Identification and Characterization of Wheat Germplasm for Salt Tolerance
Abstract
1. Introduction
2. Results
2.1. Genotypic Differences in Response to Salt Stress
2.2. Ranking of Genotypes for Salt Tolerance
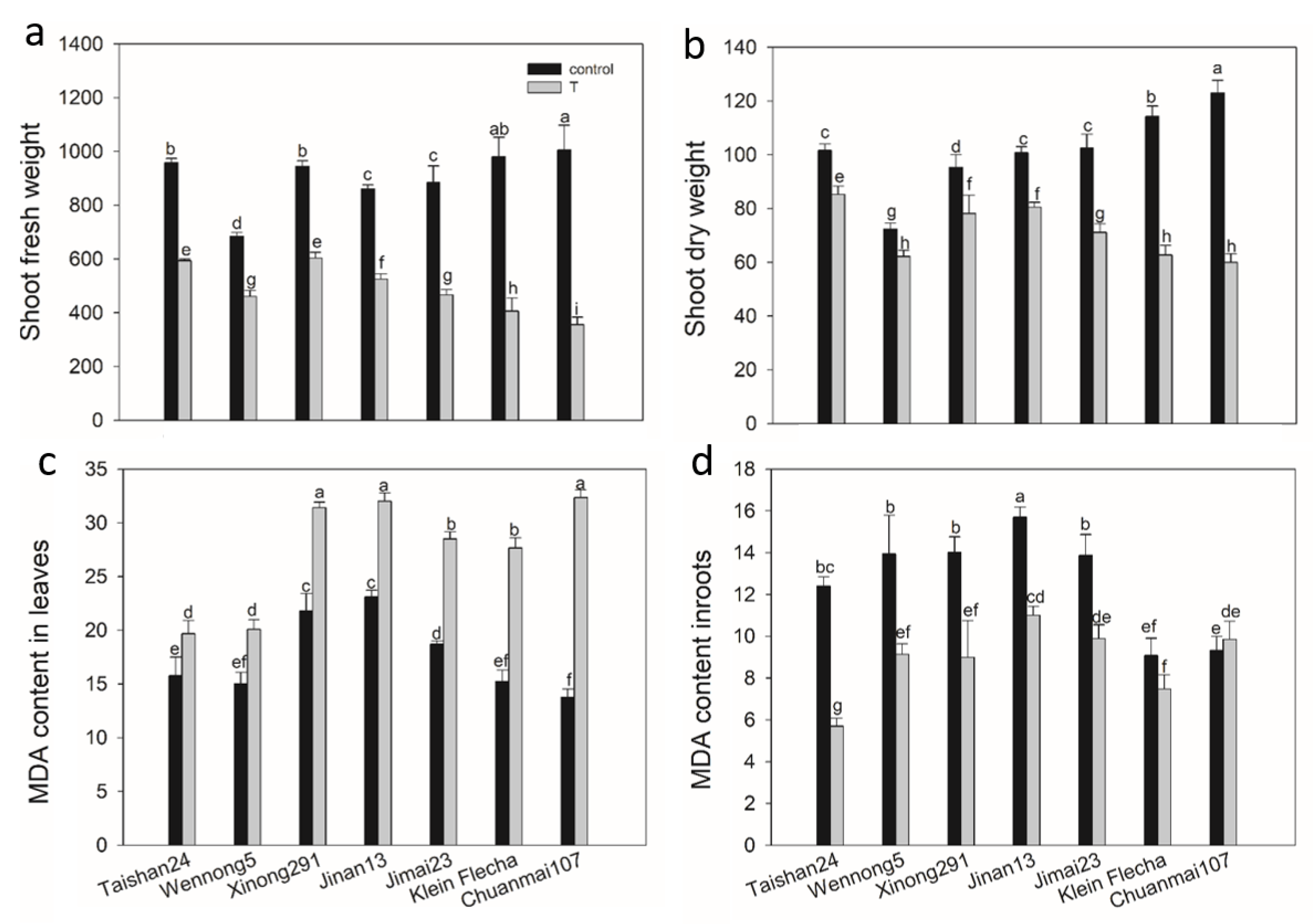
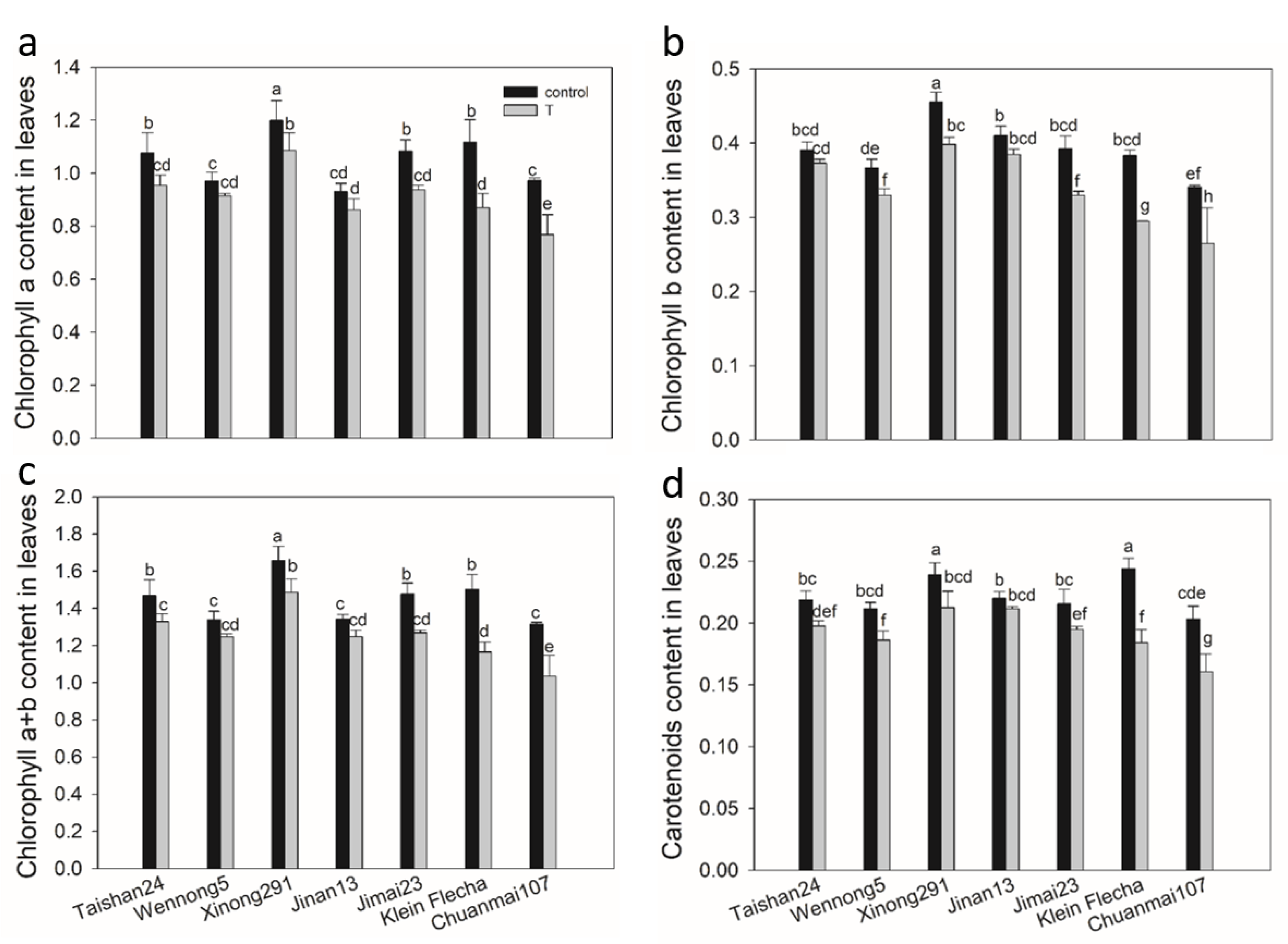
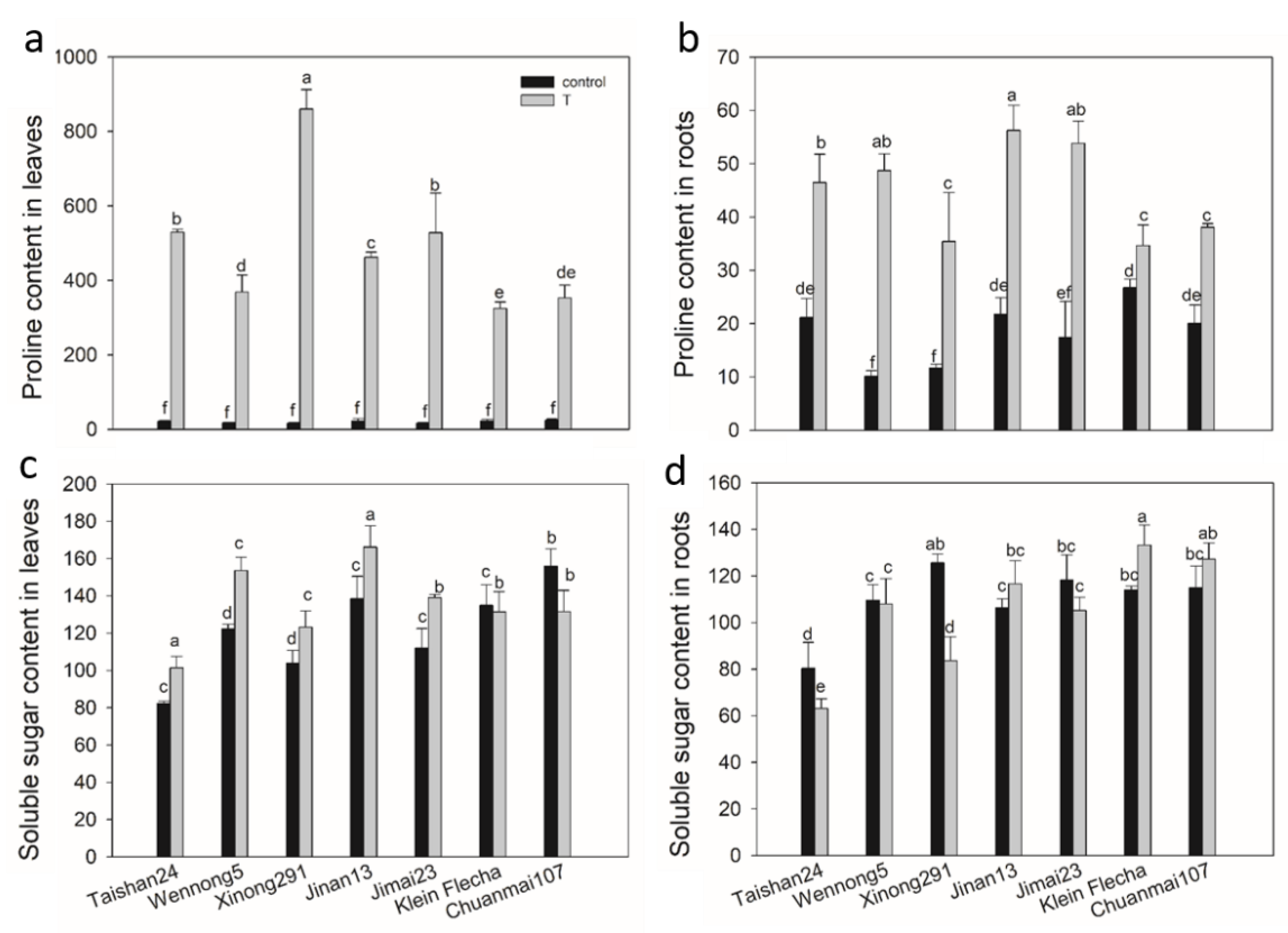
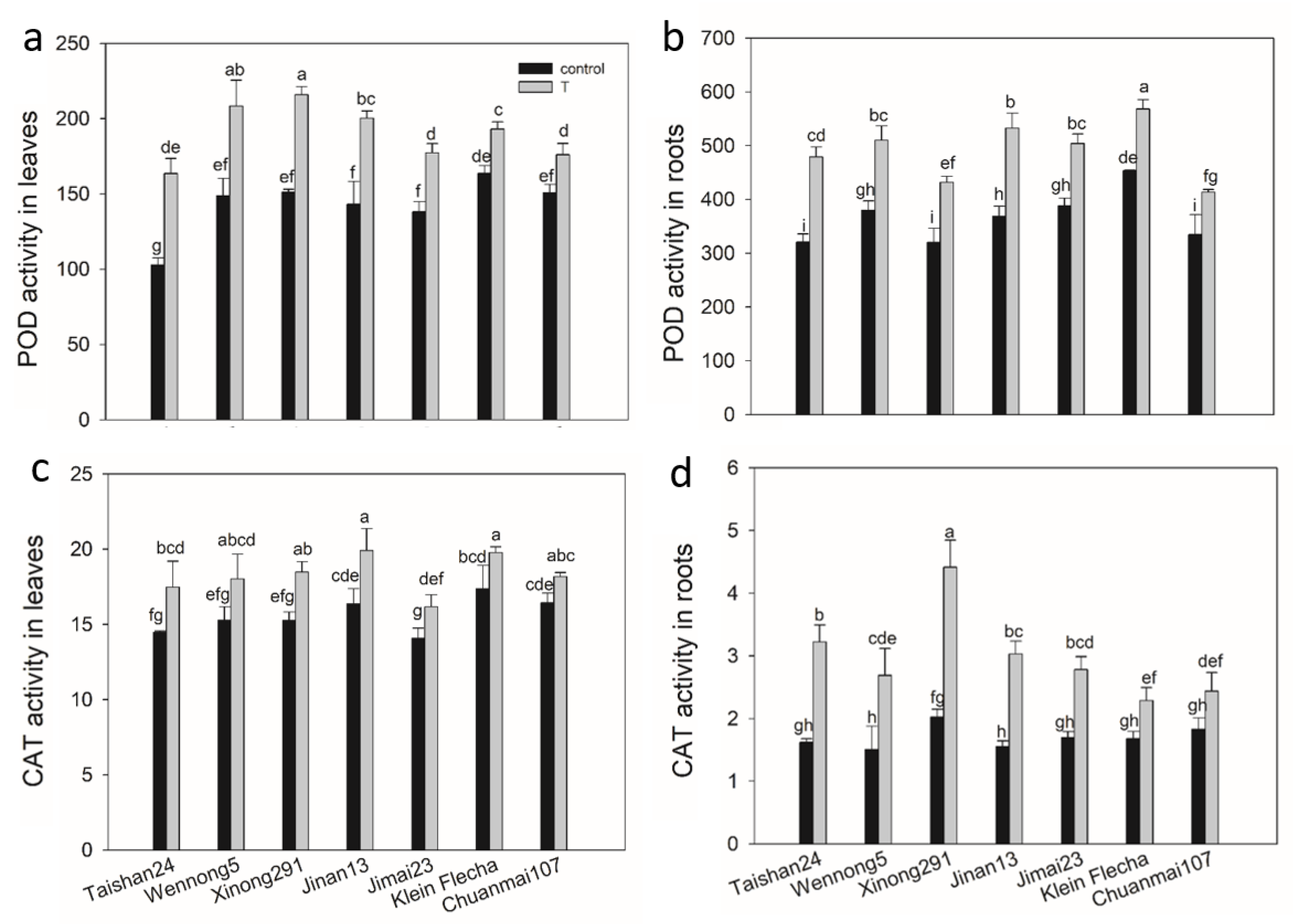
2.3. The Confirmatory Experiment
2.4. Accumulation of Osmotic Adjustment Substances
2.5. Accumulation of Na and K
2.6. The Genotypic Difference in MDA Content in Response to Salt Stress
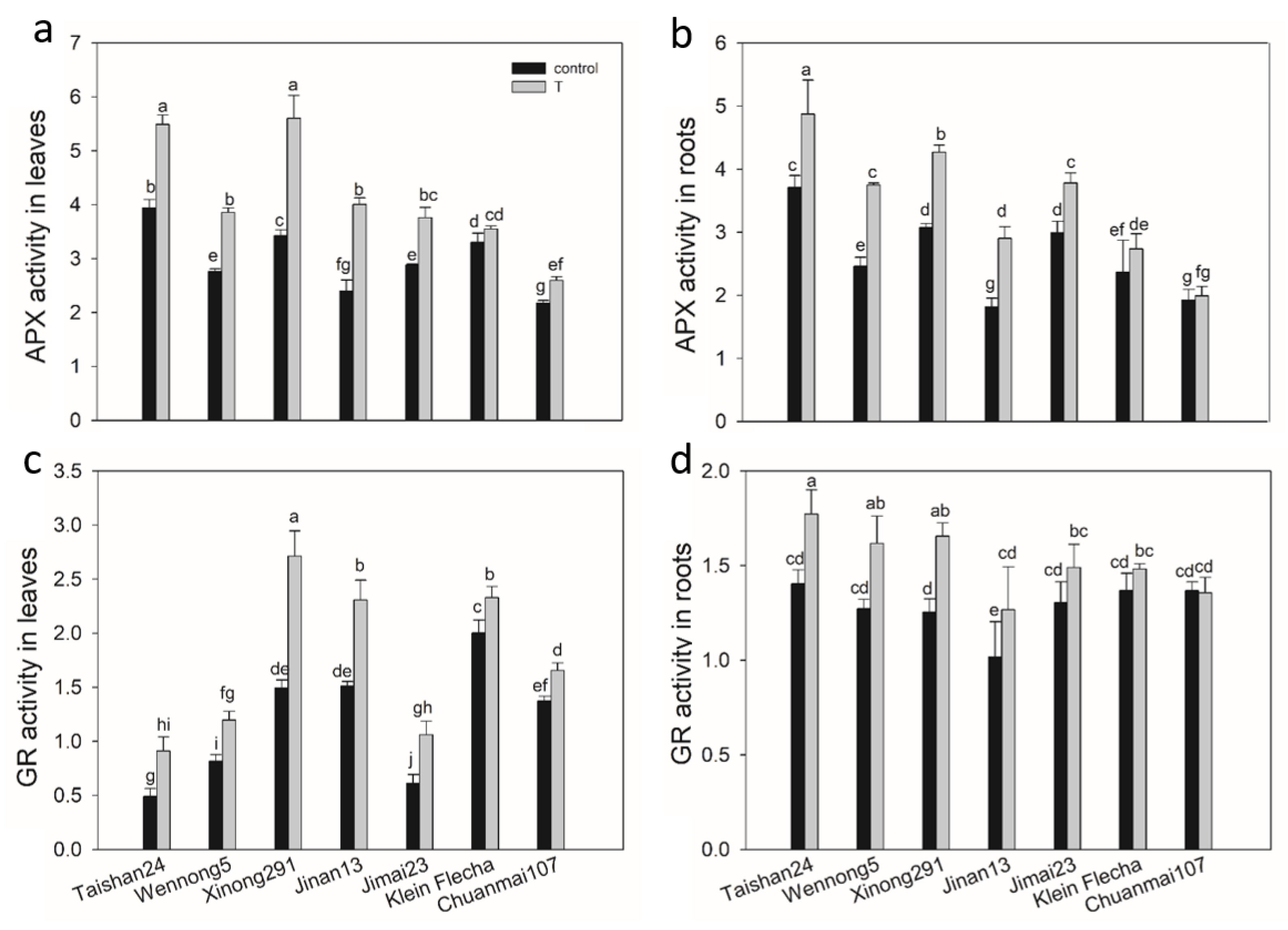
2.7. The Genotypic Difference in Antioxidant Enzyme Activity under Salt Stress
3. Discussion
3.1. Salt Tolerance Estimation for Wheat Genotypes
3.2. Physiological Analysis among Contrasting Wheat Genotypes
4. Materials and Methods
4.1. A Preliminary Screening Experiment
4.2. A Secondary Experiment for Verification
4.3. Determination of Proline and Soluble Sugar Contents
4.4. Estimation of Na and K Contents
4.5. Measuring of Antioxidant Enzyme Activities and Lipid Peroxidation
4.6. Statistical Analysis
4.7. Ranking of Genotypes for Salt Tolerance
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Ding, T.L.; Yang, Z.; Wei, X.C.; Yuan, F.; Yin, S.S.; Wang, B.S. Evaluation of salt-tolerant germplasm and screening of the salt-tolerance traits of sweet sorghum in the germination stage. Funct. Plant. Biol. 2018, 45, 1073–1081. [Google Scholar] [CrossRef]
- Li, W.H.; Zhang, H.Z.; Zeng, Y.L.; Xiang, L.J.; Lei, Z.H.; Huang, Q.X.; Li, T.Y.; Shen, F.; Cheng, Q. A salt tolerance evaluation method for sunflower (Helianthus annuus L.) at the seed germination stage. Sci. Rep. 2020, 10, 106261. [Google Scholar] [CrossRef]
- Song, J.; Wang, B.S. Using euhalophytes to understand salt tolerance and to develop saline agriculture: Suaeda salsa as a promising model. Ann. Bot. 2015, 115, 541–553. [Google Scholar] [CrossRef]
- Oyiga, B.C.; Sharma, R.C.; Shen, J.; Baum, M.; Ogbonnaya, F.C.; Léon, J.; Ballvora, A. Identification and characterization of salt tolerance of wheat germplasm using a multivariable screening approach. J. Agron. Crop. Sci. 2016, 202, 472–485. [Google Scholar] [CrossRef]
- Chatrath, R.; Mishra, B.; Ferrara, G.O.; Singh, S.K.; Joshi, A.K. Challenges to wheat production in South Asia. Euphytica 2007, 157, 447–456. [Google Scholar] [CrossRef]
- Asada, K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006, 141, 391–396. [Google Scholar] [CrossRef]
- Zeng, Y.; Li, L.; Yang, R.; Yi, X.; Zhang, B. Contribution and distribution of inorganic ions and organic compounds to the osmotic adjustment in Halostachys caspica response to salt stress. Sci. Rep. 2015, 5, 13639. [Google Scholar] [CrossRef]
- Zhu, J.K. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 2003, 6, 441–445. [Google Scholar] [CrossRef]
- de Freitas, P.A.F.; de Carvalho, H.H.; Costa, J.H.; Miranda, R.S.; Saraiva, K.; de Oliveira, F.D.B.; Coelho, D.G.; Prisco, J.T.; Gomes-Filho, E. Salt acclimation in sorghum plants by exogenous proline: Physiological and biochemical changes and regulation of proline metabolism. Plant Cell Rep. 2019, 38, 403–416. [Google Scholar] [CrossRef]
- Liang, W.; Ma, X.; Wan, P.; Liu, L. Plant salt-tolerance mechanism: A review. Biochem. Bioph. Res. Co. 2018, 495, 286–291. [Google Scholar] [CrossRef]
- Tang, X.; Mu, X.; Shao, H.; Wang, H.; Brestic, M. Global plant-responding mechanisms to salt stress: Physiological and molecular levels and implications in biotechnology. Crit. Rev. Biotechnol. 2015, 35, 425–437. [Google Scholar] [CrossRef]
- Arifa, Y.; Singha, P.; Siddiquia, H.; Bajguzb, A.; Hayata, S. Salinity induced physiological and biochemical changes in plants: An omic approach towards salt stress tolerance. Plant Physiol. Bioch. 2020, 156, 64–77. [Google Scholar] [CrossRef]
- Gupta, B.; Huang, B. Mechanism of salinity tolerance in plants: Physiological, biochemical, and molecular characterization. Int. J. Genom. 2014, 2014, 701596. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Ann. Rev. Plant. Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Gapińska, M.; Skłodowska, M.; Gabara, B. Effect of short-and long-term salinity on the activities of antioxidative enzymes and lipid peroxidation in tomato roots. Acta Physiol. Plant 2008, 30, 11–18. [Google Scholar] [CrossRef]
- Vranová, E.; Inzé, D.; Van Breusegem, F. Signal transduction during oxidative stress. J. Exp. Bot. 2002, 53, 1227–1236. [Google Scholar] [CrossRef]
- Hernández, J.A.; Almansa, M.S. Short-term effects of salt stress on antioxidant systems and leaf water relations of pea leaves. Physiol. Plant. 2002, 115, 251–257. [Google Scholar] [CrossRef]
- Toyooka, K.; Goto, Y.; Asatsuma, S.; Koizumi, M.; Mitsui, T.; Matsuoka, K. A mobile secretory vesicle cluster involved in mass transport from the golgi to the plant cell exterior. Plant Cell 2009, 21, 1212–1229. [Google Scholar] [CrossRef]
- Zeng, L.; Shannon, M.C.; Grieve, C.M. Evaluation of salt tolerance in rice genotypes by multiple agronomic parameters. Euphytica 2002, 127, 235–245. [Google Scholar] [CrossRef]
- Füzy, A.; Kovács, R.; Cseresnyés, I.; Parádi, I.; Szili‑Kovács, T.; Kelemen, B.; Rajkai, K.; Takács, T. Selection of plant physiological parameters to detect stress effects in pot experiments using principal component analysis. Acta Physiol. Plant. 2019, 41, 56. [Google Scholar] [CrossRef]
- Munns, R.; James, R.A.; Lauchli, A. Approaches to increasing the salt tolerance of wheat and other cereals. J. Exp. Bot. 2006, 57, 1025–1043. [Google Scholar] [CrossRef]
- Munns, R.; James, R.A. Screening methods for salinity tolerance: A case study with tetraploid wheat. Plant Soil 2003, 253, 201–218. [Google Scholar] [CrossRef]
- Genc, Y.; Mcdonald, G.K.; Tester, M. Reassessment of tissue Na+ concentration as a criterion for salinity tolerance in bread wheat. Plant Cell Environ. 2007, 30, 1486–1498. [Google Scholar] [CrossRef]
- Zeng, L.; Poss, J.A.; Wilson, C.; Draz, A.E.; Gregorio, G.B.; Grieve, C.M. Evaluation of salt tolerance in rice genotypes by physiological characters. Euphytica 2003, 129, 281–292. [Google Scholar] [CrossRef]
- El-Hendawy, S.E.; Hu, Y.; Yakout, G.M.; Awad, A.M.; Hafiz, S.E.; Schmidhalter, U. Evaluating salt tolerance of wheat genotypes using multiple parameters. Europ. J. Agron. 2005, 22, 243–253. [Google Scholar] [CrossRef]
- Hasan, A.; Hafiz, H.R.; Siddiqui, N.; Khatun, M.; Islam, R.; Mamun, A. Evaluation of wheat genotypes for salt tolerance based on some physiological traits. J. Crop. Sci. Biotech. 2015, 18, 333–340. [Google Scholar] [CrossRef]
- Genc, Y.; Tester, M.; McDonald, G.K. Calcium requirement of wheat in saline and non-saline conditions. Plant Soil 2010, 327, 331–345. [Google Scholar] [CrossRef]
- Zhao, J.; Sun, H.; Dai, H.; Zhang, G.; Wu, F. Difference in response to drought stress among Tibet wild barley genotypes. Euphytica 2010, 172, 395–403. [Google Scholar] [CrossRef]
- Molina, A.; Bueno, P.; Marin, M.C.; Rodriguez-Rosales, M.P.; Belver, A.; Venema, K.; Donaire, J.P. Involvement of endogenous salicylic acid content, lipoxygenase and antioxidant enzyme activities in the response of tomato cell suspension culture to NaCl. New Phytol. 2002, 156, 409–415. [Google Scholar] [CrossRef]
- van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Ann. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Gerona, M.E.B.; Deocampo, M.P.; Egdane, J.A.; Ismail, A.M.; Dionisio-Sese, M.L. Physiological responses of contrasting rice genotypes to salt stress at reproductive stage. Rice Sci. 2019, 26, 207–219. [Google Scholar] [CrossRef]
- Szira, F.; Bálint, A.F.; Börner, A.; Galiba, G. Evaluation of drought-related traits and screening methods at different developmental stages in spring barley. J. Agron. Crop. Sci. 2008, 194, 334–342. [Google Scholar] [CrossRef]
- Anjum, S.A.; Wang, L.; Farooq, M.; Khan, I.; Xue, L. Methyl jasmonate-induced alteration in lipid peroxidation, antioxidative defense system and yield in soybean under drought. J. Agron. Crop. Sci. 2011, 197, 296–301. [Google Scholar] [CrossRef]
- Tsai, Y.C.; Chen, K.C.; Cheng, T.S.; Lee, C.; Lin, S.H.; Tung, C.W. Chlorophyll fluorescence analysis in diverse rice varieties reveals the positive correlation between the seedlings salt tolerance and photosynthetic efficiency. BMC Plant Biol. 2019, 19, 403. [Google Scholar] [CrossRef]
- Wang, P.; Li, X.; Tian, L.; Gu, Z.; Yang, R. Low salinity promotes the growth of broccoli sprouts by regulating hormonal homeostasis and photosynthesis. Hort. Environ. Biotech. 2018, 60, 19–30. [Google Scholar] [CrossRef]
- Tariq, A.; Masroor, M.; Khan, A.; Jaime, A.; da Teixeira, S.; Mohd, I.; Naeem, M. Role of salicylic acid in promoting salt stress tolerance and enhanced artemisinin production in Artemisia annua L. J. Plant Growth Regul. 2011, 30, 425–435. [Google Scholar]
- Liu, X.; Chi, H.; Yue, M.; Zhang, X.F.; Li, W.J.; Jia, E.P. The regulation of exogenous jasmonic acid on UV-B stress tolerance in wheat. J. Plant Growth Regul. 2012, 31, 436–447. [Google Scholar] [CrossRef]
- Horie, T.; Kaneko, T.; Sugimoto, G.; Sasano, S.; Panda, S.K.; Shibasaka, M.; Katsuhara, M. Mechanisms of water transport mediated by PIP aquaporins and their regulation via phosphorylation events under salinity stress in barley roots. Plant Cell Physiol. 2011, 52, 663–675. [Google Scholar] [CrossRef]
- Garg, A.K.; Kim, J.K.; Owens, T.G.; Ranwala, A.P.; Choi, Y.D.; Kochian, L.V.; Wu, R.J. Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc. Natl. Acad. Sci. USA 2002, 99, 15898–15903. [Google Scholar] [CrossRef]
- Hmida-Sayari, A.; Gargouri-Bouzid, R.; Bidani, A.; Jaoua, L.; Savouré, A.; Jaoua, S. Overexpression of Δ1-pyrroline-5-carboxylate synthetase increases proline production and confers salt tolerance in transgenic potato plants. Plant Sci. 2005, 169, 746–752. [Google Scholar] [CrossRef]
- Silveira, J.A.G.; Viégas, R.D.A.; da Rocha, I.M.A.; Moreira, A.C.D.O.M.; Moreira, R.D.A.; Oliveira, J.T.A. Proline accumulation and glutamine synthetase activity are increased by salt-induced proteolysis in cashew leaves. J. Plant Physiol. 2003, 160, 115–123. [Google Scholar] [CrossRef]
- Singh, M.; Kumar, J.; Singh, S.; Singh, V.P.; Prasad, S.M. Roles of osmoprotectants in improving salinity and drought tolerance in plants: A review. Rev. Environ. Sci. Biotechnol. 2015, 14, 407–426. [Google Scholar] [CrossRef]
- Guo, R.; Yang, Z.; Li, F.; Yan, C.; Zhong, X.; Liu, Q.; Xia, X.; Li, H.; Zhao, L. Comparative metabolic responses and adaptive strategies of wheat (Triticum aestivum) to salt and alkali stress. BMC Plant Biol. 2015, 15, 170. [Google Scholar] [CrossRef]
- Kerepesi, I.; Galiba, G. Osmotic and salt stress-induced alteration in soluble carbohydrate content in wheat seedlings. Crop. Sci. 2000, 40, 482–487. [Google Scholar] [CrossRef]
- Poustini, K.; Siosemardeh, A. Ion distribution in wheat cultivars in response to salinity stress. Field Crop. Res. 2004, 85, 125–133. [Google Scholar] [CrossRef]
- Shabala, S.; Shabala, S.; Cuin, T.A.; Pang, J.Y.; Percey, W.; Chen, Z.H.; Conn, S.; Eing, C.; Wegner, L.H. Xylem ionic relations and salinity tolerance in barley. Plant J. 2010, 61, 839–853. [Google Scholar] [CrossRef]
- Blumwald, E. Sodium transport and salt tolerance in plants. Curr. Opin. Cell Biol. 2000, 12, 431–434. [Google Scholar] [CrossRef]
- Li, J.R.; Liu, M. Biological features and regulatory mechanisms of salt tolerance in plants. J. Cell Biochem. 2019, 120, 10914–10920. [Google Scholar] [CrossRef]
- Attia, H.; Al-Yasi, H.; Alamer, K.; Ali, E.; Hassan, F.; Elshazly, S.; Hessini, K. Induced anti-oxidation efficiency and others by salt stress in Rosa damascena Miller. Sci Hortic. 2020, 274, 109681. [Google Scholar] [CrossRef]
- Luna, C.; Seffino, L.G.; Arias, C.; Taleisnik, E. Oxidative stress indicators as selection tools for salt tolerance in Chloris gayana. Plant Breed. 2000, 119, 341–345. [Google Scholar] [CrossRef]
- Shalata, A.; Mittova, V.; Volokita, M.; Guy, M.; Tal, M. Response of the cultivated tomato and its wild salt-tolerant relative Lycopersicon pennellii to salt-dependent oxidative stress: The root antioxidative system. Physiol. Plant. 2001, 112, 487–494. [Google Scholar] [CrossRef]
- Meloni, D.A.; Oliva, M.A.; Martinez, C.A.; Cambraia, J. Photosynthesis and activity of superoxide dismutase, peroxidase and glutathione reductase in cotton under salt stress. Environ. Exp. Bot. 2003, 49, 69–76. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Xie, Z.X.; Duan, L.S.; Tian, X.L.; Wang, B.M.; Eneji, A.E.; Li, Z.H. Coronatine alleviates salinity stress in cotton by improving the antioxidative defense system and radical-scavenging activity. J. Plant Physiol. 2008, 165, 375–384. [Google Scholar] [CrossRef]
- de Azevedo Neto, A.D.; Prisco, J.T.; Enéas-Filho, J.; de Abreu, C.E.B.; Gomes-Filho, E. Effect of salt stress on antioxidative enzymes and lipid peroxidation in leaves and roots of salt-tolerant and salt-sensitive maize genotypes. Environ. Exp. Bot. 2006, 56, 87–94. [Google Scholar] [CrossRef]
- Zhang, Q.; Dai, W. Plant response to salinity stress. In Stress Physiology of Woody Plants; Dai, W., Ed.; CRC Press: Boca Raton, FL, USA, 2019; pp. 155–173. [Google Scholar]
- Nelson, M.; Dempster, W.F.; Silverstone, S.; Alling, A.; Allen, J.P.; van Thillo, M. Crop yield and light/energy efficiency in a closed ecological system: Laboratory Biosphere experiments with wheat and sweet potato. Adv. Space Res. 2005, 35, 1539–1543. [Google Scholar] [CrossRef]
- Zhang, R.; Liu, J.X.; Chai, Z.Z.; Chen, S.; Bai, Y.; Zong, Y.; Chen, K.L.; Li, J.Y.; Jiang, L.J.; Gao, C.X. Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing. Nat. Plants 2019, 5, 480–485. [Google Scholar] [CrossRef]
- Shan, C.J.; Zhang, S.L.; Ou, X.Q. The roles of H2S and H2O2 in regulating AsA-GSH cycle in the leaves of wheat seedlings under drought stress. Protoplasm 2018, 255, 1257–1262. [Google Scholar] [CrossRef]
- Wu, D.Z.; Sato, K.; Ma, J.F. Genome-wide association mapping of cadmium accumulation in different organs of barley. New Phytol. 2015, 208, 817–829. [Google Scholar] [CrossRef]
- Houshmand, S.; Arzani, A.; Maibody, S.A.M.; Feizi, M. Evaluation of salt-tolerant genotypes of durum wheat derived from in vitro and field experiments. Field Crops Res. 2005, 91, 345–354. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Method Enzymol. 1987, 148, 350–382. [Google Scholar]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Giannakoula, A.; Moustakas, M.; Mylona, P.; Ioannis, P.; Traianos, Y. Aluminium tolerance in maize is correlated with increased levels of mineral nutrients, carbohydrates and proline and decreased levels of lipid peroxidation and Al accumulation. J. Plant Physiol. 2008, 165, 385–396. [Google Scholar] [CrossRef]
- Kumar, G.M.; Knowles, N.R. Changes in lipid peroxidation and lipolytic and free-radical scavenging enzyme activities during aging and sprouting of potato (Solanum tuberosum) seed-tubers. Plant Physiol. 1993, 102, 115–124. [Google Scholar] [CrossRef]
- Hammerschmidt, R.; Nuckles, E.M.; Kuc, J. Association of enhanced peroxidase activity with induced systemic resistance of cucumber to Colletotrchum lagenarium. Physiol. Mol. Plant Pathol. 1982, 20, 73–82. [Google Scholar]
- Shi, Q.; Ding, F.; Wang, X.; Wei, M. Exogenous nitric oxide protect cucumber roots against oxidative stress induced by salt stress. Plant Physiol. Bioch. 2007, 45, 542–550. [Google Scholar] [CrossRef]
- Nakano, Y.; Asada, K. Hydrogen peroxide is scavenged by ascorbate specific peroxidase in spinach chloroplasts. Plant Cell Physiol. 1981, 22, 867–880. [Google Scholar]
- Wheeler, C.R.; Salzman, J.A.; Elsayed, N.M.; Omaye, S.T.; Korte, D.W., Jr. Automated assays for superoxide dismutase, catalase, glutathione peroxidase, and glutathione reductase activity. Anal. Biochem. 1990, 184, 193–199. [Google Scholar] [CrossRef]
| Trait | Minimum | Maximum | Mean | CV (%) | AmongGenoTypes | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | T | T/C | C | T | T/C | C | T | T/C | C | T | T/C | ||
| SDW (mg) | 83.50 | 43.00 | 0.37 | 213.75 | 129.75 | 0.94 | 145.89 | 89.09 | 0.62 | 16.65 | 14.38 | 14.16 | ** |
| RDW (mg) | 33.00 | 8.50 | 0.17 | 104.50 | 50.75 | 1.00 | 63.71 | 32.52 | 0.52 | 19.75 | 21.46 | 21.54 | ** |
| SFW (mg) | 659.50 | 231.50 | 0.22 | 2069.75 | 810.00 | 0.78 | 1278.17 | 541.85 | 0.43 | 17.66 | 19.00 | 17.90 | ** |
| RFW (mg) | 502.25 | 131.00 | 0.14 | 1759.00 | 825.25 | 1.05 | 1066.10 | 484.70 | 0.46 | 21.41 | 25.77 | 26.16 | ** |
| SH (cm) | 16.50 | 12.25 | 0.62 | 27.40 | 22.53 | 1.03 | 22.02 | 17.42 | 0.79 | 9.75 | 9.51 | 7.54 | ** |
| RL (cm) | 23.58 | 7.73 | 0.31 | 48.73 | 29.58 | 0.90 | 34.71 | 21.22 | 0.62 | 12.33 | 13.96 | 13.48 | ** |
| Genotype | K Content | Na Content | Na/K | ||
|---|---|---|---|---|---|
| C | T | T/C | T | T | |
| Taishan24 | 69.97h | 68.46i | 0.98 | 24.67f | 0.36 |
| Wennong5 | 71.36g | 85.70a | 1.20 | 66.11b | 0.77 |
| Xinong291 | 71.53g | 74.98de | 1.05 | 37.79d | 0.50 |
| Jinan13 | 74.58ef | 73.35f | 0.98 | 32.50e | 0.44 |
| Jimai23 | 76.03cd | 78.84b | 1.04 | 38.06d | 0.48 |
| Klein Flecha | 69.65hi | 62.15j | 0.89 | 52.50c | 0.84 |
| Chuanmai107 | 76.43c | 59.43k | 0.78 | 68.88a | 1.16 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quan, X.; Liang, X.; Li, H.; Xie, C.; He, W.; Qin, Y. Identification and Characterization of Wheat Germplasm for Salt Tolerance. Plants 2021, 10, 268. https://doi.org/10.3390/plants10020268
Quan X, Liang X, Li H, Xie C, He W, Qin Y. Identification and Characterization of Wheat Germplasm for Salt Tolerance. Plants. 2021; 10(2):268. https://doi.org/10.3390/plants10020268
Chicago/Turabian StyleQuan, Xiaoyan, Xiaoli Liang, Hongmei Li, Chunjuan Xie, Wenxing He, and Yuxiang Qin. 2021. "Identification and Characterization of Wheat Germplasm for Salt Tolerance" Plants 10, no. 2: 268. https://doi.org/10.3390/plants10020268
APA StyleQuan, X., Liang, X., Li, H., Xie, C., He, W., & Qin, Y. (2021). Identification and Characterization of Wheat Germplasm for Salt Tolerance. Plants, 10(2), 268. https://doi.org/10.3390/plants10020268




