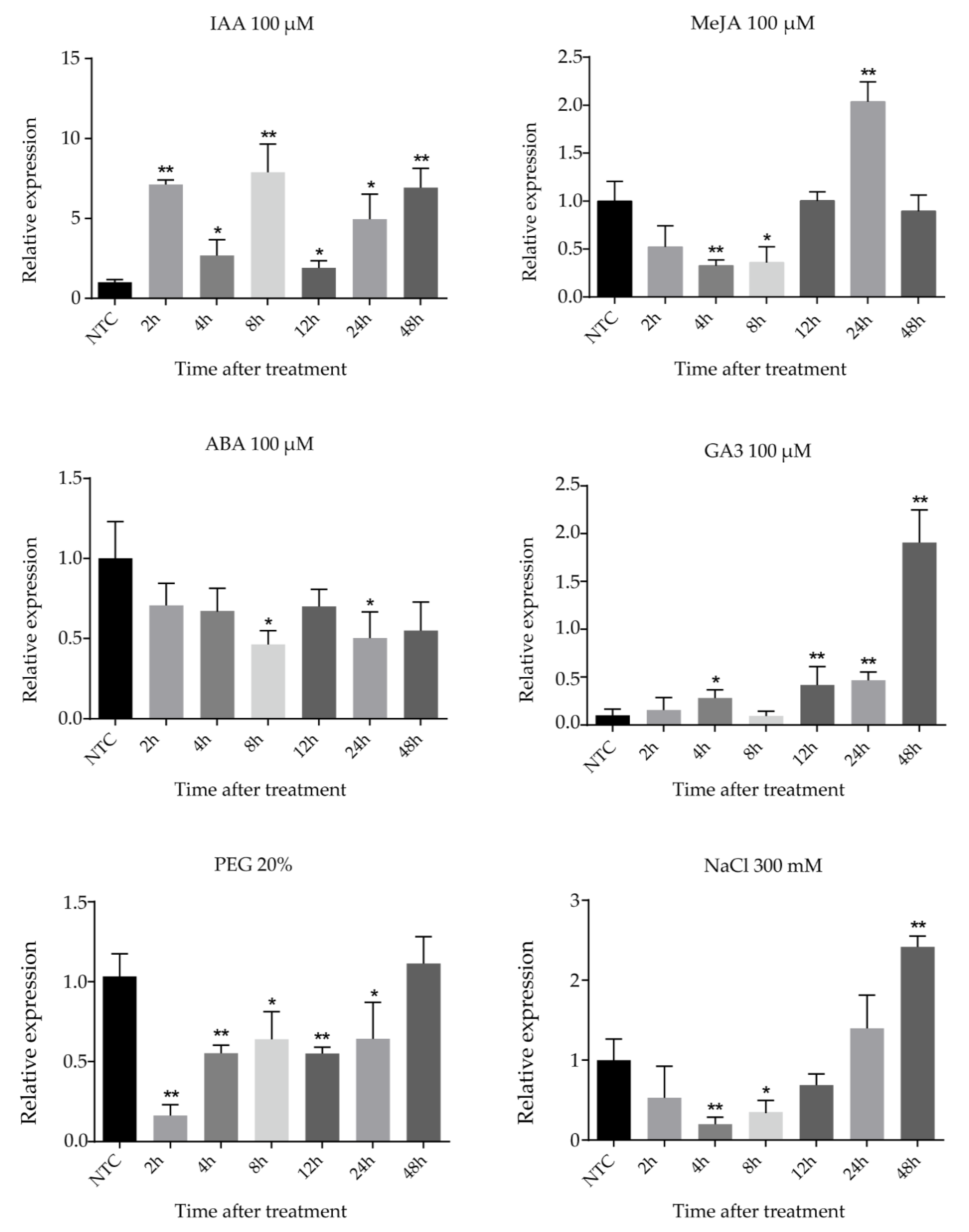
2.6. Expression Patterns of FtsZ2-1 Gene in Response to Various Stresses
3. Discussion
4. Materials and Methods
4.1. Identification of the Tubulin Gene Family
4.2. Sequence Analysis and Structural Characterization
4.3. Chromosomal Localization and Gene Duplication
4.4. Phylogenetic Analysis of Cassava Tubulin Genes Family
4.5. RNA-Seq Data Analysis of Tubulin Genes
4.6. Plant Materials and Treatments
4.7. RNA Extraction and qRT-PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Steven, R.; Ludwig, D.G. Characterization of the a-tubulin gene family of Arabidopsis thaliana. Genetics 1987, 84, 5833–5837. [Google Scholar]
- Sutkovic, J.; Gawwad, M.R. In silico prediction of three-dimensional structure and interactome analysis of Tubulin α subfamily of Arabidopsis thaliana.pdf. Netw. Biol. 2014, 4, 47–57. [Google Scholar]
- Gavazzi, F.; Pigna, G.; Braglia, L.; Giani, S.; Breviario, D.; Morello, L. Evolutionary characterization and transcript profiling of beta-tubulin genes in flax (Linum usitatissimum L.) during plant development. BMC Plant Biol. 2017, 17, 237. [Google Scholar] [CrossRef]
- Lowe, J.; Li, H.; Downing, K.H.; Nogales, E. Refined structure of αβ-tubulin at 3.5 resolution 1. J. Mol. Biol. 2001, 313, 1045–1057. [Google Scholar] [CrossRef] [PubMed]
- Serna, M.; Carranza, G.; Martín-Benito, J.; Janowski, R.; Canals, A.; Coll, M.; Zabala, J.C.; Valpuesta, J.M. The structure of the complex between α-tubulin, TBCE and TBCB reveals a tubulin dimer dissociation mechanism. J. Cell Sci. 2015, 128, 1824–1834. [Google Scholar] [CrossRef]
- Oakley, R.V.; Wang, Y.S.; Ramakrishna, W.; Harding, S.A.; Tsai, C.J. Differential expansion and expression of alpha- and beta-tubulin gene families in Populus. Plant Physiol. 2007, 145, 961–973. [Google Scholar] [CrossRef] [PubMed]
- Pydiura, N.; Pirko, Y.; Galinousky, D.; Postovoitova, A.; Yemets, A.; Kilchevsky, A.; Blume, Y. Genome-wide identification, phylogenetic classification, and exon-intron structure characterization of the tubulin and actin genes in flax (Linum usitatissimum). Cell Biol. Int. 2019, 43, 1010–1019. [Google Scholar] [CrossRef] [PubMed]
- Dryková, D.; Cenklová, V.; Sulimenko, V.; Volc, J.i.; Dráber, P.; Binarová, P. Plant γ-tubulin interacts with αβ-tubulin dimers and forms membrane-associated complexes. Plant Cell 2003, 15, 465–480. [Google Scholar] [CrossRef] [PubMed]
- Kállai, B.M.; Kourová, H.; Chumová, J.; Papdi, C.; Binarová, P. γ-Tubulin interacts with E2FA, E2FB and E2FC transcription factors, regulates proliferation and endocycle in Arabidopsis. J. Exp. Bot. 2019, 71, 498–531. [Google Scholar] [CrossRef]
- Miao, H.; Guo, R.; Chen, J.; Wang, Q.; Lee, Y.J.; Liu, B. The gamma-tubulin complex protein GCP6 is crucial for spindle morphogenesis but not essential for microtubule reorganization in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 27115–27123. [Google Scholar] [CrossRef]
- Silflow, C.D.; Oppenheimer, D.G.; Kopozak, S.D.; Ploense, S.E.; Ludwig, S.R.; Haas, N.; Snustad, D.P. Plant tubulin genes structure and differential expression during development. Genetics 1987, 8, 435–460. [Google Scholar] [CrossRef]
- Bao, Y.; Kost, B.; Chua, N.H. Reduced expression of α-tubulin genes in Arabidopsis thaliana specifically affects root growth and morphology, root hair development and root gravitropism. Plant J. 2001, 28, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Jan, A.; Komatsu, S. Functional analysis of OsTUB8, an anther-specific β-tubulin in rice. Plant Sci. 2007, 172, 832–838. [Google Scholar] [CrossRef]
- Du, S.; Lutkenhaus, J. Assembly and activation of the Escherichia coli divisome. Mol. Microbiol. 2017, 105, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Erickson, H.P.; Anderson, D.E.; Osawa, M. FtsZ in bacterial cytokinesis: Cytoskeleton and force generator all in one. Microbiol. Mol. Biol. Rev. 2010, 74, 504–528. [Google Scholar] [CrossRef] [PubMed]
- Lwe, J.; Amos, L.A. Crystal structure of the bacterial cell-division protein FtsZ. Nature 1998, 391, 203–206. [Google Scholar] [CrossRef]
- Ma, X.; Ehrhardt, D.W.; Margolin, W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using greenfluorescentprotein. Proc. Natl. Acad. Sci. USA 1996, 93, 12998–13003. [Google Scholar] [CrossRef]
- Miyagishima, S.Y.; Takahara, M.; Mori, T.; Kuroiwa, H.; Kuroiwa, H.T. Plastid division is driven by a complex mechanism that involves differential transition of the bacterial and eukaryotic division rings. Plant Cell 2001, 13, 2257–2268. [Google Scholar] [CrossRef]
- Stanislav Vitha, R.S.; Katherine, W.O. FtsZ ring formation at the chloroplast division site in plants. J. Cell Biol. 2001, 153, 111–120. [Google Scholar] [CrossRef]
- Chen, C.; Maccready, J.S.; Ducat, D.C.; Osteryoung, K.W. The molecular machinery of chloroplast division. Plant Physiol. 2018, 176, 138–151. [Google Scholar] [CrossRef]
- Grosche, C.; Rensing, S.A. Three rings for the evolution of plastid shape: A tale of land plant FtsZ. Protoplasma 2017, 254, 1879–1885. [Google Scholar] [CrossRef]
- Osteryoung, K.W.; Vierling, E. Conserved cell and organelle division. Nature 1995, 376, 473–474. [Google Scholar] [CrossRef]
- Terbush, A.D.; Yoshida, Y.; Osteryoung, K.W. FtsZ in chloroplast division: Structure, function and evolution. Curr. Opin. Cell Biol. 2013, 25, 461–470. [Google Scholar] [CrossRef]
- Osteryoung, K.W.; Stokes, K.D.; Rutherford, S.M.; Percival, A.L.; Lee, W.Y. Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell 1998, 10, 1991–2004. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, A.J.; Glynn, J.M.; Olson, B.J.; Stokes, K.D.; Osteryoung, K.W. Arabidopsis FtsZ2-1 and FtsZ2-2 are functionally redundant, but FtsZ-based plastid division is not essential for chloroplast partitioning or plant growth and development. Mol. Plant 2009, 2, 1211–1222. [Google Scholar] [CrossRef] [PubMed]
- McAndrew, R.S.; Olson, B.J.; Kadirjan-Kalbach, D.K.; Chi-Ham, C.L.; Vitha, S.; Froehlich, J.E.; Osteryoung, K.W. In vivo quantitative relationship between plastid division proteins FtsZ1 and FtsZ2 and identification of ARC6 and ARC3 in a native FtsZ complex. Biochem. J. 2008, 412, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Yoder, D.W.; Kadirjan-Kalbach, D.; Olson, B.J.; Miyagishima, S.Y.; Deblasio, S.L.; Hangarter, R.P.; Osteryoung, K.W. Effects of mutations in Arabidopsis FtsZ1 on plastid division, FtsZ ring formation and positioning, and FtsZ filament morphology in vivo. Plant Cell Physiol. 2007, 48, 775–791. [Google Scholar] [CrossRef] [PubMed]
- Leech, R.M.; Baker, N.R. Growth and functioning of leaves. In Development of Photosynthetic Capacity in Leaves, Proceedings of the 13th International Botanical Congress, University of Sydney, Sydney, Austrilia, 18–20 August 1981; Cambridge University Press: Cambridge, UK, 1983. [Google Scholar]
- Yong, L.; Binbin, R.; Lei, D.; Qirong, S.; Shaobing, P.; Shiwei, G.; Ive, D.S. Does chloroplast size influence photosynthetic nitrogen Use efficiency. PLoS ONE 2013, 8, e62036. [Google Scholar]
- Nogales, E.; Wolf, S.G.; Downing, K.H. Structure of the alpha beta tubulin dimer by electron crystallography. Nature 1998, 391, 199–203. [Google Scholar] [CrossRef]
- Nogales, E.; Downing, K.H.; Amos, L.A.; Lowe, J. Tubulin and FtsZ form a distinct family of GTPases. Nat. Struct. Mol. Biol. 1998, 5, 451–458. [Google Scholar] [CrossRef]
- Carlier, M.F.; Pantaloni, D. Kinetic analysis of guanosine 5′-triphosphate hydrolysis associated with tubulin polymerization. Biochemistry 1981, 20, 1918–1924. [Google Scholar] [CrossRef] [PubMed]
- Krol, E.; de Sousa Borges, A.; da Silva, I.; Polaquini, C.R.; Regasini, L.O.; Ferreira, H.; Scheffers, D.J. Antibacterial activity of alkyl gallates is a combination of direct targeting of FtsZ and permeabilization of bacterial membranes. Front. Microbiol. 2015, 6, 390–401. [Google Scholar] [CrossRef] [PubMed]
- Yutin, N.; Koonin, E.V. Archaeal origin of tubulin. Biol. Direct 2012, 7, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Dufour, D.L. Cyanide content of cassava (Manihot esculenta, Euphorbiaceae) cultivars used by tukanoan indians in northwest amazonia. Econ. Bot. 1988, 42, 255–266. [Google Scholar] [CrossRef]
- Westby, A.; Hillocks, R.J.; Thresh, J.M. Cassava utilization, storage and small-scale processing. Cassava Biol. Prod. Util. 2002, 14, 281–300. [Google Scholar]
- Jiao, J.; Li, J.; Bai, Y. Uncertainty analysis in the life cycle assessment of cassava ethanol in China. J. Clean. Prod. 2019, 206, 438–451. [Google Scholar] [CrossRef]
- Okogbenin, E.; Setter, T.L.; Ferguson, M.; Mutegi, R.; Ceballos, H.; Olasanmi, B.; Fregene, M. Phenotypic approaches to drought in cassava: Review. Front. Physiol. 2013, 4, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.E.; Dong, M.Y.; Fan, X.W.; Nong, L.L.; Li, Y.Z. A study on cassava tolerance to and growth responses under salt stress. Environ. Exp. Bot. 2018, 155, 429–440. [Google Scholar] [CrossRef]
- Gleadow, R.; Pegg, A.; Blomstedt, C.K. Resilience of cassava (Manihot esculenta Crantz) to salinity: Implications for food security in low-lying regions. J. Exp. Bot. 2016, 67, 5403–5413. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.; Liu, X.; Guo, J.; Chen, Y.; Li, S.; Wang, C.; Huang, W.; Min, Y. Genome-wide analysis of Dynamin gene family in cassava (Manihot esculenta Crantz) and transcriptional regulation of family members ARC5 in hormonal treatments. Int. J. Mol. Sci. 2019, 20, 5094. [Google Scholar] [CrossRef]
- Wei, Y.; Shi, H.; Xia, Z.; Tie, W.; Ding, Z.; Yan, Y.; Wang, W.; Hu, W.; Li, K. Genome-wide identification and expression Analysis of the WRKY gene family in cassava. Front. Plant Sci. 2016, 7, 25–42. [Google Scholar] [CrossRef]
- Rauwane, M.; Ntushelo, K. Understanding biotic stress and hormone aignalling in cassava (Manihot esculenta): Potential for using hyphenated analytical techniques. Appl. Sci. 2020, 10, 8152. [Google Scholar] [CrossRef]
- Blume, Y.B.; Krasylenko, Y.A.; Yemets, A.I. Effects of phytohormones on the cytoskeleton of the plant cell. Russ. J. Plant Physiol. 2012, 59, 515–529. [Google Scholar] [CrossRef]
- Fan, S.; Chang, Y.; Liu, G.; Shang, S.; Tian, L.; Shi, H. Molecular functional analysis of auxin/indole-3-acetic acid proteins (Aux/IAAs) in plant disease resistance in cassava. Physiol. Plant. 2020, 168, 88–97. [Google Scholar] [CrossRef]
- Li, K.; Zhu, W.; Zeng, K.; Zhang, Z.; Ye, J.; Ou, W.; Rehman, S.; Heuer, B.; Chen, S. Proteome characterization of cassava (Manihot esculenta Crantz) somatic embryos, plantlets and tuberous roots. Proteome Sci. 2010, 8, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.A.; Jo, H.Y.; Han, I.S. Auxin activates promoter of a soybean β-tubulin, tubB1 gene. Korean J. Genet. 2005, 27, 383–388. [Google Scholar]
- Yang, H.; Deng, Z.; Liu, H.; Dai, L.J.; Li, D.J. Cloning, expression and bioinformatics analysis of HbTUA2 gene in Hevea brasiliensis (rubber tree). Plant Physiol. J. 2017, 53, 52–62. [Google Scholar]
- Radchuk, V.V. The transcriptome of the tubulin gene family in plants. In The Plant Cytoskeleton: A Key Tool for Agro-Biotechnology; Springer: Dordrecht, The Netherland, 2009; pp. 219–241. [Google Scholar]
- Wilson, M.C.; Mutka, A.M.; Hummel, A.W.; Berry, J.; Chauhan, R.D.; Vijayaraghavan, A.; Taylor, N.J.; Voytas, D.F.; Chitwood, D.H.; Bart, R.S. Gene expression atlas for the food security crop cassava. New Phytol. 2017, 213, 1632–1641. [Google Scholar] [CrossRef]
- Baranova, E.N.; Christov, N.K.; Kurenina, L.V.; Khaliluev, M.R.; Todorovska, E.G.; Smirnova, E.A. Formation of atypical tubulin structures in plant cells as a nonspecific response to abiotic stress. Bulg. J. Agric. Sci. 2016, 22, 987–992. [Google Scholar]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R.; et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar] [CrossRef]
- Zhang, Y.M.; Chen, M.; Sun, L.; Wang, Y.; Yin, J.; Liu, J.; Sun, X.Q.; Hang, Y.Y. Genome-wide identification and evolutionary analysis of NBS-LRR genes from Dioscorea rotundata. Front. Genet. 2020, 11, 484–494. [Google Scholar] [CrossRef]
- Li, H.L.; Guo, D.; Yang, Z.P.; Tang, X.; Peng, S.Q. Genome-wide identification and characterization of WRKY gene family in Hevea brasiliensis. Genomics 2014, 104, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Tyagi, A.K.; Khurana, J.P. Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics 2006, 88, 360–371. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Wang, Y.; Chai, F.; Li, S.; Xin, H.; Liang, Z. Genome-wide identification and characterization of the 14-3-3 family in Vitis vinifera L. during berry development and cold- and heat-stress response. BMC Genom. 2018, 19, 579–592. [Google Scholar] [CrossRef]
- Brand, L.H.; Fischer, N.M.; Harter, K.; Kohlbacher, O.; Wanke, D. Elucidating the evolutionary conserved DNA-binding specificities of WRKY transcription factors by molecular dynamics and in vitro binding assays. Nucleic Acids Res. 2013, 41, 9764–9778. [Google Scholar] [CrossRef] [PubMed]
- Fiume, E.; Christou, P.; Gian, S.; Breviario, D. Introns are key regulatory elements of rice tubulin expression. Planta 2004, 218, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.F.; Chen, J.; Chen, Y.F.; Wu, L.J.; Xie, D.X. Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res. 2012, 19, 153–164. [Google Scholar] [CrossRef]
- Kawagoe, Y. Septum Formation in Amyloplasts Produces Compound Granules in the Rice Endosperm and is Regulated by Plastid Division Proteins. Plant Cell Physiol. 2010, 51, 1469–1479. [Google Scholar]
- Tangphatsornruang, S.; Uthaipaisanwong, P.; Sangsrakru, D.; Chanprasert, J.; Yoocha, T.; Jomchai, N.; Tragoonrung, S. Characterization of the complete chloroplast genome of Hevea brasiliensis reveals genome rearrangement, RNA editing sites and phylogenetic relationships. Gene 2011, 475, 104–112. [Google Scholar] [CrossRef]
- Xianyun, M.U.; Shen, X.; Yuanmi, W.U.; Zhu, Y.; Dong, S.; Xia, X.; Lin, Q.; Fan, P.; Liu, Y.; Zhang, Z. Plastid phylogenomic study of grape species and its implications for evolutionary study and conservation of Vitis. Phytotaxa 2018, 364, 71–80. [Google Scholar]
- Chothia, C.; Gough, J.; Vogel, C.; Teichmann, S.A. Evolution of the protein repertoire. Science 2003, 300, 1701–1703. [Google Scholar] [CrossRef]
- Ohno, S.; Wolf, U.; Atkin, N.B. Evolution from fish to mammals by gene duplication. Hereditas 1968, 59, 169–187. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10–30. [Google Scholar] [CrossRef]
- Holub, E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Zhao, X.Q.; Wang, J.; Wong, K.S.; Yu, J. KaKs_Calculator: Calculating Ka and Ks through model selection and model averaging. Genom. Proteom. Bioinform. 2006, 4, 259–263. [Google Scholar] [CrossRef]
- Geng, M.T.; Min, Y.; Yao, Y.; Chen, X.; Fan, J.; Yuan, S.; Wang, L.; Sun, C.; Zhang, F.; Shang, L.; et al. Isolation and characterization of ftsz genes in cassava. Genes 2017, 8, 391. [Google Scholar] [CrossRef]
- Kadirjan-Kalbach, D.K.; Turmo, A.; Wang, J.; Smith, B.C.; Chen, C.; Porter, K.J.; Childs, K.L.; DellaPenna, D.; Osteryoung, K.W. Allelic variation in the chloroplast division gene FtsZ2-2 leads to natural variation in chloroplast size. Plant Physiol. 2019, 181, 1059–1074. [Google Scholar] [CrossRef]
- Hu, W.; Yang, H.; Yan, Y.; Wei, Y.; Tie, W.; Ding, Z.; Zuo, J.; Peng, M.; Li, K. Genome-wide characterization and analysis of bZIP transcription factor gene family related to abiotic stress in cassava. Sci. Rep. 2016, 6, 22783–22794. [Google Scholar] [CrossRef]
- Sojikul, P.; Saithong, T.; Kalapanulak, S.; Pisuttinusart, N.; Limsirichaikul, S.; Tanaka, M.; Utsumi, Y.; Sakurai, T.; Seki, M.; Narangajavana, J. Genome-wide analysis reveals phytohormone action during cassava storage root initiation. Plant Mol. Biol. 2015, 88, 531–543. [Google Scholar] [CrossRef]
- Wang, S.Y.; Faust, M.; Line, M.J. Apical dominance in apple (malus domestica borkh): The possible role of indole-3-acetic acid (IAA). Sci. Hortic. 1994, 60, 167–172. [Google Scholar] [CrossRef]
- Bertini, L.; Proietti, S.; Focaracci, F.; Sabatini, B.; Caruso, C. Epigenetic control of defense genes following MeJA-induced priming in rice (O. sativa). J. Plant Physiol. 2018, 228, 166–177. [Google Scholar]
- Zhao, H.; Wu, C.; Yan, Y.; Tie, W.; Ding, Z.; Liu, G.; Yan, W.; Li, Y.; Wang, W.; Peng, M.; et al. Genomic analysis of the core components of ABA signaling reveals their possible role in abiotic stress response in cassava. Environ. Exp. Bot. 2019, 167, 103855. [Google Scholar] [CrossRef]
- Quarrie, S.A. Genotypic differences in leaf water potential, abscisic acid and proline concentrations in spring wheat during drought stress. Ann. Bot. 1980, 46, 383–394. [Google Scholar] [CrossRef]
- Schaffer, M.A.; Fischer, R.L. Analysis of mRNAs that accumulate in response to low temperature identifies a thiol protease gene in tomato. Plant Physiol. 1988, 87, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Heng-Rui, L.I.; Jian-Kun, Z.; Wen-Wu, Q.; Lian-Jun, L.; Chong, P.; Zhen-Hua, L.; Qiu-Lian, N.; Jia-Wen, L.; Zhou-Mei, M.O.; Mei-Ying, L.U. Relationship between endogenous gibberellin and starch accumulation in cassava tuber. Hubei Agric. Sci. 2015, 54, 1628–1632. [Google Scholar]
- Kang, M.S. Water and Agricultural Sustainability Strategies; CRC Press: Leiden, The Netherlands; Balkema: Leiden, The Netherlands, 2010; pp. 169–278. [Google Scholar]
- Potter, S.C.; Aurelien, L.; Eddy, S.R.; Youngmi, P.; Rodrigo, L.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Zhao, P.; Wang, D.; Wang, R.; Kong, N.; Zhang, C.; Yang, C.; Wu, W.; Ma, H.; Chen, Q. Genome-wide analysis of the potato Hsp20 gene family: Identification, genomic organization and expression profiles in response to heat stress. BMC Genom. 2018, 19, 61–73. [Google Scholar] [CrossRef]
- Guo, A.Y.; Zhu, Q.H.; Chen, X.; Luo, J.C. GSDS: A gene structure display server. Hereditas 2007, 29, 1023–1026. [Google Scholar] [CrossRef]
- Tao, P.; Zhong, X.; Li, B.; Wang, W.; Yue, Z.; Lei, J.; Guo, W.; Huang, X. Genome-wide identification and characterization of aquaporin genes (AQPs) in Chinese cabbage (Brassica rapa ssp. pekinensis). Mol. Genet. Genom. 2014, 289, 1131–1145. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. Hered 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Xie, T.; Chen, C.; Luan, A.; Long, J.; Li, C.; Ding, Y.; He, Y. Genome-wide organization and expression profiling of the R2R3-MYB transcription factor family in pineapple (Ananas comosus). BMC Genom. 2017, 18, 503–518. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit Incorporating gamma-series methods and sliding window Strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.M.; Mcgettigan, P.A.; Mcwilliam, H.; Valentin, F.; Wallace, I.M.W.; Wilm, A.; Lopez, R.; et al. Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1807–1817. [Google Scholar] [CrossRef]
- Chuang, K.S.; Jan, M.L.; Wu, J.; Lu, J.C.; Chen, S.; Hsu, C.H.; Fu, Y.K. A maximum likelihood expectation maximization algorithm with thresholding. Comput. Med. Imaging Graph. 2005, 29, 571–578. [Google Scholar] [CrossRef]
- Howe, E.A.; Sinha, R.; Schlauch, D.; Quackenbush, J. RNA-Seq analysis in MeV. Bioinformatics 2011, 27, 3209–3210. [Google Scholar] [CrossRef]
- Salcedo, A.; Zambrana, C.; Siritunga, D. Comparative expression analysis of reference genes in field-grown cassava. Trop. Plant Biol. 2014, 7, 53–64. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]



| Gene Name | Gene ID | p-Value | Size (aa) | MW (KDa) | pI | Predicted Subcellular Location | Chr. |
|---|---|---|---|---|---|---|---|
| MeTubulin1 | Manes.08G061700 | 5.6 × 10−114 | 454 | 50.31 | 4.76 | cytoplasm | 8 |
| MeTubulin2 | Manes.08G024800 | 2.6 × 10−43 | 492 | 51.13 | 5.56 | chloroplast thylakoid membrane | 8 |
| MeTubulin3 | Manes.02G136200 | 3.4 × 10−114 | 453 | 50.06 | 4.77 | cytoplasm | 2 |
| MeTubulin4 | Manes.02G123600 | 2.8 × 10−114 | 454 | 50.35 | 4.75 | cytoplasm | 2 |
| MeTubulin5 | Manes.05G200200 | 2.7 × 10−114 | 453 | 50.08 | 4.77 | cytoplasm | 5 |
| MeTubulin6 | Manes.05G025900 | 6 × 10−94 | 481 | 53.40 | 5.70 | cytoplasm | 5 |
| MeTubulin7 | Manes.05G147000 | 1.5 × 10−112 | 456 | 50.38 | 4.74 | cytoplasm | 5 |
| MeTubulin8 | Manes.13G106300 | 8.6 × 10−94 | 424 | 45.79 | 4.93 | cytoplasm | 13 |
| MeTubulin9 | Manes.09G100400 | 2.2 × 10−113 | 451 | 49.93 | 4.85 | cytoplasm | 9 |
| MeTubulin10 | Manes.09G055800 | 1.6 × 10−42 | 493 | 51.31 | 5.96 | chloroplast stroma | 9 |
| MeTubulin11 | Manes.09G140200 | 1.5 × 10−98 | 456 | 49.69 | 5.00 | cytoplasm | 9 |
| MeTubulin12 | Manes.06G058600 | 8 × 10−112 | 451 | 49.89 | 4.78 | cytoplasm | 6 |
| MeTubulin13 | Manes.06G007200 | 2 × 10−114 | 454 | 50.39 | 4.72 | cytoplasm | 6 |
| MeTubulin14 | Manes.06G147900 | 9 × 10−115 | 454 | 50.33 | 4.75 | cytoplasm | 6 |
| MeTubulin15 | Manes.15G108500 | 3.3 × 10−99 | 456 | 49.70 | 5.00 | cytoplasm | 15 |
| MeTubulin16 | Manes.01G249000 | 7.2 × 10−94 | 480 | 53.24 | 5.67 | cytoplasm | 1 |
| MeTubulin17 | Manes.01G061400 | 4.2 × 10−112 | 453 | 50.12 | 4.68 | cytoplasm | 1 |
| MeTubulin18 | Manes.01G166100 | 2.6 × 10−114 | 454 | 50.35 | 4.74 | cytoplasm | 1 |
| MeTubulin19 | Manes.14G124600 | 1.1 × 10−113 | 451 | 49.84 | 4.78 | cytoplasm | 14 |
| MeTubulin20 | Manes.03G135500 | 6.1 × 10−38 | 421 | 43.29 | 6.93 | chloroplast stroma | 3 |
| MeTubulin21 | Manes.03G098100 | 2.8 × 10−100 | 457 | 49.74 | 4.90 | cytoplasm | 3 |
| MeTubulin22 | Manes.10G087300 | 3.7 × 10−100 | 457 | 49.57 | 4.93 | cytoplasm | 10 |
| MeTubulin23 | Manes.10G087200 | 3.8 × 10−73 | 421 | 45.61 | 4.74 | cytoplasm | 10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Cao, P.; Wang, C.; Guo, J.; Zang, Y.; Wu, K.; Ran, F.; Liu, L.; Wang, D.; Min, Y. Genome-Wide Analysis of Tubulin Gene Family in Cassava and Expression of Family Member FtsZ2-1 during Various Stress. Plants 2021, 10, 668. https://doi.org/10.3390/plants10040668
Li S, Cao P, Wang C, Guo J, Zang Y, Wu K, Ran F, Liu L, Wang D, Min Y. Genome-Wide Analysis of Tubulin Gene Family in Cassava and Expression of Family Member FtsZ2-1 during Various Stress. Plants. 2021; 10(4):668. https://doi.org/10.3390/plants10040668
Chicago/Turabian StyleLi, Shuangbao, Peng Cao, Congcong Wang, Jianchun Guo, Yuwei Zang, Kunlin Wu, Fangfang Ran, Liangwang Liu, Dayong Wang, and Yi Min. 2021. "Genome-Wide Analysis of Tubulin Gene Family in Cassava and Expression of Family Member FtsZ2-1 during Various Stress" Plants 10, no. 4: 668. https://doi.org/10.3390/plants10040668
APA StyleLi, S., Cao, P., Wang, C., Guo, J., Zang, Y., Wu, K., Ran, F., Liu, L., Wang, D., & Min, Y. (2021). Genome-Wide Analysis of Tubulin Gene Family in Cassava and Expression of Family Member FtsZ2-1 during Various Stress. Plants, 10(4), 668. https://doi.org/10.3390/plants10040668






