The Diverse Salt-Stress Response of Arabidopsis ctr1-1 and ein2-1 Ethylene Signaling Mutants Is Linked to Altered Root Auxin Homeostasis
Abstract
1. Introduction
2. Results
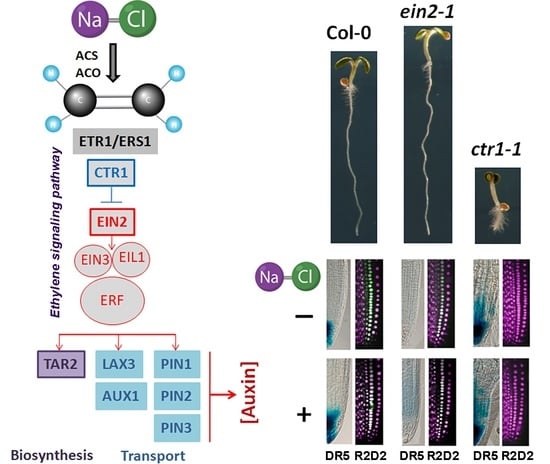
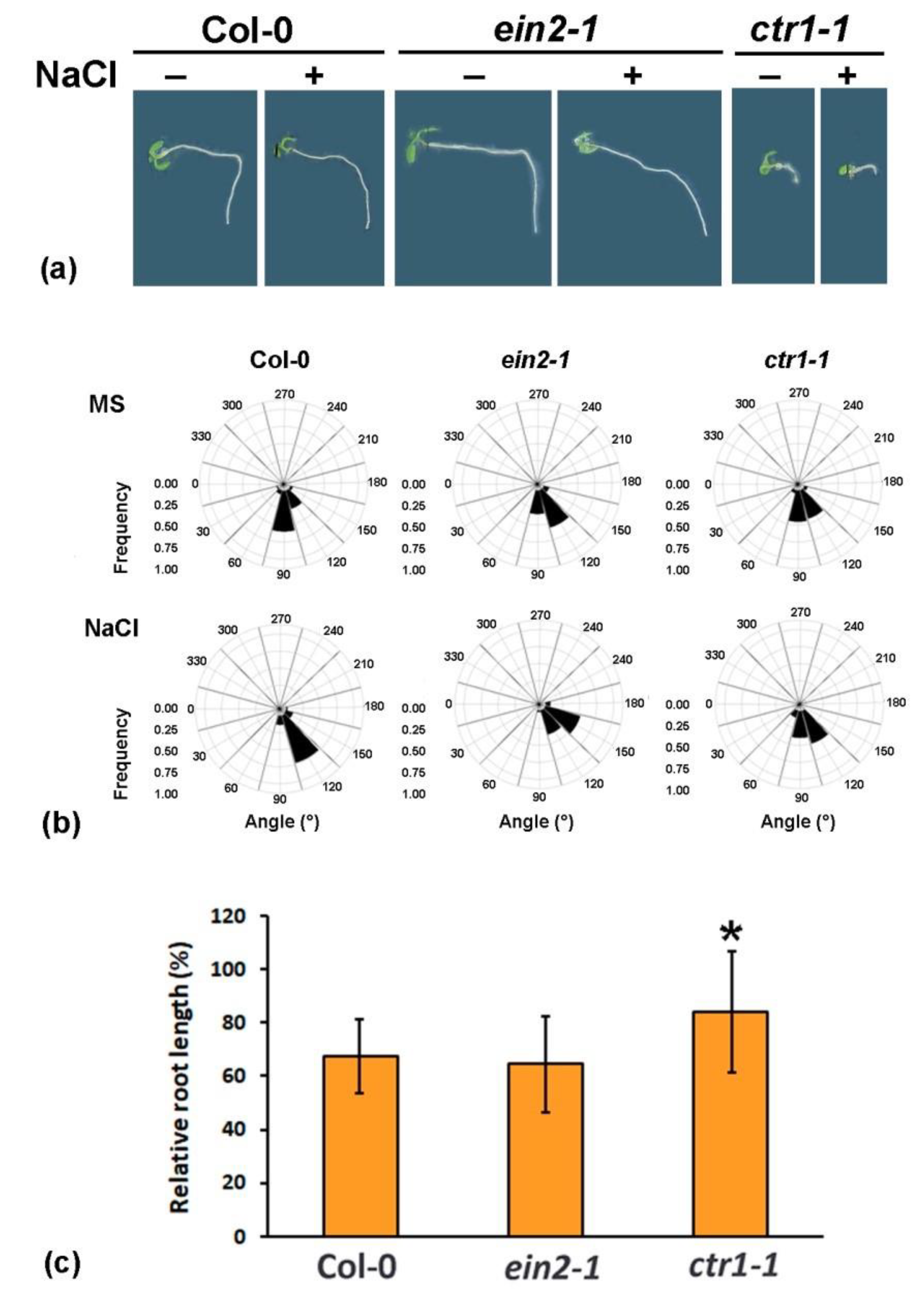
2.1. Altered Gravity Bending of the Ethylene-Signaling Mutants at High Salinity
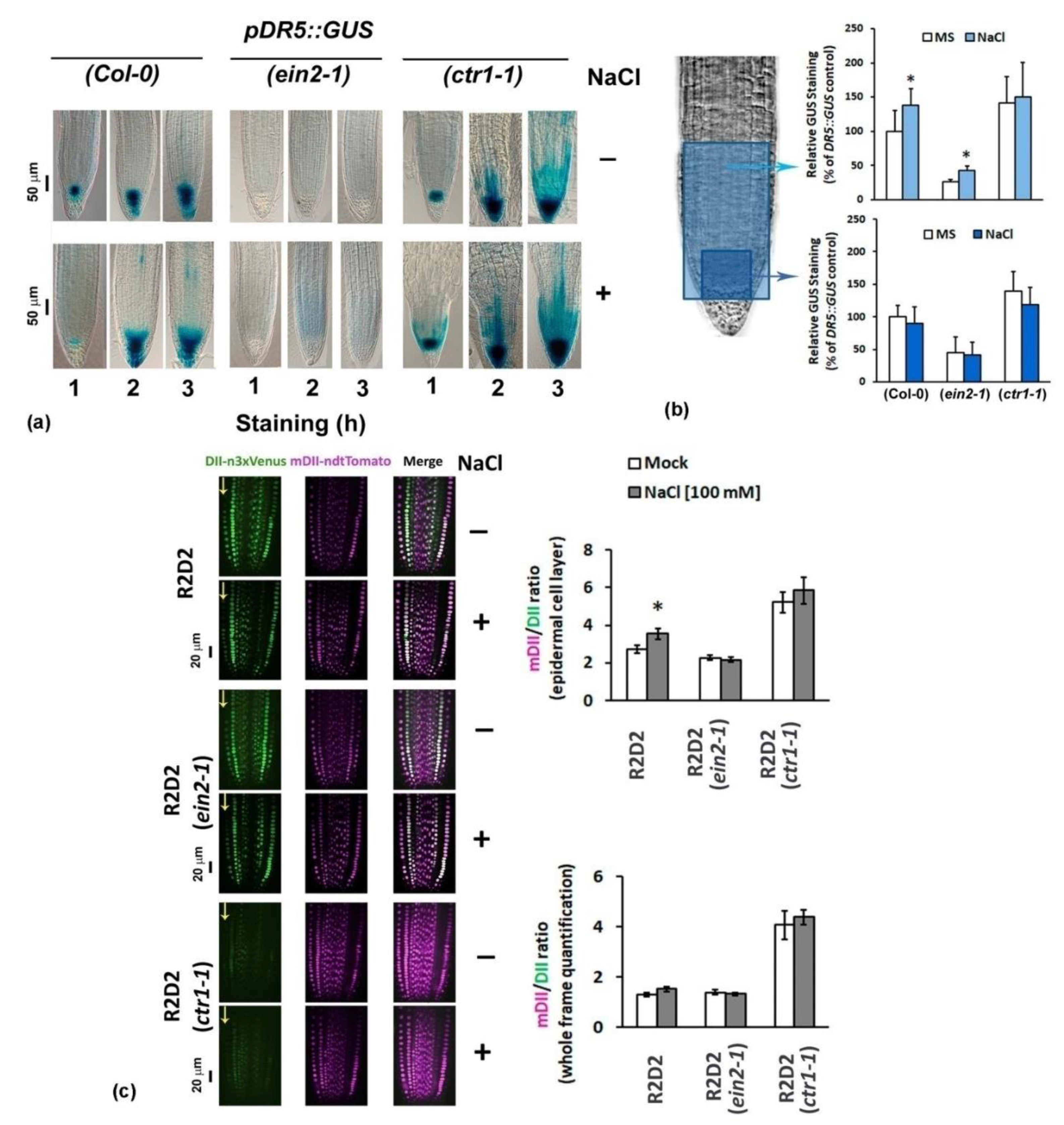
2.2. Differential Performance of the Ethylene-Signaling Mutants under Salt Stress Corresponds to Severely Altered Auxin Content in the Primary Root
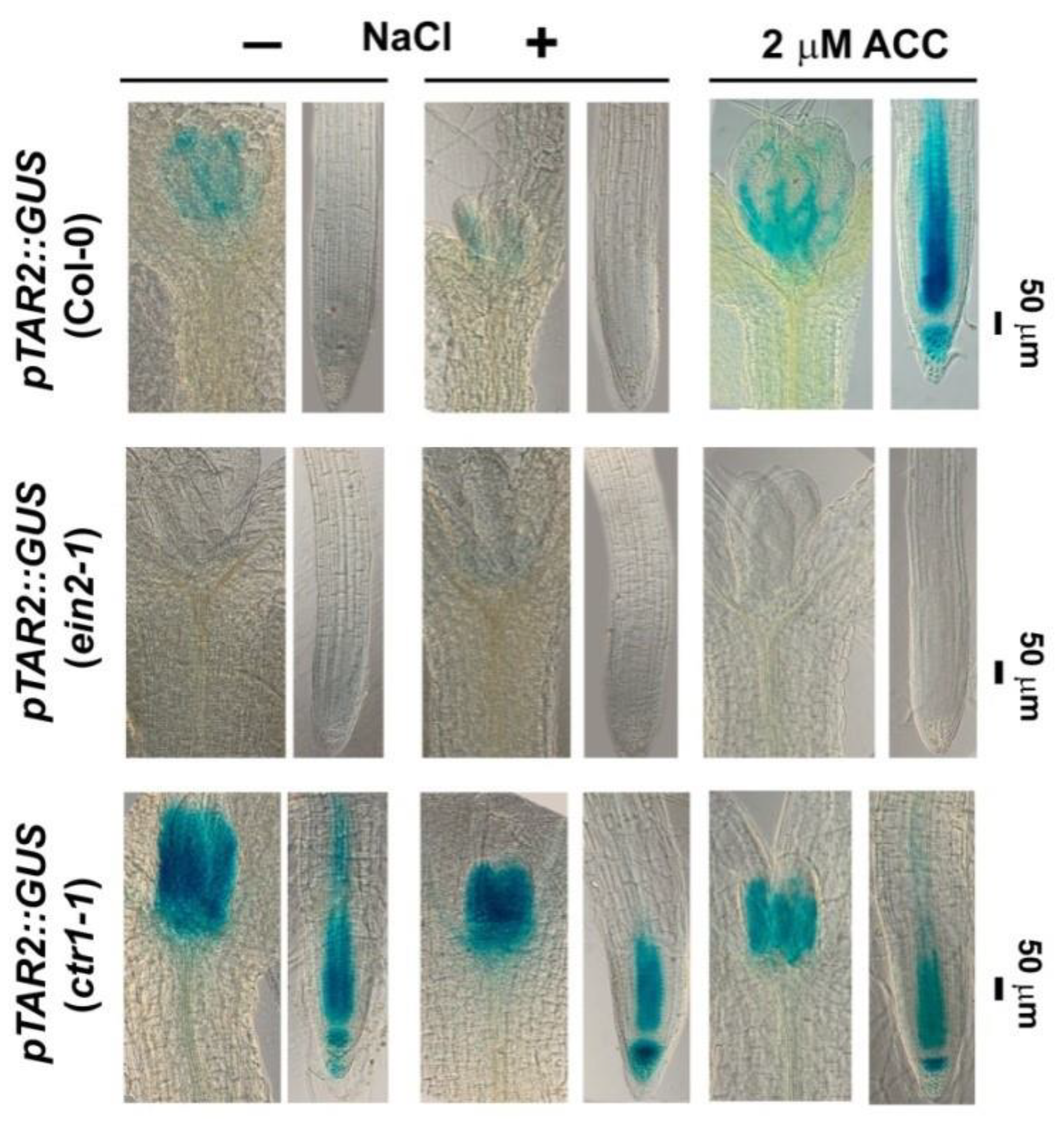
2.3. Constitutive Mutant ctr1-1 Has Increased Local Auxin Biosynthesis
2.4. Stabilized Polar AuxinTtransport in Salt-Treated ctr1-1 Plants
2.4.1. In Silico Characterization of Auxin Transport Genes Reveals Regulation by Ethylene-Related Transcription Factors
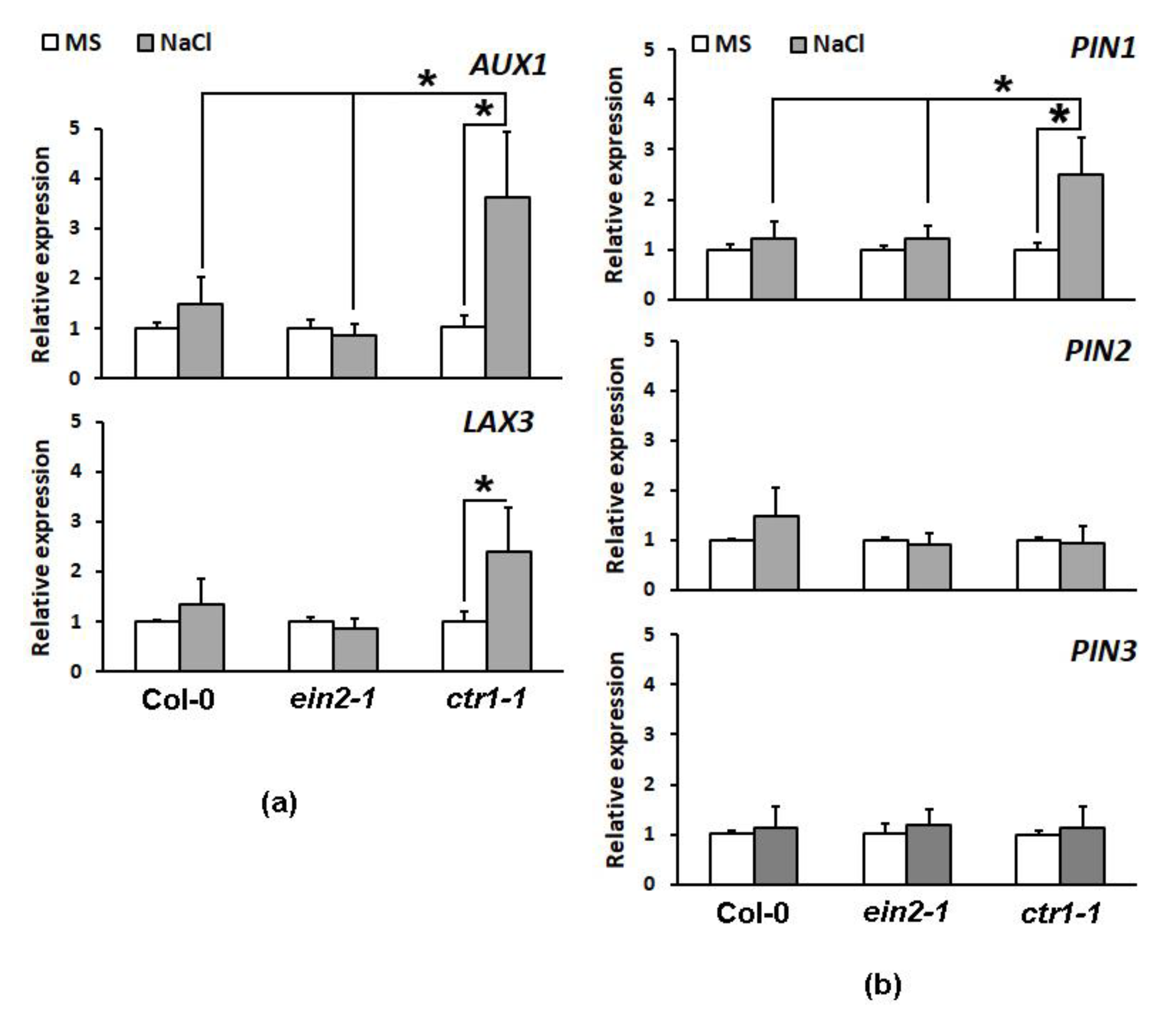
2.4.2. Expression Profiling of Auxin Transporter Genes
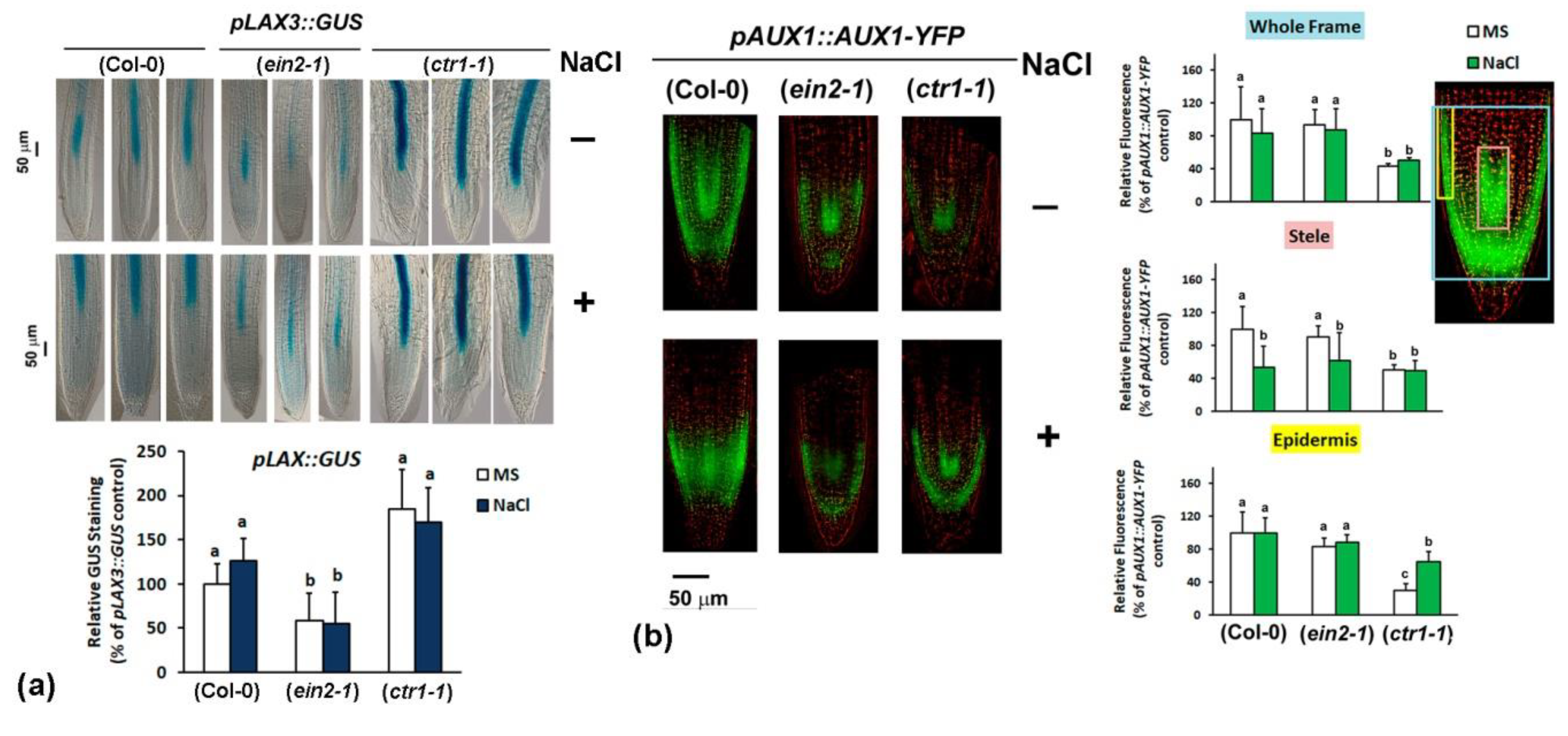
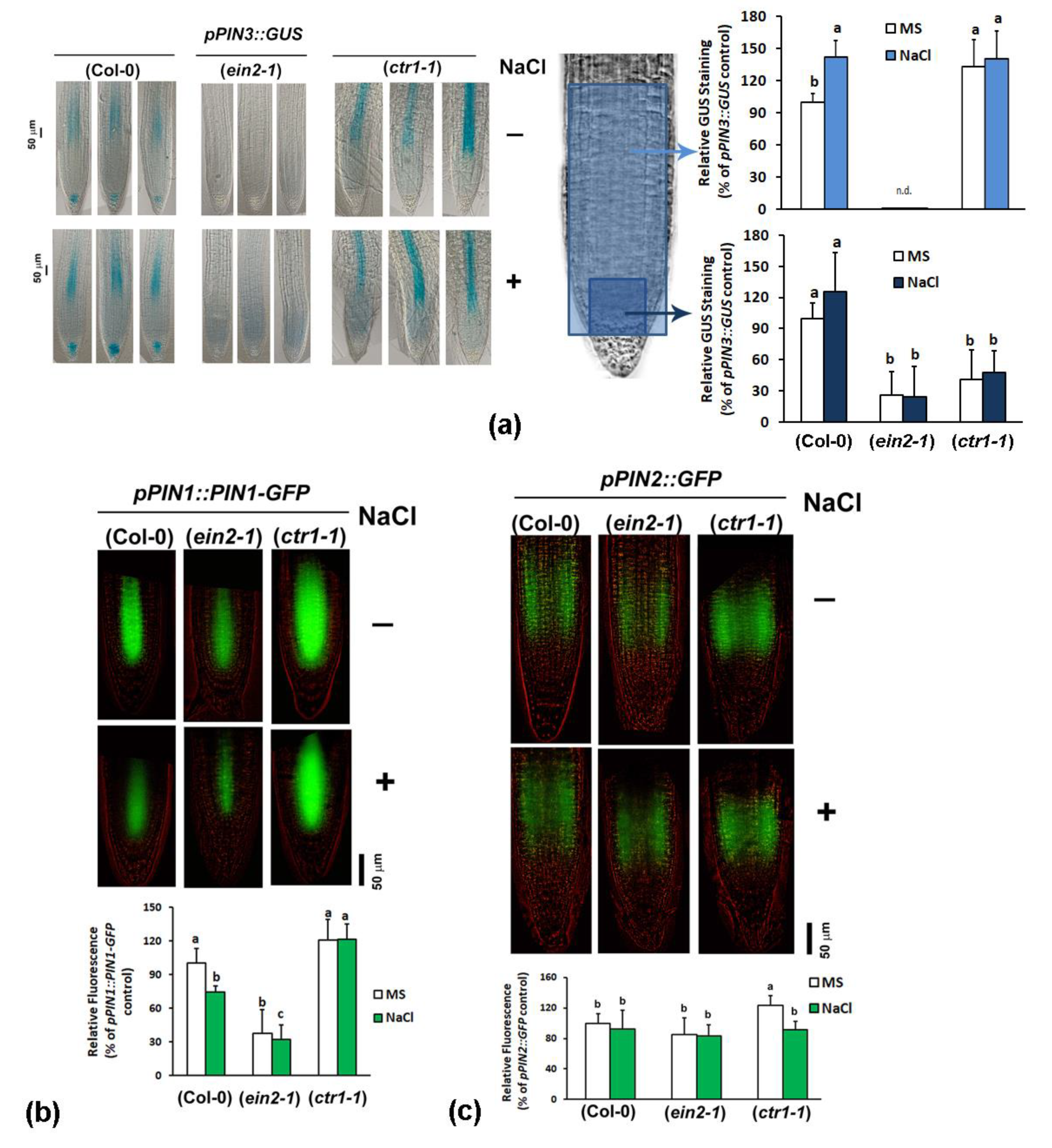
2.4.3. Analyses of Auxin Transport Reporters in Ethylene Mutant Backgrounds
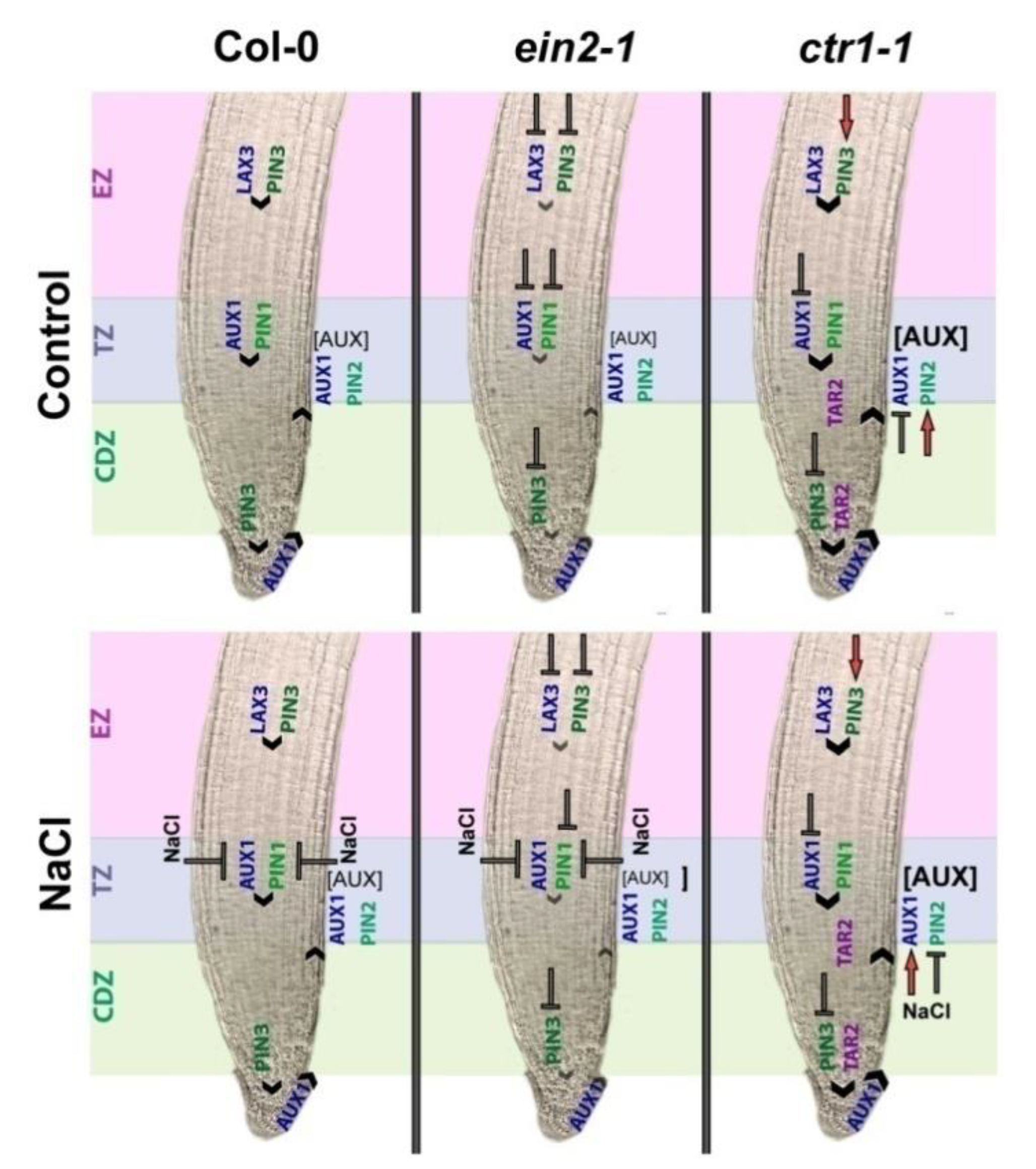
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. GUS Staining and Quantification
4.3. Fluorescent and Confocal Microscopic Observations
4.4. TF DEACoN and AthaMap Analyses
4.5. Real-Time Quantitative RT-PCR Analyses
4.6. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Kaya, C.; Tuna, A.L.; Yokaş, I. The Role of Plant Hormones in Plants Under Salinity Stress. In Sabkha Ecosystems; Springer International Publishing: Berlin/Heidelberg, Germany, 2009; Volume 44, pp. 45–50. [Google Scholar] [CrossRef]
- Golldack, D.; Li, C.; Mohan, H.; Probst, N. Tolerance to drought and salt stress in plants: Unraveling the signaling networks. Front. Plant Sci. 2014, 5, 151. [Google Scholar] [CrossRef] [PubMed]
- Abeles, F.; Morgan, P.; Saltveit, M. Ethylene in Plant Biology, 2nd ed.; Academic Press: San Diego, CA, USA, 1992; pp. 147–155. [Google Scholar]
- Etao, J.-J.; Echen, H.-W.; Ema, B.; Ezhang, W.-K.; Echen, S.-Y.; Ezhang, J.-S. The Role of Ethylene in Plants Under Salinity Stress. Front. Plant Sci. 2015, 6, 1059. [Google Scholar] [CrossRef]
- Achard, P.; Chen, D.; Steele, A.D.; Lindquist, S.; Guarente, L. Integration of Plant Responses to Environmentally Activated Phytohormonal Signals. Science 2006, 311, 91–94. [Google Scholar] [CrossRef]
- Cao, W.-H.; Liu, J.; He, X.-J.; Mu, R.-L.; Zhou, H.-L.; Chen, S.-Y.; Zhang, J.-S. Modulation of Ethylene Responses Affects Plant Salt-Stress Responses. Plant Physiol. 2006, 143, 707–719. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, C.; Li, K.; Sun, F.; Hu, H.; Li, X.; Zhao, Y.; Han, C.; Zhang, W.; Duan, Y.; et al. Arabidopsis EIN2 modulates stress response through abscisic acid response pathway. Plant Mol. Biol. 2007, 64, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, P.; Gao, Q.; Liu, X.; Kuang, T.; Shen, S.; He, Y. Proteomic analysis of the response to high-salinity stress in Physcomitrella patens. Planta 2008, 228, 167–177. [Google Scholar] [CrossRef]
- Jiang, C.; Belfield, E.J.; Cao, Y.; Smith, J.A.C.; Harberd, N.P. An Arabidopsis Soil-Salinity–Tolerance Mutation Confers Ethylene-Mediated Enhancement of Sodium/Potassium Homeostasis. Plant Cell 2013, 25, 3535–3552. [Google Scholar] [CrossRef] [PubMed]
- Li, C.-H.; Wang, G.; Zhao, J.-L.; Zhang, L.-Q.; Ai, L.-F.; Han, Y.-F.; Sun, D.-Y.; Zhang, S.-W.; Sun, Y. The Receptor-Like Kinase SIT1 Mediates Salt Sensitivity by Activating MAPK3/6 and Regulating Ethylene Homeostasis in Rice. Plant Cell 2014, 26, 2538–2553. [Google Scholar] [CrossRef] [PubMed]
- Vaseva, I.I.; Qudeimat, E.; Potuschak, T.; Du, Y.; Genschik, P.; Vandenbussche, F.; Van Der Straeten, D. The plant hormone ethylene restricts Arabidopsis growth via the epidermis. Proc. Natl. Acad. Sci. USA 2018, 115, E4130–E4139. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The Salt Overly Sensitive (SOS) Pathway: Established and Emerging Roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Kieber, J.J.; Rothenberg, M.; Roman, G.; Feldmann, K.A.; Ecker, J.R. CTR1, a negative regulator of the ethylene response pathway in arabidopsis, encodes a member of the Raf family of protein kinases. Cell 1993, 72, 427–441. [Google Scholar] [CrossRef]
- Alonso, J.M.; Hirayama, T.; Roman, G.; Nourizadeh, S.; Ecker, J.R. EIN2, a Bifunctional Transducer of Ethylene and Stress Responses in Arabidopsis. Science 1999, 284, 2148–2152. [Google Scholar] [CrossRef]
- Li, W.; Ma, M.; Feng, Y.; Li, H.; Wang, Y.; Ma, Y.; Li, M.; An, F.; Guo, H. EIN2-Directed Translational Regulation of Ethylene Signaling in Arabidopsis. Cell 2015, 163, 670–683. [Google Scholar] [CrossRef]
- Merchante, C.; Brumos, J.; Yun, J.; Hu, Q.; Spencer, K.R.; Enríquez, P.; Binder, B.M.; Heber, S.; Stepanova, A.N.; Alonso, J.M. Gene-Specific Translation Regulation Mediated by the Hormone-Signaling Molecule EIN2. Cell 2015, 163, 684–697. [Google Scholar] [CrossRef]
- Peng, J.; Li, Z.; Wen, X.; Li, W.; Shi, H.; Yang, L.; Zhu, H.; Guo, H. Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation in Arabidopsis. PLoS Genet. 2014, 10, e1004664. [Google Scholar] [CrossRef]
- Ulmasov, T.; Murfett, J.; Hagen, G.; Guilfoyle, T.J. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 1997, 9, 1963–1971. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.-Y.; Smet, W.; Brunoud, G.; Yoshida, S.; Vernoux, T.; Weijers, D. Reporters for sensitive and quantitative measurement of auxin response. Nat. Methods 2015, 12, 207–210. [Google Scholar] [CrossRef]
- Su, W.; Howell, S.H. A Single Genetic Locus, Ckr1, Defines Arabidopsis Mutants in which Root Growth Is Resistant to Low Concentrations of Cytokinin. Plant Physiol. 1992, 99, 1569–1574. [Google Scholar] [CrossRef] [PubMed]
- Fujita, H.; Syono, K. Genetic Analysis of the Effects of Polar Auxin Transport Inhibitors on Root Growth in Arabidopsis thaliana. Plant Cell Physiol. 1996, 37, 1094–1101. [Google Scholar] [CrossRef]
- Beaudoin, N.; Serizet, C.; Gosti, F.; Giraudat, J. Interactions between Abscisic Acid and Ethylene Signaling Cascades. Plant Cell 2000, 12, 1103–1115. [Google Scholar] [CrossRef] [PubMed]
- Ghassemian, M.; Nambara, E.; Cutler, S.; Kawaide, H.; Kamiya, Y.; McCourt, P. Regulation of Abscisic Acid Signaling by the Ethylene Response Pathway in Arabidopsis. Plant Cell 2000, 12, 1117–1126. [Google Scholar] [CrossRef]
- Müller, M.; Munné-Bosch, S. Ethylene Response Factors: A Key Regulatory Hub in Hormone and Stress Signaling. Plant Physiol. 2015, 169, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Grieneisen, V.A.; Xu, J.; Marée, A.F.M.; Hogeweg, P.; Scheres, B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nat. Cell Biol. 2007, 449, 1008–1013. [Google Scholar] [CrossRef] [PubMed]
- Chai, S.; Li, E.; Zhang, Y.; Li, S. NRT1.1-Mediated Nitrate Suppression of Root Coiling Relies on PIN2- and AUX1-Mediated Auxin Transport. Front. Plant Sci. 2020, 11, 671. [Google Scholar] [CrossRef]
- Di Mambro, R.; De Ruvo, M.; Pacifici, E.; Salvi, E.; Sozzani, R.; Benfey, P.N.; Busch, W.; Novak, O.; Ljung, K.; Di Paola, L.; et al. Auxin minimum triggers the developmental switch from cell division to cell differentiation in the Arabidopsis root. Proc. Natl. Acad. Sci. USA 2017, 114, E7641–E7649. [Google Scholar] [CrossRef] [PubMed]
- Korver, R.A.; Koevoets, I.T.; Testerink, C. Out of Shape During Stress: A Key Role for Auxin. Trends Plant Sci. 2018, 23, 783–793. [Google Scholar] [CrossRef]
- Muday, G.K. Auxins and Tropisms. J. Plant Growth Regul. 2001, 20, 226–243. [Google Scholar] [CrossRef]
- Friml, J.; Wiśniewska, J.; Benková, E.; Mendgen, K.; Palme, K. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nat. Cell Biol. 2002, 415, 806–809. [Google Scholar] [CrossRef]
- Vandenbussche, F.; Petrášek, J.; Žádníková, P.; Hoyerová, K.; Pešek, B.; Raz, V.; Swarup, R.; Bennett, M.; Zažímalová, E.; Benková, E.; et al. The auxin influx carriers AUX1 and LAX3 are involved in auxin-ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Development 2010, 137, 597–606. [Google Scholar] [CrossRef]
- Méndez-Bravo, A.; Ruiz-Herrera, L.F.; Cruz-Ramírez, A.; Guzman, P.; Martínez-Trujillo, M.; Ortiz-Castro, R.; López-Bucio, J. CONSTITUTIVE TRIPLE RESPONSE1 and PIN2 act in a coordinate manner to support the indeterminate root growth and meristem cell proliferating activity in Arabidopsis seedlings. Plant Sci. 2019, 280, 175–186. [Google Scholar] [CrossRef]
- Sun, F.; Zhang, W.; Hu, H.; Li, B.; Wang, Y.; Zhao, Y.; Li, K.; Liu, M.; Li, X. Salt Modulates Gravity Signaling Pathway to Regulate Growth Direction of Primary Roots in Arabidopsis. Plant Physiol. 2008, 146, 178–188. [Google Scholar] [CrossRef]
- Buer, C.S.; Sukumar, P.; Muday, G.K. Ethylene Modulates Flavonoid Accumulation and Gravitropic Responses in Roots of Arabidopsis. Plant Physiol. 2006, 140, 1384–1396. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, C.; Sakata, Y.; Taji, T.; Baba, T.; Tanaka, S. Unique ethylene-regulated touch responses of Arabidopsis thaliana roots to physical hardness. J. Plant Res. 2008, 121, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Galvan-Ampudia, C.S.; Julkowska, M.M.; Darwish, E.; Gandullo, J.; Korver, R.A.; Brunoud, G.; Haring, M.A.; Munnik, T.; Vernoux, T.; Testerink, C. Halotropism Is a Response of Plant Roots to Avoid a Saline Environment. Curr. Biol. 2013, 23, 2044–2050. [Google Scholar] [CrossRef]
- Brumos, J.; Zhao, C.; Gong, Y.; Soriano, D.; Patel, A.P.; Perez-Amador, M.A.; Stepanova, A.N.; Alonso, J.M. An Improved Recombineering Toolset for Plants. Plant Cell 2020, 32, 100–122. [Google Scholar] [CrossRef]
- Steffens, N.O. AthaMap: An online resource for in silico transcription factor binding sites in the Arabidopsis thaliana genome. Nucleic Acids Res. 2004, 32, 368D–372. [Google Scholar] [CrossRef]
- Philosoph-Hadas, S.; Friedman, H.; Meir, S. Gravitropic Bending and Plant Hormones. In Vitamins & Hormones; Elsevier BV: Amsterdam, The Netherlands, 2005; Volume 72, pp. 31–78. [Google Scholar]
- Paciorek, T.; Zažímalová, E.; Ruthardt, N.; Petrášek, J.; Stierhof, Y.-D.; Kleine-Vehn, J.; Morris, D.A.; Emans, N.; Jürgens, G.; Geldner, N.; et al. Auxin inhibits endocytosis and promotes its own efflux from cells. Nat. Cell Biol. 2005, 435, 1251–1256. [Google Scholar] [CrossRef]
- Dhonukshe, P.; Grigoriev, I.; Fischer, R.; Tominaga, M.; Robinson, D.G.; Hašek, J.; Paciorek, T.; Petrášek, J.; Seifertová, D.; Tejos, R.; et al. Auxin transport inhibitors impair vesicle motility and actin cytoskeleton dynamics in diverse eukaryotes. Proc. Natl. Acad. Sci. USA 2008, 105, 4489–4494. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Li, R.-J.; Han, T.-T.; Cai, W.; Fu, Z.-W.; Lu, Y.-T. Salt Stress Reduces Root Meristem Size by Nitric Oxide-Mediated Modulation of Auxin Accumulation and Signaling in Arabidopsis. Plant Physiol. 2015, 168, 343–356. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, A.N.; Robertson-Hoyt, J.; Yun, J.; Benavente, L.M.; Xie, D.-Y.; Doležal, K.; Schlereth, A.; Jürgens, G.; Alonso, J.M. TAA1-Mediated Auxin Biosynthesis Is Essential for Hormone Crosstalk and Plant Development. Cell 2008, 133, 177–191. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, A.N.; Yun, J.; Robles, L.M.; Novak, O.; He, W.; Guo, H.; Ljung, K.; Alonso, J.M. The Arabidopsis YUCCA1 Flavin Monooxygenase Functions in the Indole-3-Pyruvic Acid Branch of Auxin Biosynthesis. Plant Cell 2011, 23, 3961–3973. [Google Scholar] [CrossRef]
- Yang, Z.-B.; Geng, X.; He, C.; Zhang, F.; Wang, R.; Horst, W.J.; Ding, Z. TAA1-Regulated Local Auxin Biosynthesis in the Root-Apex Transition Zone Mediates the Aluminum-Induced Inhibition of Root Growth in Arabidopsis. Plant Cell 2014, 26, 2889–2904. [Google Scholar] [CrossRef]
- Brumos, J.; Robles, L.M.; Yun, J.; Vu, T.C.; Jackson, S.; Alonso, J.M.; Stepanova, A.N. Local Auxin Biosynthesis Is a Key Regulator of Plant Development. Dev. Cell 2018, 47, 306–318.e5. [Google Scholar] [CrossRef]
- Harkey, A.F.; Sims, K.N.; Muday, G.K. A new tool for discovering transcriptional regulators of co-expressed genes predicts gene regulatory networks that mediate ethylene-controlled root development. Silico Plants 2020, 2, 006. [Google Scholar] [CrossRef]
- Vandenbussche, F.; Vaseva, I.; Vissenberg, K.; Van Der Straeten, D. Ethylene in vegetative development: A tale with a riddle. New Phytol. 2012, 194, 895–909. [Google Scholar] [CrossRef]
- He, W.; Brumos, J.; Li, H.; Ji, Y.; Ke, M.; Gong, X.; Zeng, Q.; Li, W.; Zhang, X.; An, F.; et al. A Small-Molecule Screen Identifies l-Kynurenine as a Competitive Inhibitor of TAA1/TAR Activity in Ethylene-Directed Auxin Biosynthesis and Root Growth in Arabidopsis. Plant Cell 2011, 23, 3944–3960. [Google Scholar] [CrossRef]
- Hentrich, M.; Sánchez-Parra, B.; Alonso, M.-M.P.; Loba, V.C.; Carrillo, L.; Vicente-Carbajosa, J.; Medina, J.; Pollmann, S. YUCCA8 and YUCCA9 overexpression reveals a link between auxin signaling and lignification through the induction of ethylene biosynthesis. Plant Signal. Behav. 2013, 8, e26363. [Google Scholar] [CrossRef] [PubMed]
- Kriechbaumer, V.; Botchway, S.W.; Hawes, C. Localization and interactions between Arabidopsis auxin biosynthetic enzymes in the TAA/YUC-dependent pathway. J. Exp. Bot. 2016, 67, 4195–4207. [Google Scholar] [CrossRef] [PubMed]
- Růžička, K.; Ljung, K.; Vanneste, S.; Podhorská, R.; Beeckman, T.; Friml, J.; Benková, E. Ethylene Regulates Root Growth through Effects on Auxin Biosynthesis and Transport-Dependent Auxin Distribution. Plant Cell 2007, 19, 2197–2212. [Google Scholar] [CrossRef] [PubMed]
- Saini, K.; AbdelGawad, H.; Markakis, M.N.; Schoenaers, S.; Asard, H.; Prinsen, E.; Beemster, G.T.S.; Vissenberg, K. Perturbation of Auxin Homeostasis and Signaling by PINOID Overexpression Induces Stress Responses in Arabidopsis. Front. Plant Sci. 2017, 8, 1308. [Google Scholar] [CrossRef] [PubMed]
- Junghans, U.; Polle, A.; Duchting, P.; Weiler, E.W.; Kuhlman, B.; Gruber, F.; Teichmann, T. Adaptation to high salinity in poplar involves changes in xylem anatomy and auxin physiology. Plant Cell Environ. 2006, 29, 1519–1531. [Google Scholar] [CrossRef] [PubMed]
- Ribba, T.; Garrido-Vargas, F.; O’Brien, J.A. Auxin-mediated responses under salt stress: From developmental regulation to biotechnological applications. J. Exp. Bot. 2020, 71, 3843–3853. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Jiang, Q.; Hu, Z.; Sun, X.; Fan, S.; Zhang, H. Function of the auxin-responsive gene TaSAUR75 under salt and drought stress. Crop. J. 2018, 6, 181–190. [Google Scholar] [CrossRef]
- Swarup, R.; Perry, P.; Hagenbeek, D.; Van Der Straeten, D.; Beemster, G.T.; Sandberg, G.; Bhalerao, R.; Ljung, K.; Bennett, M.J. Ethylene Upregulates Auxin Biosynthesis in Arabidopsis Seedlings to Enhance Inhibition of Root Cell Elongation. Plant Cell 2007, 19, 2186–2196. [Google Scholar] [CrossRef] [PubMed]
- Pitts, R.J.; Cernac, A.; Estelle, M. Auxin and ethylene promote root hair elongation inArabidopsis. Plant J. 1998, 16, 553–560. [Google Scholar] [CrossRef]
- Rahman, A.; Amakawa, T.; Goto, N.; Tsurumi, S. Auxin is a Positive Regulator for Ethylene-Mediated Response in the Growth of Arabidopsis Roots. Plant Cell Physiol. 2001, 42, 301–307. [Google Scholar] [CrossRef]
- Rahman, A.; Hosokawa, S.; Oono, Y.; Amakawa, T.; Goto, N.; Tsurumi, S. Auxin and Ethylene Response Interactions during Arabidopsis Root Hair Development Dissected by Auxin Influx Modulators. Plant Physiol. 2002, 130, 1908–1917. [Google Scholar] [CrossRef]
- Zhang, S.; Huang, L.; Yan, A.; Liu, Y.; Liu, B.; Yu, C.; Zhang, A.; Schiefelbein, J.; Gan, Y. Multiple phytohormones promote root hair elongation by regulating a similar set of genes in the root epidermis in Arabidopsis. J. Exp. Bot. 2016, 67, 6363–6372. [Google Scholar] [CrossRef] [PubMed]
- Ivanchenko, M.G.; Muday, G.K.; Dubrovsky, J.G. Ethyleneauxin interactions regulate lateral root initiation and emergence inArabidopsis thaliana. Plant J. 2008, 55, 335–347. [Google Scholar] [CrossRef]
- Arif, M.R.; Islam, M.T.; Robin, A.H.K. Salinity Stress Alters Root Morphology and Root Hair Traits in Brassica napus. Plants 2019, 8, 192. [Google Scholar] [CrossRef]
- Swarup, R.; Kramer, E.M.; Perry, P.; Knox, K.; Leyser, H.M.O.; Haseloff, J.; Beemster, G.T.S.; Bhalerao, R.; Bennett, M.J. Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat. Cell Biol. 2005, 7, 1057–1065. [Google Scholar] [CrossRef] [PubMed]
- Ribaut, J.M.; Pilet, P.E. Water stress and indol-3yl-acetic acid content of maize roots. Planta 1994, 193, 502–507. [Google Scholar] [CrossRef]
- Ma, Q.; Grones, P.; Robert, S. Auxin signaling: A big question to be addressed by small molecules. J. Exp. Bot. 2017, 69, 313–328. [Google Scholar] [CrossRef]
- Benková, E.; Michniewicz, M.; Sauer, M.; Teichmann, T.; Seifertová, D.; Jürgens, G.; Friml, J. Local, Efflux-Dependent Auxin Gradients as a Common Module for Plant Organ Formation. Cell 2003, 115, 591–602. [Google Scholar] [CrossRef]
- Petrášek, J.; Mravec, J.; Bouchard, R.; Blakeslee, J.J.; Abas, M.; Seifertová, D.; Wiśniewska, J.; Tadele, Z.; Kubeš, M.; Čovanová, M.; et al. PIN Proteins Perform a Rate-Limiting Function in Cellular Auxin Efflux. Science 2006, 312, 914–918. [Google Scholar] [CrossRef] [PubMed]
- Swarup, R.; Kargul, J.; Marchant, A.; Zadik, D.; Rahman, A.; Mills, R.; Yemm, A.; May, S.; Williams, L.; Millner, P.; et al. Structure-Function Analysis of the Presumptive Arabidopsis Auxin Permease AUX1. Plant Cell 2004, 16, 3069–3083. [Google Scholar] [CrossRef]
- Swarup, K.; Benková, E.; Swarup, R.; Casimiro, I.; Péret, B.; Yang, Y.; Parry, G.; Nielsen, E.; De Smet, I.; Vanneste, S.; et al. The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 2008, 10, 946–954. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vaseva, I.I.; Mishev, K.; Depaepe, T.; Vassileva, V.; Van Der Straeten, D. The Diverse Salt-Stress Response of Arabidopsis ctr1-1 and ein2-1 Ethylene Signaling Mutants Is Linked to Altered Root Auxin Homeostasis. Plants 2021, 10, 452. https://doi.org/10.3390/plants10030452
Vaseva II, Mishev K, Depaepe T, Vassileva V, Van Der Straeten D. The Diverse Salt-Stress Response of Arabidopsis ctr1-1 and ein2-1 Ethylene Signaling Mutants Is Linked to Altered Root Auxin Homeostasis. Plants. 2021; 10(3):452. https://doi.org/10.3390/plants10030452
Chicago/Turabian StyleVaseva, Irina I., Kiril Mishev, Thomas Depaepe, Valya Vassileva, and Dominique Van Der Straeten. 2021. "The Diverse Salt-Stress Response of Arabidopsis ctr1-1 and ein2-1 Ethylene Signaling Mutants Is Linked to Altered Root Auxin Homeostasis" Plants 10, no. 3: 452. https://doi.org/10.3390/plants10030452
APA StyleVaseva, I. I., Mishev, K., Depaepe, T., Vassileva, V., & Van Der Straeten, D. (2021). The Diverse Salt-Stress Response of Arabidopsis ctr1-1 and ein2-1 Ethylene Signaling Mutants Is Linked to Altered Root Auxin Homeostasis. Plants, 10(3), 452. https://doi.org/10.3390/plants10030452