Cytochrome c Deficiency Differentially Affects the In Vivo Mitochondrial Electron Partitioning and Primary Metabolism Depending on the Photoperiod
Abstract
1. Introduction
2. Results
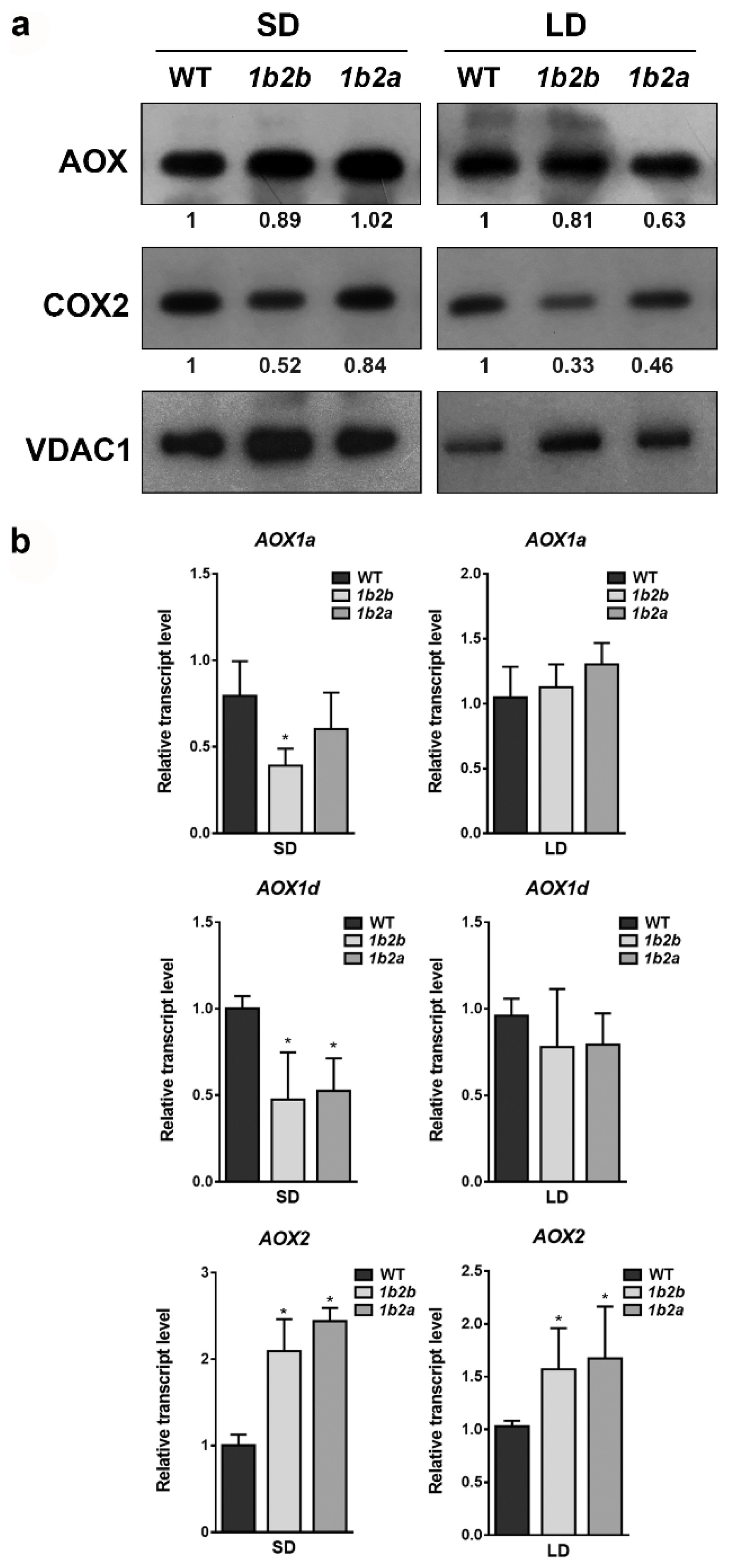
2.1. Differences in the Relative Expression of Respiratory Chain Components between Cytc Mutants and WT Plants under Short- and Long-Day Photoperiods
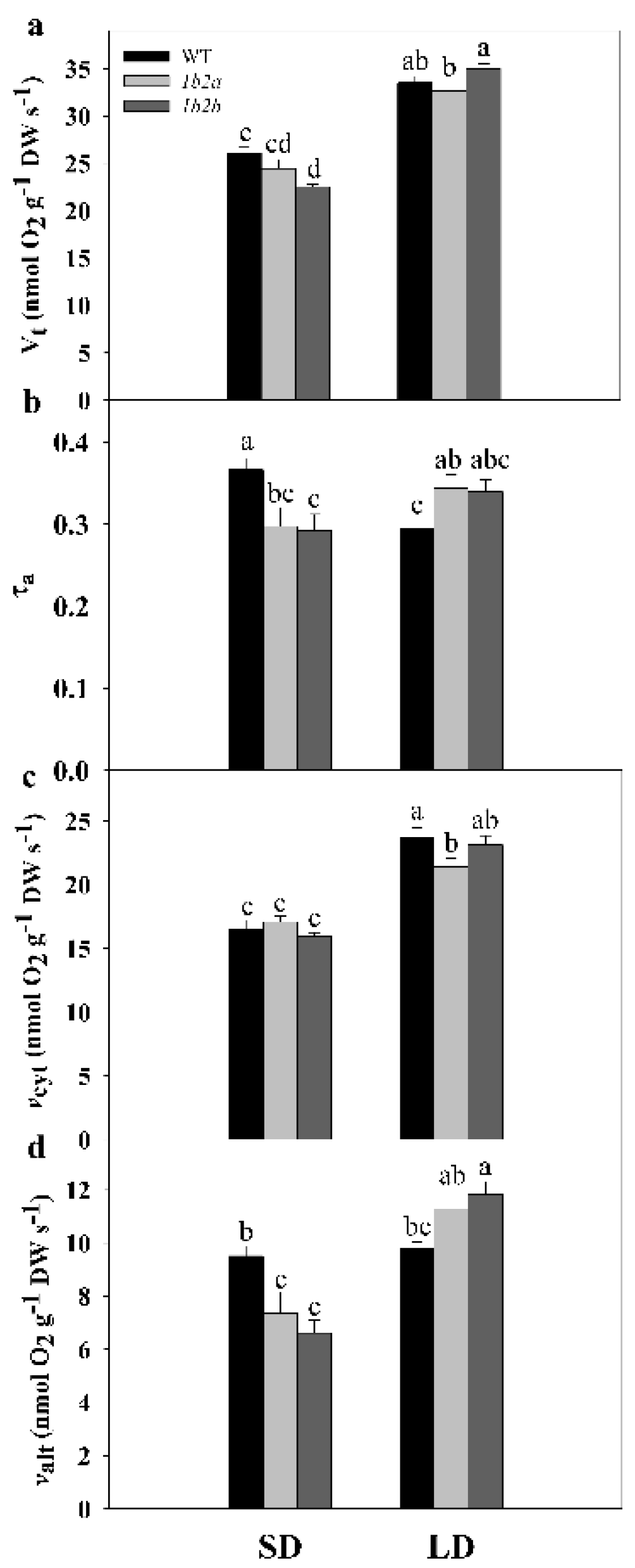
2.2. Respiration, Electron Partitioning to the AOX Pathway, and Photosynthesis in Cytc Mutants and WT Plants under Short- and Long-Day Photoperiods
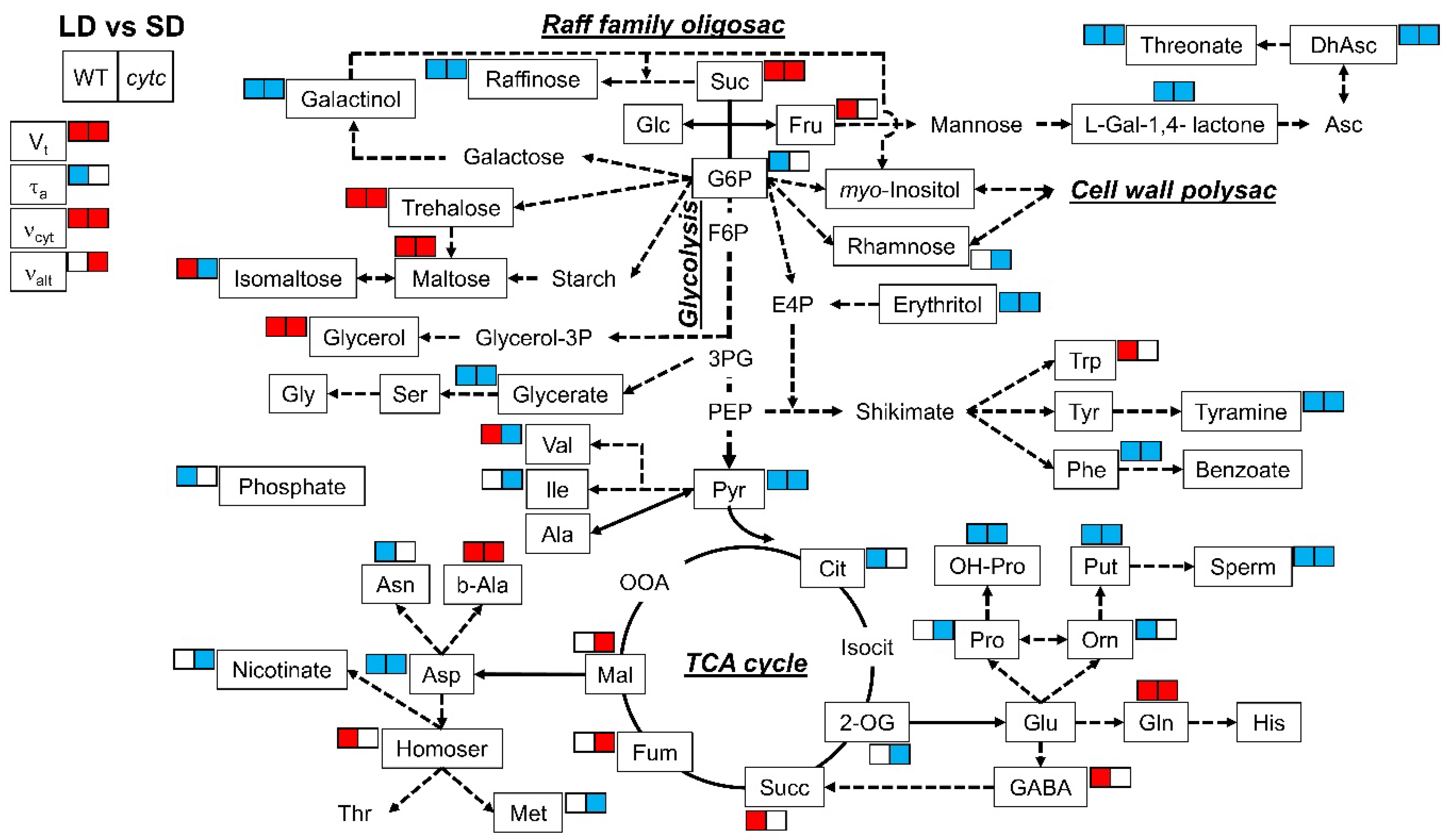
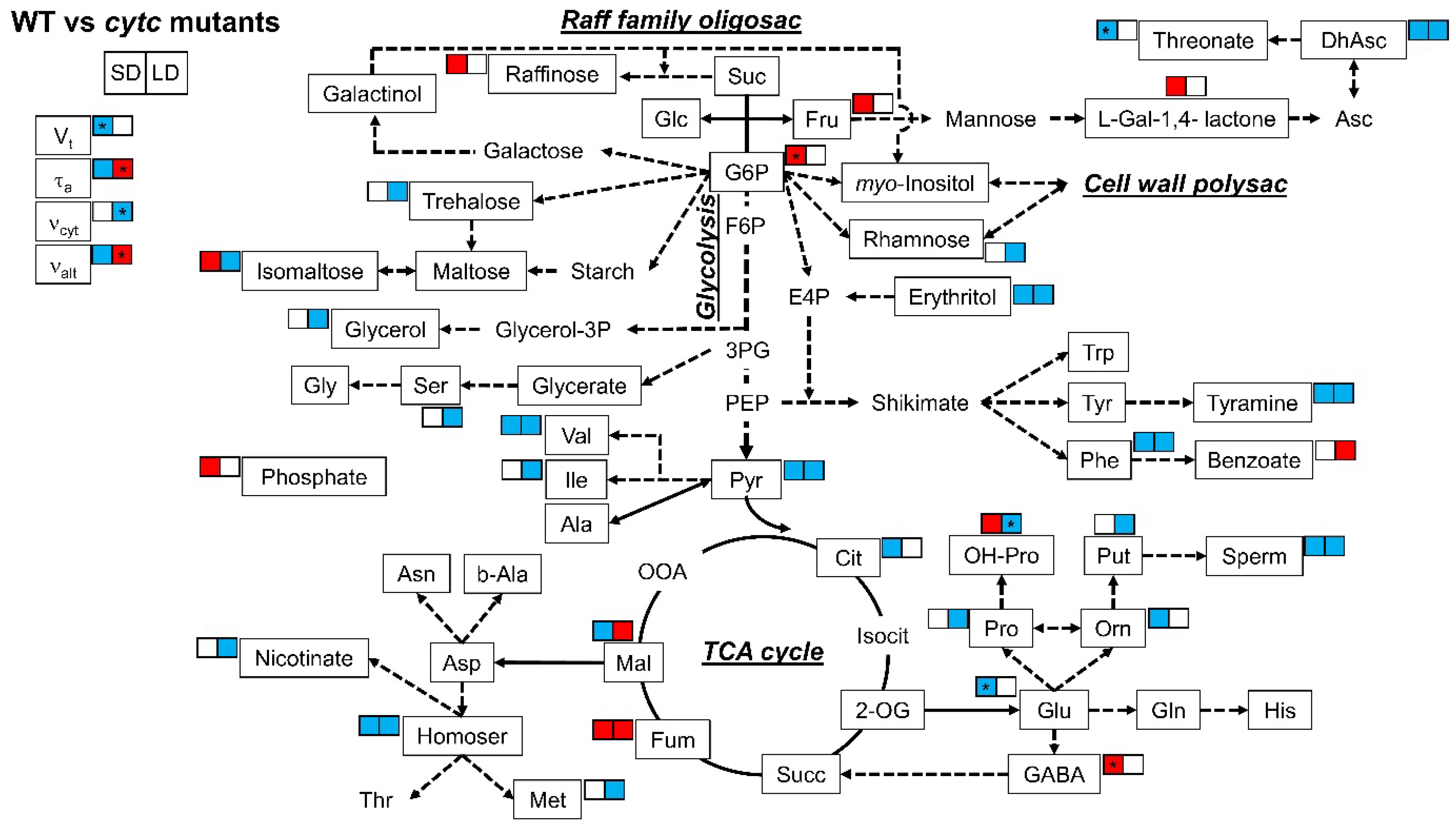
2.3. Metabolite Profiling in Cytc Mutants and WT Plants under Short- and Long-Day Photoperiods
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Plant Material and Growth Conditions
5.2. Protein and Transcript Levels
5.3. Respiration and Oxygen Isotope Discrimination Measurements
5.4. Leaf Gas Exchange and Chlorophyll Fluorescence Measurements
5.5. Metabolite Profiling
5.6. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shameer, S.; Ratcliffe, R.G.; Sweetlove, L.J. Leaf energy balance requires mitochondrial respiration and export of chloroplast NADPH in the light. Plant Physiol. 2019, 180, 1947–1961. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, B.M.; Asao, S.; Millar, A.H.; Atkin, O. Core principles which explain variation in respiration across biological scales. New Phytol. 2019, 222, 670–686. [Google Scholar] [CrossRef] [PubMed]
- Vanlerberghe, G.C.; Dahal, K.; Alber, N.A.; Chadee, A. Photosynthesis, respiration and growth: A carbon and energy balancing act for alternative oxidase. Mitochondrion 2020, 52, 197–211. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Nguyen, H.; Lam, H.-M. Legumes—The art and science of environmentally sustainable agriculture. Plant Cell Environ. 2019, 42, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Del-Saz, N.F.; Ribas-Carbo, M.; McDonald, A.E.; Lambers, H.; Fernie, A.R.; Florez-Sarasa, I. An in vivo perspective of the role (s) of the alternative oxidase pathway. Trends Plant Sci. 2018, 23, 206–219. [Google Scholar] [CrossRef]
- Millar, A.H.; Whelan, J.; Soole, K.L.; Day, D.A. Organization and regulation of mitochondrial respiration in plants. Annu. Rev. Plant Biol. 2011, 62, 79–104. [Google Scholar] [CrossRef] [PubMed]
- Racca, S.; Welchen, E.; Gras, D.E.; Tarkowská, D.; Turečková, V.; Maurino, V.G.; Gonzalez, D.H. Interplay between cytochrome c and gibberellins during Arabidopsis vegetative development. Plant J. 2018, 94, 105–121. [Google Scholar] [CrossRef]
- Welchen, E.; Gonzalez, D.H. Cytochrome c, a hub linking energy, redox, stress and signaling pathways in mitochondria and other cell compartments. Physiol. Plant 2016, 157, 310–321. [Google Scholar] [CrossRef]
- Ow, Y.L.; Green, D.R.; Hao, Z.; Mak, T.W. Cytochrome c: Functions beyond respiration. Nat. Rev. Mol. Cell Biol. 2008, 9, 532–542. [Google Scholar] [CrossRef]
- Martínez-Fábregas, J.; Díaz-Moreno, I.; González-Arzola, K.; Janocha, S.; Navarro, J.A.; Hervás, M.; Bernhardt, R.; Díaz-Quintana, A.; De la Rosa, M.Á. New Arabidopsis thaliana cytochrome c partners: A look into the elusive role of cytochrome c in programmed cell death in plants. Mol. Cell. Proteom. 2013, 12, 3666–3676. [Google Scholar] [CrossRef]
- Bihlmaier, K.; Mesecke, N.; Terzyiska, N.; Bien, M.; Hell, K.; Herrmann, J.M. The disulfide relay system of mitochondria is connected to the respiratory chain. J. Cell Biol. 2007, 179, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Bartoli, C.G.; Pastori, G.M.; Foyer, C.H. Ascorbate biosynthesis in mitochondria is linked to the electron transport chain between complexes III and IV. Plant Physiol. 2000, 123, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Engqvist, M.; Drincovich, M.F.; Flügge, U.I.; Maurino, V.G. Two D-2-hydroxy-acid dehydrogenases in Arabidopsis thaliana with catalytic capacities to participate in the last reactions of the methylglyoxal and beta-oxidation pathways. J. Biol. Chem. 2009, 284, 25026–25037. [Google Scholar] [CrossRef] [PubMed]
- Welchen, E.; Schmitz, J.; Fuchs, P.; García, L.; Wagner, S.; Wienstroer, J.; Schertl, P.; Braun, H.P.; Schwarzländer, M.; Gonzalez, D.H.; et al. D-Lactate dehydrogenase links methylglyoxal degradation and electron transport through cytochrome c. Plant Physiol. 2016, 172, 901–912. [Google Scholar]
- Ribas-Carbo, M.; Berry, J.A.; Yakir, D.; Giles, L.; Robinson, S.A.; Lennon, A.M.; Siedow, J.N. Electron partitioning between the cytochrome and alternative pathways in plant mitochondria. Plant Physiol. 1995, 109, 829–837. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.J.; Igamberdiev, A.U.; Mur, L.A. NO and ROS homeostasis in mitochondria: A central role for alternative oxidase. New Phytol. 2012, 195, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Del Río, L.A. ROS and RNS in plant physiology: An overview. J. Exp. Bot. 2015, 66, 2827–2837. [Google Scholar] [CrossRef]
- Considine, M.J.; Sandalio, L.M.; Foyer, C.H. Unravelling how plants benefit from ROS and NO reactions, while resisting oxidative stress. Ann Bot. 2015, 116, 469–473. [Google Scholar] [CrossRef]
- Mansilla, N.; Welchen, E.; Gonzalez, D.H. Arabidopsis SCO proteins oppositely influence cytochrome c oxidase levels and gene expression during salinity stress. Plant Cell Physiol. 2019, 60, 2769–2784. [Google Scholar] [CrossRef]
- Kromer, S. Respiration during photosynthesis. Annu. Rev. Plant Biol. 1995, 46, 45–70. [Google Scholar] [CrossRef]
- Nunes-Nesi, A.; Sulpice, R.; Gibon, Y.; Fernie, A.R. The enigmatic contribution of mitochondrial function in photosynthesis. J Exp. Bot. 2008, 59, 1675–1684. [Google Scholar] [CrossRef]
- Tcherkez, G.; Gauthier, P.; Buckley, T.N.; Busch, F.A.; Barbour, M.M.; Bruhn, D.; Heskel, M.A.; Gong, X.Y.; Crous, K.Y.; Griffin, K.; et al. Leaf day respiration: Low CO2 flux but high significance for metabolism and carbon balance. New Phytol. 2017, 216, 986–1001. [Google Scholar] [CrossRef]
- Noguchi, K.; Yoshida, K. Interaction between photosynthesis and respiration in illuminated leaves. Mitochondrion 2008, 8, 87–99. [Google Scholar] [CrossRef]
- Rasmusson, A.G.; Escobar, M.A.; Hao, M.; Podgórska, A.; Szal, B. Mitochondrial NAD(P)H oxidation pathways and nitrate/ammonium redox balancing in plants. Mitochondrion 2020, 53, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Voon, C.P.; Guan, X.; Yang, Y.; Gardeström, P.; Lim, B.L. In planta study of photosynthesis and photorespiration using NADPH and NADH/NAD+ fluorescent protein sensors. Nat. Commun. 2020, 11, 3238. [Google Scholar] [CrossRef] [PubMed]
- Juszczuk, I.M.; Szal, B.; Rychter, A.M. Oxidation-reduction and reactive oxygen species homeostasis in mutant plants with respiratory chain complex I dysfunction. Plant Cell Environ. 2011, 35, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Kühn, K.; Obata, T.; Feher, K.; Bock, R.; Fernie, A.R.; Meyer, E.H. Complete mitochondrial complex I deficiency induces an up-regulation of respiratory fluxes that is abolished by traces of functional complex I. Plant Physiol. 2015, 168, 1537–1549. [Google Scholar] [CrossRef]
- Pétriacq, P.; de Bont, L.; Genestout, L.; Hao, J.; Laureau, C.; Florez-Sarasa, I.; Rzigui, T.; Queval, G.; Gilard, F.; Mauve, C.; et al. Photoperiod affects the phenotype of mitochondrial complex I mutants. Plant Physiol. 2017, 173, 434–455. [Google Scholar] [CrossRef]
- Lothier, J.; De Paepe, R.; Tcherkez, G. Mitochondrial complex I dysfunction increases CO2 efflux and reconfigures metabolic fluxes of day respiration in tobacco leaves. New Phytol. 2019, 221, 750–763. [Google Scholar] [CrossRef]
- Del-Saz, N.F.; Ribas-Carbo, M.; Martorell, G.; Fernie, A.R.; Florez-Sarasa, I. Measurements of electron partitioning between cytochrome and alternative oxidase pathways in plant tissues. In Plant Respiration and Internal Oxygen: Methods and Protocols; Gupta, J.K., Ed.; Humana Press: New York, NY, USA, 2017; pp. 203–217. [Google Scholar]
- Priault, P.; Vidal, G.; De Paepe, R.; Ribas-Carbo, M. Leaf age-related changes in respiratory pathways are dependent on complex I activity in Nicotiana sylvestris. Physiol. Plant 2007, 129, 152–162. [Google Scholar] [CrossRef]
- Vidal, G.; Ribas-Carbo, M.; Garmier, M.; Dubertret, G.; Rasmusson, A.G.; Mathieu, C.; Foyer, C.H.; De Paepe, R. Lack of respiratory chain complex I impairs alternative oxidase engagement and modulates redox signaling during elicitor-induced cell death in tobacco. Plant Cell 2007, 19, 640–655. [Google Scholar] [CrossRef]
- Galle, A.; Florez-Sarasa, I.; Thameur, A.; de Paepe, R.; Flexas, J.; Ribas-Carbo, M. Effects of drought stress and subsequent rewatering on photosynthetic and respiratory pathways in Nicotiana sylvestris wild type and the mitochondrial complex 1-deficient CMSII mutant. J. Exp. Bot. 2010, 61, 765–775. [Google Scholar] [CrossRef] [PubMed]
- Szal, B.; Dabrowska, Z.; Malmberg, G.; Gardeström, P.; Rychter, A.M. Changes in energy status of leaf cells as a consequence of mitochondrial genome rearrangement. Planta 2008, 227, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Cardol, P. Mitochondrial NADH: Ubiquinone oxidoreductase (complex I) in eukaryotes: A highly conserved subunit composition highlighted by mining of protein databases. Biochim. Biophys. Acta 2011, 1807, 1390–1397. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gibon, Y.; Pyl, E.-T.; Sulpice, R.; Lunn, J.E.; Höhne, M.; Günther, M.; Stitt, M. Adjustment of growth, starch turnover, protein content and central metabolism to a decrease of the carbon supply when Arabidopsis is grown in very short photoperiods. Plant Cell Environ. 2009, 32, 859–874. [Google Scholar] [CrossRef]
- Sulpice, R.; Flis, A.; Ivakov, A.A.; Apelt, F.; Krohn, N.; Encke, B.; Abel, C.; Feil, R.; Lunn, J.E.; Stitt, M. Arabidopsis coordinates the diurnal regulation of carbon allocation and growth across a wide range of photoperiods. Mol. Plant 2014, 7, 137–155. [Google Scholar] [CrossRef]
- Welchen, E.; Hildebrandt, T.M.; Lewejohann, D.; Gonzalez, D.H.; Braun, H.P. Lack of cytochrome c in Arabidopsis decreases stability of Complex IV and modifies redox metabolism without affecting Complexes I and III. Biochim. Biophys. Acta 2012, 1817, 990–1001. [Google Scholar] [CrossRef]
- Flis, A.; Mengin, V.; Ivakov, A.A.; Mugford, S.T.; Hubberten, H.-M.; Encke, B.; Krohn, N.; Hohne, M.; Feil, R.; Hoefgen, R.; et al. Multiple circadian clock outputs regulate diel turnover of carbon and nitrogen reserves. Plant Cell Environ. 2019, 42, 549–573. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, C.M.; Lunn, J.E. A Tale of Two Sugars: Trehalose 6-Phosphate and Sucrose. Plant Physiol. 2016, 172, 7–27. [Google Scholar] [CrossRef] [PubMed]
- Caspar, T.; Huber, S.C.; Somerville, C. Alterations in growth, photosynthesis, and respiration in a starchless mutant of Arabidopsis thaliana (L.) deficient in chloroplast phosphoglucomutase activity. Plant Physiol. 1985, 79, 11–17. [Google Scholar] [CrossRef]
- Florez-Sarasa, I.; Araújo, W.L.; Wallström, S.V.; Rasmusson, A.G.; Fernie, A.R.; Ribas-Carbo, M. Light-responsive metabolite and transcript levels are maintained following a dark-adaptation period in leaves of Arabidopsis thaliana. New Phytol. 2012, 195, 136–148. [Google Scholar] [CrossRef]
- O’Leary, B.M.; Lee, C.P.; Atkin, O.K.; Cheng, R.; Brown, T.B.; Millar, A.H. Variation in leaf respiration rates at night correlates with carbohydrate and amino acid supply. Plant Physiol. 2017, 174, 2261–2273. [Google Scholar] [CrossRef]
- Florez-Sarasa, I.; Ostaszewska, M.; Galle, A.; Flexas, J.; Rychter, A.M.; Ribas-Carbo, M. Changes of alternative oxidase activity, capacity and protein content in leaves of Cucumis sativus wild-type and MSC16 mutant grown under different light intensities. Physiol Plant. 2009, 137, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.M.; Stitt, M. Coordination of carbon supply and plant growth. Plant Cell Environ. 2007, 30, 1126–1149. [Google Scholar] [CrossRef] [PubMed]
- Stitt, M.; Gibon, Y. Why measure enzyme activities in the era of systems biology? Trends Plant Sci. 2014, 19, 256–265. [Google Scholar] [CrossRef]
- Baerenfaller, K.; Massonnet, C.; Hennig, L.; Russenberger, D.; Sulpice, R.; Walsh, S.; Stitt, M.; Granier, C.; Gruissem, W. A long photoperiod relaxes energy management in Arabidopsis leaf six. Curr. Opin. Plant 2015, 2, 34–45. [Google Scholar] [CrossRef]
- Lu, Y.; Gehan, J.P.; Sharkey, T.D. Daylength and circadian effects on starch degradation and maltose metabolism. Plant Physiol. 2005, 138, 2280–2291. [Google Scholar] [CrossRef]
- Sweetlove, L.J.; Beard, K.F.; Nunes-Nesi, A.; Fernie, A.R.; Ratcliffe, R.G. Not just a circle: Flux modes in the plant TCA cycle. Trends Plant Sci. 2010, 15, 462–470. [Google Scholar] [CrossRef] [PubMed]
- Florez-Sarasa, I.; Ribas-Carbo, M.; Del-Saz, N.F.; Schwahn, K.; Nikoloski, Z.; Fernie, A.R.; Flexas, J. Unravelling the in vivo regulation and metabolic role of the alternative oxidase pathway in C3 species under photoinhibitory conditions. New Phytol. 2016, 212, 66–79. [Google Scholar] [CrossRef]
- Fait, A.; Fromm, H.; Walter, D.; Galili, G.; Fernie, A.R. Highway or byway: The metabolic role of the GABA shunt in plants. Trends Plant Sci. 2008, 13, 14–19. [Google Scholar] [CrossRef]
- Bandehagh, A.; Taylor, N.L. Can alternative metabolic pathways and shunts overcome salinity induced inhibition of central carbon metabolism in crops? Front Plant Sci. 2020, 11, 1072. [Google Scholar] [CrossRef] [PubMed]
- Florez-Sarasa, I.; Obata, T.; Del-Saz, N.F.; Reichheld, J.P.; Meyer, E.H.; Rodriguez-Concepcion, M.; Ribas-Carbo, M.; Fernie, A.R. The lack of mitochondrial thioredoxin TRXo1 affects in vivo alternative oxidase activity and carbon metabolism under different light conditions. Plant Cell Physiol. 2019, 60, 2369–2381. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, K.; Nakajima, N.; Terashima, I. Acclimation of leaf respiratory properties in Alocasia odora following reciprocal transfers of plants between high- and low-light environments. Plant Cell Environ. 2001, 24, 831–839. [Google Scholar] [CrossRef]
- Hachiya, T.; Treashima, I.; Nochuchi, K. Increase in respiratory cost at high temperature is attributed to high protein turnover cost in Petunia x hybrida petals. Plant Cell Environ. 2007, 30, 1269–1283. [Google Scholar] [CrossRef]
- Ishihara, H.; Moraes, T.A.; Pyl, E.-T.; Schulze, W.X.; Obata, T.; Scheffel, A.; Fernie, A.R.; Sulpice, R.; Stitt, M. Growth rate correlates negatively with protein turnover in Arabidopsis accessions. Plant J. 2017, 91, 416–429. [Google Scholar] [CrossRef] [PubMed]
- Kirma, M.; Araújo, W.L.; Fernie, A.R.; Galili, G. The multifaceted role of aspartate-family amino acids in plant metabolism. J. Exp. Bot. 2012, 63, 4995–5001. [Google Scholar] [CrossRef]
- Daloso, D.M.; Müller, K.; Obata, T.; Florian, A.; Tohge, T.; Bottcher, A.; Riondet, C.; Bariat, L.; Carrari, F.; Nunes-Nesi, A.; et al. Thioredoxin, a master regulator of the tricarboxylic acid cycle in plant mitochondria. Proc. Natl. Acad. Sci. USA 2015, 112, 1392–1400. [Google Scholar] [CrossRef]
- Fonseca-Pereira, P.; Daloso, D.M.; Gago, J.; Nunes-Nesi, A.; Araújo, W.L. On the role of the plant mitochondrial thioredoxin system during abiotic stress. Plant Signal. Behav. 2019, 14, 6. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Dudareva, N. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef]
- Tohge, T.; Watanabe, M.; Hoefgen, R.; Fernie, A.R. Shikimate and phenylalanine biosynthesis in the green lineage. Front. Plant Sci. 2013, 4, 62. [Google Scholar] [CrossRef] [PubMed]
- Florez-Sarasa, I.D.; Bouma, T.J.; Medrano, H.; Azcon-Bieto, J.; Ribas-Carbo, M. Contribution of the cytochrome and alternative pathways to growth respiration and maintenance respiration in Arabidopsis thaliana. Physiol. Plant 2007, 129, 143–151. [Google Scholar] [CrossRef]
- Vanlerberghe, G.C. Alternative oxidase: A mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. Int. J. Mol. Sci. 2013, 14, 6805–6847. [Google Scholar] [CrossRef] [PubMed]
- Florez-Sarasa, I.; Fernie, A.R.; Gupta, K.J. Does the alternative respiratory pathway offer protection against the adverse effects resulting from climate change? J. Exp. Bot. 2020, 71, 465–469. [Google Scholar] [CrossRef]
- Alegre, M.L.; Steelheart, C.; Baldet, P.; Rothan, C.; Just, D.; Okabe, Y.; Ezura, H.; Smirnoff, N.; Gergoff Grozeff, G.E.; Bartoli, C.G. Deficiency of GDP-L-galactose phosphorylase, an enzyme required for ascorbic acid synthesis, reduces tomato fruit yield. Planta 2020, 22, 251–254. [Google Scholar] [CrossRef]
- Bartoli, C.G.; Yu, J.; Gómez, F.; Fernández, L.; McIntosh, L.; Foyer, C.H. Inter-relationships between light and respiration in the control of ascorbic acid synthesis and accumulation in Arabidopsis thaliana leaves. J. Exp. Bot. 2006, 57, 1621–1631. [Google Scholar] [CrossRef]
- Millar, A.H.; Wiskich, J.T.; Whelan, J.; Day, D.A. Organic acid activation of the alternative oxidase of plant mitochondria. FEBS Lett. 1993, 329, 259–262. [Google Scholar] [CrossRef]
- Selinski, J.; Hartmann, A.; Deckers-Hebestreit, G.; Day, D.A.; Whelan, J.; Scheibe, R. Alternative oxidase isoforms are differentially activated by tricarboxylic acid cycle intermediates. Plant Physiol. 2018, 176, 1423–1432. [Google Scholar] [CrossRef]
- Martínez-García, J.F.; Monte, E.; Quail, P.H. A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J. 1999, 20, 251–257. [Google Scholar] [CrossRef]
- Schägger, H. Tricine–SDS-PAGE. Nat. Protoc. 2006, 1, 16–22. [Google Scholar] [CrossRef]
- O’Connell, J. The basics of RT-PCR. Some practical considerations. Methods Mol. Biol. 2002, 193, 19–25. [Google Scholar] [PubMed]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Charrier, B.; Champion, A.; Henry, Y.; Kreis, M. Expression profiling of the whole Arabidopsis shaggy-like kinase multigene family by real-time reverse transcriptase-polymerase chain reaction. Plant Physiol. 2002, 130, 577–590. [Google Scholar] [CrossRef] [PubMed]
- Guy, R.D.; Berry, J.A.; Fogel, M.L.; Hoering, T.C. Differential fractionation of oxygen isotopes by cyanide-resistant and cyanide-sensitive respiration in plants. Planta 1989, 177, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Florez-Sarasa, I.; Flexas, J.; Rasmusson, A.G.; Umbach, A.L.; Siedow, J.N.; Ribas-Carbo, M. In vivo cytochrome and alternative pathway respiration in leaves of Arabidopsis thaliana plants with altered alternative oxidase under different light conditions. Plant Cell Environ. 2011, 34, 1373–1383. [Google Scholar] [CrossRef]
- Lisec, J.; Schauer, N.; Kopka, J.; Willmitzer, L.; Fernie, A.R. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protoc. 2006, 1, 387–396. [Google Scholar] [CrossRef]
- Luedemann, A.; Strassburg, K.; Erban, A.; Kopka, J. TagFinder for the quantitative analysis of gas chromatography-mass spectrometry (GC-MS)-based metabolite profiling experiments. Bioinformatics 2008, 24, 732–737. [Google Scholar] [CrossRef] [PubMed]
- Kopka, J.; Schauer, N.; Krueger, S.; Birkemeyer, C.; Usadel, B.; Bergmuller, E.; Dörmann, P.; Weckwerth, W.; Gibon, Y.; Stitt, M.; et al. GMD@CSB.DB: The Golm Metabolome Database. Bioinformatics 2005, 21, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Fernie, A.R.; Aharoni, A.; Willmitzer, L.; Stitt, M.; Tohge, T.; Kopka, J.; Carroll, A.J.; Saito, K.; Fraser, P.D.; DeLuca, V. Recommendations for reporting metabolite data. Plant Cell 2011, 23, 2477–2482. [Google Scholar] [CrossRef] [PubMed]
| Parameter | Photoperiod | Genotype | Photoperiod × Genotype |
|---|---|---|---|
| Vt | *** | ns | ** |
| τa | ns | ns | *** |
| vcyt | *** | ns | * |
| valt | *** | ns | *** |
| ETR | ns | ns | ns |
| Metabolite | SD | LD | ||||
|---|---|---|---|---|---|---|
| WT | 1b2a | 1b2b | WT | 1b2a | 1b2b | |
| Amino acids | ||||||
| Alanine | 1 ± 0.23 ab | 1.06 ± 0.02 ab | 0.87 ± 0.03 b | 1.24 ± 0.02 a | 1.25 ± 0.03 a | 0.74 ± 0.02 b |
| Asparagine | 1 ± 0.02 a | 1.02 ± 0.05 a | 0.96 ± 0.06 a | 0.6 ± 0.02 b | 0.98 ± 0.03 a | 0.69 ± 0.03 b |
| Aspartate | 1 ± 0.09 a | 1.1 ± 0.06 a | 0.97 ± 0.09 a | 0.42 ± 0.01 b | 0.35 ± 0 b | 0.38 ± 0.01 b |
| Glutamate | 1 ± 0.04 ab | 0.87 ± 0.03 bc | 0.83 ± 0.06 c | 1.03 ± 0.03 a | 1.04 ± 0.01 a | 0.92 ± 0.05 abc |
| Glutamine | 1 ± 0.03 de | 0.89 ± 0.03 e | 1.12 ± 0.06 cd | 1.27 ± 0.08 bc | 1.98 ± 0.07 a | 1.37 ± 0.08 b |
| Glycine | 1 ± 0.02 a | 0.95 ± 0.05 a | 0.73 ± 0.03 b | 1.05 ± 0.03 a | 0.98 ± 0.02 a | 0.62 ± 0.05 b |
| Histidine | 1 ± 0.22 | 1.02 ± 0.09 | 0.8 ± 0.06 | 0.93 ± 0.05 | 0.76 ± 0.01 | 1 ± 0.05 |
| Homoserine | 1 ± 0.04 b | 0.82 ± 0.05 c | 0.8 ± 0.02 c | 1.21 ± 0.05 a | 0.95 ± 0 b | 0.81 ± 0.01 c |
| Isoleucine | 1 ± 0.05 a | 0.91 ± 0.02 a | 0.95 ± 0.04 a | 0.99 ± 0.03 a | 0.67 ± 0.03 b | 0.64 ± 0.05 b |
| Methionine | 1 ± 0.05 a | 1.03 ± 0.04 a | 0.91 ± 0.04 a | 1.03 ± 0.06 a | 0.27 ± 0.06 c | 0.63 ± 0.05 b |
| Ornithine | 1 ± 0.03 a | 0.76 ± 0.02 b | 0.75 ± 0.03 b | 0.77 ± 0.02 b | 0.48 ± 0.02 c | 0.81 ± 0.05 b |
| Phenylalanine | 1 ± 0.01 a | 0.84 ± 0.04 b | 0.85 ± 0.02 b | 0.86 ± 0.05 b | 0.67 ± 0 c | 0.48 ± 0.02 d |
| Proline | 1 ± 0.03 b | 1.16 ± 0.06 a | 1.08 ± 0.03 ab | 0.97 ± 0.06 b | 0.58 ± 0.01 c | 0.56 ± 0.05 c |
| Tryptophan | 1 ± 0.03 cd | 0.89 ± 0.03 d | 1.24 ± 0.12 bc | 1.34 ± 0.08 b | 0.74 ± 0.01 d | 3.19 ± 0.14 a |
| Tyrosine | 1 ± 0.02 ab | 0.87 ± 0.05 b | 1.06 ± 0.06 a | 0.93 ± 0.06 ab | 0.92 ± 0.01 ab | 1.03 ± 0.04 a |
| Serine | 1 ± 0.02 ab | 0.95 ± 0.04 ab | 1.01 ± 0.03 ab | 1.04 ± 0.04 a | 0.92 ± 0.01 bc | 0.84 ± 0.02 c |
| Valine | 1 ± 0.01 b | 0.89 ± 0.01 c | 0.86 ± 0.02 c | 1.13 ± 0.06 a | 0.67 ± 0.01 d | 0.53 ± 0.02 e |
| Organic acids | ||||||
| Benzoate | 1 ± 0.07 bc | 1.06 ± 0.04 bc | 1.03 ± 0.05 bc | 0.92 ± 0.06 c | 1.2 ± 0.11 ab | 1.27 ± 0.06 a |
| Citrate | 1 ± 0.07 a | 0.65 ± 0.03 b | 0.77 ± 0.06 b | 0.69 ± 0.03 b | 0.71 ± 0.04 b | 1 ± 0.07 a |
| Dehydroascorbate | 1 ± 0.02 a | 0.81 ± 0.05 b | 0.73 ± 0.05 b | 0.74 ± 0.03 b | 0.4 ± 0 c | 0.29 ± 0.01 c |
| Fumarate | 1 ± 0.02 c | 1.2 ± 0.04 b | 1.2 ± 0.04 b | 1.08 ± 0.03 bc | 1.54 ± 0.11 a | 1.46 ± 0.09 a |
| 2-Oxoglutarate | 1 ± 0.14 a | 1.04 ± 0.16 a | 0.91 ± 0.09 a | 0.69 ± 0.08 ab | 0.47 ± 0.03 b | 0.48 ± 0.09 b |
| Glycerate | 1 ± 0.03 a | 1 ± 0.04 a | 1.08 ± 0.03 a | 0.83 ± 0.05 b | 0.77 ± 0.01 b | 0.88 ± 0.03 b |
| Malate | 1 ± 0.06 c | 0.8 ± 0.07 d | 0.7 ± 0.04 d | 0.97 ± 0.06 c | 1.56 ± 0.02 a | 1.23 ± 0.04 b |
| Nicotinate | 1 ± 0.04 a | 1.11 ± 0.08 a | 1.11 ± 0.05 a | 0.99 ± 0.05 a | 0.76 ± 0.04 b | 0.59 ± 0.02 c |
| Phosphate | 1 ± 0.03 b | 1.21 ± 0.08 a | 1.2 ± 0.06 a | 0.4 ± 0.03 c | 0.55 ± 0 c | 1.14 ± 0.03 ab |
| Pyruvate | 1 ± 0.03 a | 0.76 ± 0.04 b | 0.71 ± 0.04 b | 0.41 ± 0.02 c | 0.22 ± 0.01 d | 0.18 ± 0.01 d |
| Threonate | 1 ± 0.05 a | 0.94 ± 0.07 ab | 0.83 ± 0.01 b | 0.51 ± 0.02 c | 0.45 ± 0.01 c | 0.53 ± 0.02 c |
| Succinate | 1 ± 0.08 c | 0.98 ± 0.07 c | 1.09 ± 0.01 c | 1.48 ± 0.09 b | 2.4 ± 0.03 a | 1.13 ± 0.04 c |
| Sugars and sugar alcohol | ||||||
| Erythritol | 1 ± 0.03 a | 0.85 ± 0.04 b | 0.82 ± 0.03 b | 0.8 ± 0.03 b | 0.43 ± 0.01 c | 0.38 ± 0.03 c |
| Fructose | 1 ± 0.04 b | 1.2 ± 0.05 a | 1.25 ± 0.04 a | 1.2 ± 0.06 a | 1.26 ± 0.01 a | 0.84 ± 0.03 c |
| Galactinol | 1 ± 0.04 a | 1.1 ± 0.04 a | 0.86 ± 0.07 b | 0.14 ± 0 d | 0.12 ± 0 d | 0.32 ± 0.02 c |
| Glucose | 1 ± 0.02 b | 1.01 ± 0.02 b | 1.04 ± 0.03 ab | 1.05 ± 0.02 ab | 1.13 ± 0.08 a | 0.9 ± 0.02 c |
| Glucose-6-P | 1 ± 0.07 b | 1.32 ± 0.12 a | 1.08 ± 0.09 ab | 0.52 ± 0.02 c | 0.48 ± 0.01 c | 1.02 ± 0.08 b |
| Glycerol | 1 ± 0.06 c | 0.93 ± 0.01 c | 0.94 ± 0.03 c | 1.8 ± 0.12 a | 1.43 ± 0.14 b | 1.41 ± 0.05 b |
| Myo-inositol | 1 ± 0.03 | 1 ± 0.03 | 0.99 ± 0.02 | 0.87 ± 0.14 | 0.95 ± 0.01 | 0.84 ± 0.05 |
| Isomaltose | 1 ± 0.01 b | 1.39 ± 0.05 a | 1.44 ± 0.04 a | 1.3 ± 0.03 a | 0.47 ± 0.03 c | 0.99 ± 0.08 b |
| maltose | 1 ± 0.09 d | 1.65 ± 0.08 c | 1.13 ± 0.06 d | 2.31 ± 0.17 b | 2.96 ± 0.02 a | 1.64 ± 0.04 c |
| Raffinose | 1 ± 0.05 b | 1.23 ± 0.11 a | 1.41 ± 0.05 a | 0.68 ± 0.06 c | 0.26 ± 0 d | 0.57 ± 0.06 c |
| Rhamnose | 1 ± 0.06 a | 1.06 ± 0.04 a | 1.08 ± 0.04 a | 1.05 ± 0.02 a | 0.5 ± 0 b | 0.48 ± 0.02 b |
| Sucrose | 1 ± 0.07 c | 0.9 ± 0.1 c | 0.9 ± 0.09 c | 2.65 ± 0.1 a | 2.66 ± 0.02 a | 2.08 ± 0.15 b |
| Trehalose | 1 ± 0.04 cd | 0.83 ± 0.04 d | 0.85 ± 0.02 d | 1.88 ± 0.09 a | 1.39 ± 0.01 b | 1.13 ± 0.08 c |
| Other metabolites | ||||||
| GABA | 1 ± 0.04 d | 1.15 ± 0.03 cd | 1.24 ± 0.07 c | 1.54 ± 0.06 b | 1.97 ± 0.1 a | 1.07 ± 0.09 cd |
| Beta-alanine | 1 ± 0.06 cd | 1.06 ± 0.06 c | 0.99 ± 0.01 c | 1.3 ± 0.06 b | 1.53 ± 0.02 a | 1.22 ± 0.06 b |
| Galactono-1,4-lactone | 1 ± 0.03 b | 1.12 ± 0.05 a | 1.19 ± 0.02 a | 0.59 ± 0.01 c | 0.52 ± 0.03 c | 0.53 ± 0.02 c |
| 4-Hydroxy-proline | 1 ± 0.08 b | 1.18 ± 0.07 a | 1.17 ± 0.05 a | 0.51 ± 0.02 c | 0.43 ± 0.04 cd | 0.33 ± 0.01 d |
| Putrescine | 1 ± 0.06 a | 0.98 ± 0.04 a | 0.82 ± 0.02 b | 0.77 ± 0.06 b | 0.64 ± 0.02 c | 0.47 ± 0.04 d |
| Spermidine | 1 ± 0.03 a | 0.81 ± 0.03 b | 0.76 ± 0.02 b | 0.65 ± 0.04 c | 0.13 ± 0 d | 0.13 ± 0 d |
| Tyramine | 1 ± 0.04 a | 0.87 ± 0.03 b | 0.85 ± 0.02 b | 0.85 ± 0.03 b | 0.57 ± 0.02 c | 0.43 ± 0.02 d |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Florez-Sarasa, I.; Welchen, E.; Racca, S.; Gonzalez, D.H.; Vallarino, J.G.; Fernie, A.R.; Ribas-Carbo, M.; Del-Saz, N.F. Cytochrome c Deficiency Differentially Affects the In Vivo Mitochondrial Electron Partitioning and Primary Metabolism Depending on the Photoperiod. Plants 2021, 10, 444. https://doi.org/10.3390/plants10030444
Florez-Sarasa I, Welchen E, Racca S, Gonzalez DH, Vallarino JG, Fernie AR, Ribas-Carbo M, Del-Saz NF. Cytochrome c Deficiency Differentially Affects the In Vivo Mitochondrial Electron Partitioning and Primary Metabolism Depending on the Photoperiod. Plants. 2021; 10(3):444. https://doi.org/10.3390/plants10030444
Chicago/Turabian StyleFlorez-Sarasa, Igor, Elina Welchen, Sofia Racca, Daniel H. Gonzalez, José G. Vallarino, Alisdair R. Fernie, Miquel Ribas-Carbo, and Nestor Fernandez Del-Saz. 2021. "Cytochrome c Deficiency Differentially Affects the In Vivo Mitochondrial Electron Partitioning and Primary Metabolism Depending on the Photoperiod" Plants 10, no. 3: 444. https://doi.org/10.3390/plants10030444
APA StyleFlorez-Sarasa, I., Welchen, E., Racca, S., Gonzalez, D. H., Vallarino, J. G., Fernie, A. R., Ribas-Carbo, M., & Del-Saz, N. F. (2021). Cytochrome c Deficiency Differentially Affects the In Vivo Mitochondrial Electron Partitioning and Primary Metabolism Depending on the Photoperiod. Plants, 10(3), 444. https://doi.org/10.3390/plants10030444







