Genome-Wide Identification and Characterization of Caffeic Acid O-Methyltransferase Gene Family in Soybean
Abstract
1. Introduction
2. Results
2.1. Identification of COMT Gene Family in Soybean
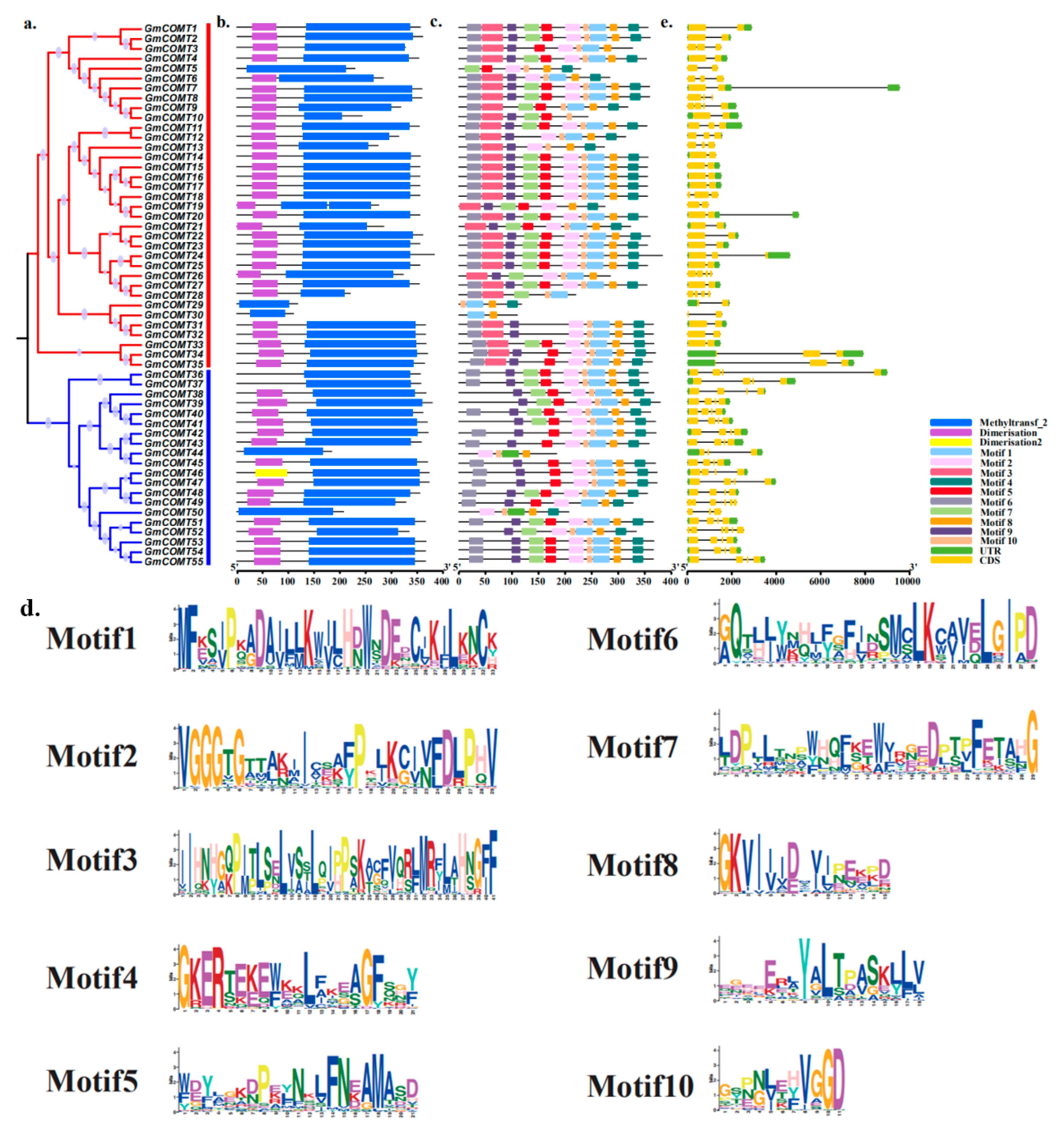
2.2. Phylogenetic Analysis of the COMT Gene Family in Soybean
2.3. Evolutionary Conservation Analysis of COMTs in Soybean
2.4. Chromosomal Location and Collinearity Analysis of COMT Gene FAMILY in Soybean
2.5. The Cis-Regulatory Elements in the Promoter of GmCOMTs Genes in Soybean
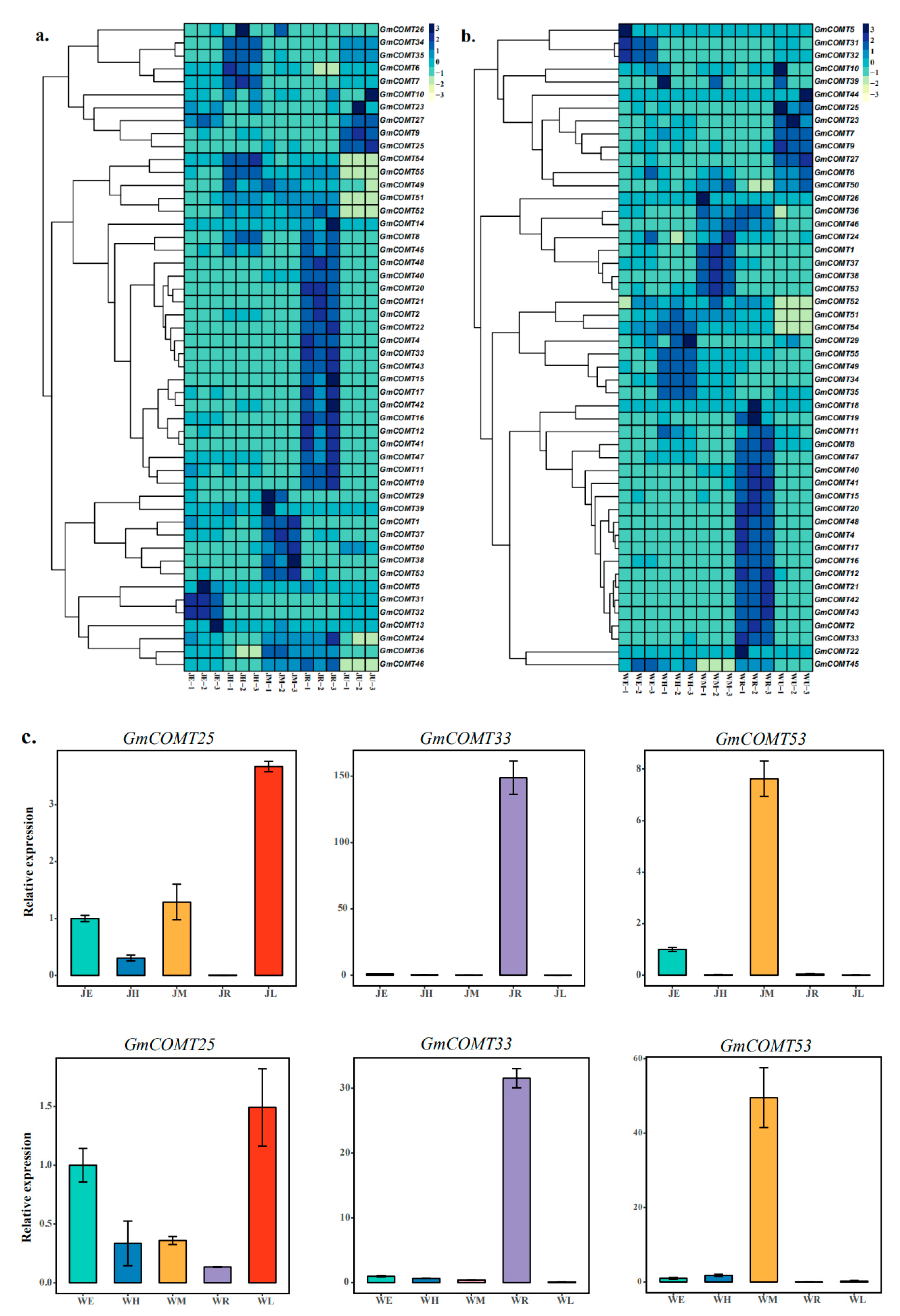
2.6. Tissue Specificity of GmCOMTs Genes Expression
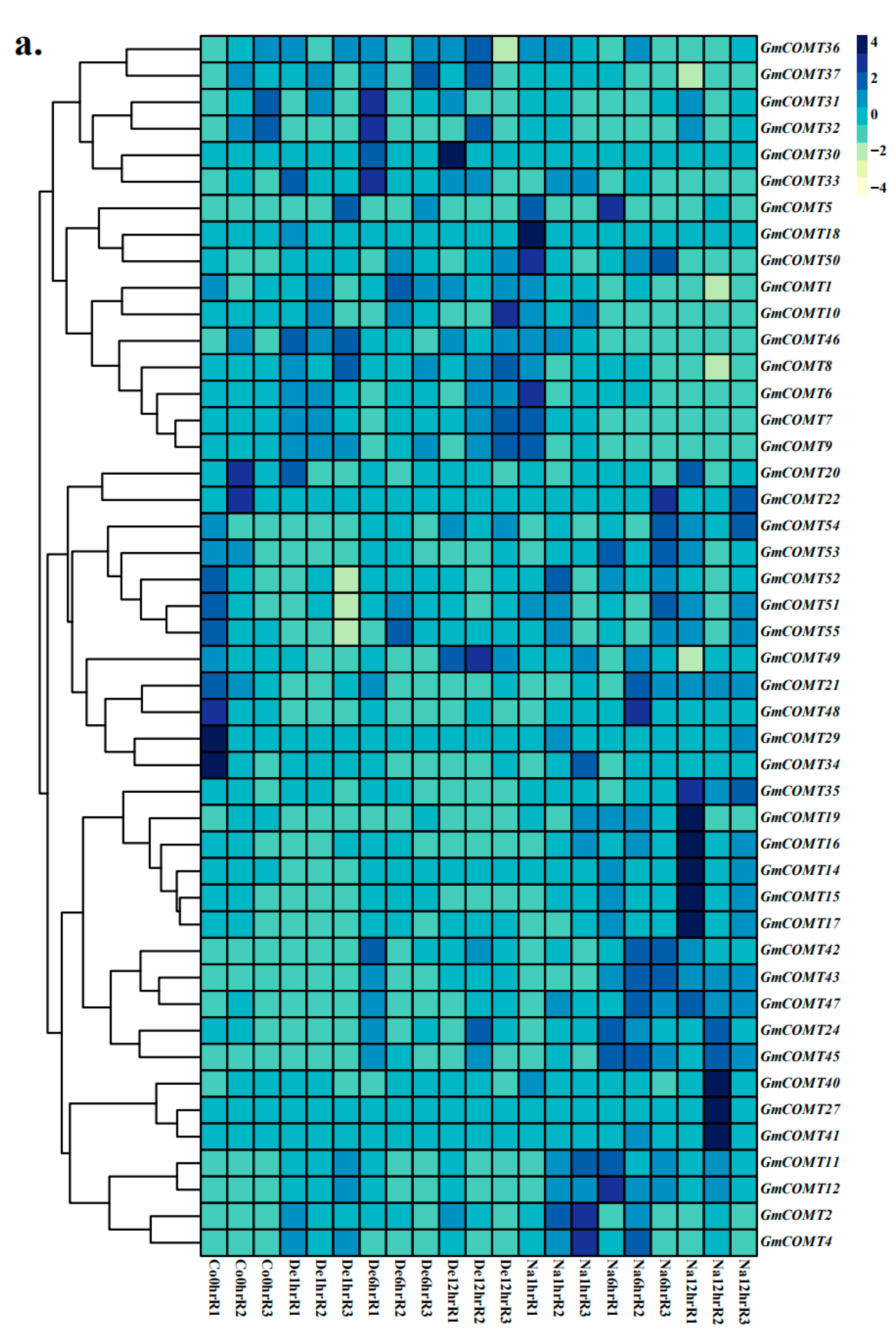
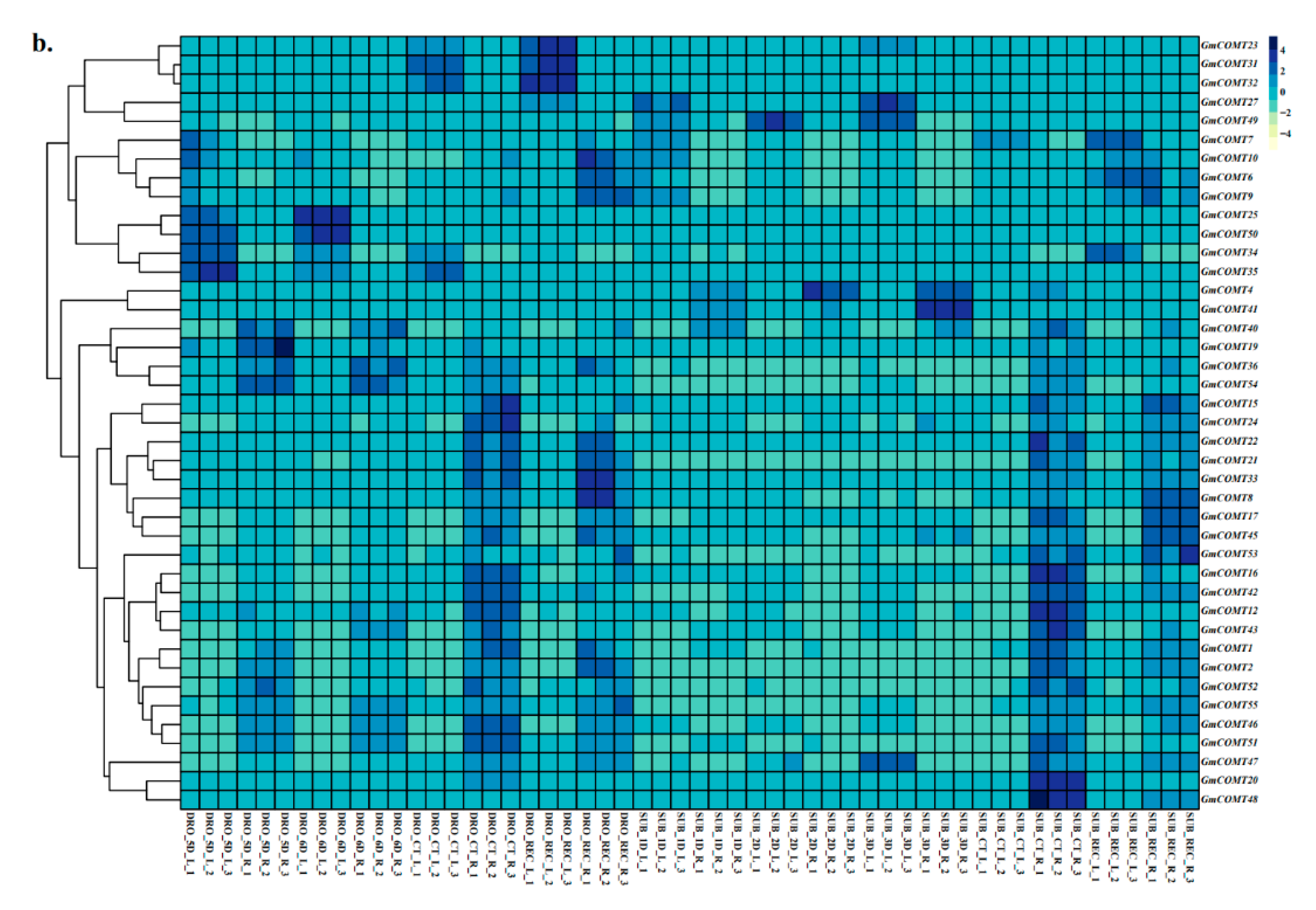
2.7. Expression Patterns of COMT Genes under Abiotic Stresses
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Identification of COMT Gene Family in Soybean
4.3. Phylogenetic Analysis of COMT Proteins in Soybean
4.4. Gene Structure, Motif and Domain Analyses for the GmCOMT Genes
4.5. Chromosomal Location, Collinearity Analysis, Gene Duplication Events, and Ka/Ks Analysis of GmCOMT Genes
4.6. Prediction of Cis-Acting Elements of COMT Genes in Soybean
4.7. Expression Pattern Analysis
4.8. Genomic RNA Extraction and Quantitative RT-PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pagano, M.C.; Miransari, M. The importance of soybean production worldwide. In Abiotic and Biotic Stresses in Soybean Production; Academic Press: Cambridge, MA, USA, 2016; Volume 1, pp. 1–26. [Google Scholar]
- Koberg, M.; Abu-Much, R.; Gedanken, A. Optimization of bio-diesel production from soybean and wastes of cooked oil: Combining dielectric microwave irradiation and a SrO catalyst. Bioresour. Technol. 2011, 102, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.L.; Liu, W.G.; Yuan, J.; Jiang, T.; Yang, W.Y. Relationship between lignin synthesis and lodging resistance at seedlings stage in soybean intercropping system. Acta Agron. Sin. 2015, 41, 1098–1104. [Google Scholar] [CrossRef]
- Klis, F.M.; Mol, P.; Hellingwerf, K.; Brul, S. Dynamics of cell wall structure in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2002, 26, 239–256. [Google Scholar] [CrossRef]
- Canut, H.; Albenne, C.; Jamet, E. Isolation of the cell wall. Methods Mol. Biol. 2017, 1511, 171–185. [Google Scholar] [PubMed]
- Underwood, W. The plant cell wall: A dynamic barrier against pathogen invasion. Front. Plant Sci. 2012, 3, 85. [Google Scholar] [CrossRef] [PubMed]
- Ralph, J.; Lundquist, K.; Brunow, G.; Lu, F.; Boerjan, W. Lignins: Natural polymers from oxidative coupling of 4-hydroxyphenyl- propanoids. Phytochem. Rev. 2004, 3, 29–60. [Google Scholar] [CrossRef]
- Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and biological functions in plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef] [PubMed]
- Ithal, N.; Recknor, J.; Dan, N.; Maier, T.; Mitchum, M.G. Developmental transcript profiling of cyst nematode feeding cells in soybean roots. Mol. Plant Microbe Interact. 2007, 20, 510–525. [Google Scholar] [CrossRef]
- Peng, D.; Chen, X.; Yin, Y.; Lu, K.; Yang, W.; Tang, Y.; Wang, Z. Lodging resistance of winter wheat (Triticum aestivum L.): Lignin accumulation and its related enzymes activities due to the application of paclobutrazol or gibberellin acid. Field Crop. Res. 2014, 157, 1–7. [Google Scholar] [CrossRef]
- Moura, J.C.M.S.; Bonine, C.; Viana, J.; Dornelas, M.C.; Mazzafera, P. Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 2010, 52, 360–376. [Google Scholar] [CrossRef]
- Xiang, D.B.; Guo, K.; Lei, T.; Yu, X.B.; Luo, Q.M.; Yang, W.Y. Effects of phosphorus and potassium on stem characteristics and lodging resistance of relay cropping soybean. Chin. J. Oil Crop Sci. 2010, 32, 395–402. [Google Scholar]
- Tripathi, S.C.; Sayre, K.D.; Kaul, J.N.; Narang, R.S. Growth and morphology of spring wheat (Triticum aestivum L.) culms and their association with lodging: Effects of genotypes, N levels and ethephon. Field Crop. Res. 2003, 84, 271–290. [Google Scholar] [CrossRef]
- Vanholme, R.; Meester, B.D.; Ralph, J.; Boerjan, W. Lignin biosynthesis and its integration into metabolism. Curr. Opin. Biotechnol. 2019, 56, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Yan, C.; Qiu, J.; Zhang, N.; Lin, Y.; Cai, Y. Structural characterization and deposition of stone cell lignin in Dangshan Su pear. Sci. Hortic. 2013, 155, 123–130. [Google Scholar] [CrossRef]
- Abdelaziz, O.Y.; Brink, D.P.; Prothmann, J.; Ravi, K.; Gorwa-Grauslund, M.F. Biological valorization of low molecular weight lignin. Biotechnol. Adv. 2016, 34, 1318–1346. [Google Scholar] [CrossRef]
- Vanholme, R.; Demedts, B.; Morreel, K.; Ralph, J.; Boerjan, W. Lignin biosynthesis and structure. Plant Physiol. 2010, 153, 895–905. [Google Scholar] [CrossRef]
- Trabucco, G.M.; Matos, D.A.; Lee, S.J.; Saathoff, A.J.; Hazen, S.P. Functional characterization of cinnamyl alcohol dehydrogenase and caffeic acid O-methyltransferase in Brachypodium distachyon. BMC Biotechnol. 2013, 13, 61. [Google Scholar] [CrossRef] [PubMed]
- Faraji, M.; Fonseca, L.L.; Escamilla-Trevio, L.; Barros-Rios, J.; Voit, E.O. Mathematical models of lignin biosynthesis. Biotechnol. Biofuels 2018, 11, 34. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Pei, J.; Li, Y.; Sun, H. Genome-wide identification and characterization of COMT gene family during the development of blueberry fruit. BMC Plant Biol. 2021, 21, 5. [Google Scholar] [CrossRef]
- Zägel, P.; Dell’Orco, D.; Koch, K.W. The dimerization domain in outer segment guanylate cyclase is a Ca2+-sensitive control switch module. Biochemistry 2013, 52, 5065–5074. [Google Scholar] [CrossRef]
- Huang, S.; Ding, J.; Deng, D.; Tang, W.; Sun, H.; Liu, D.; Zhang, L.; Niu, X.; Zhang, X.; Meng, M.; et al. Draft genome of the kiwifruit Actinidia chinensis. Nat. Commun. 2013, 4, 2640. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Zhang, G.; Yu, J.H.; Li, Y.Y.; Liao, H. Molecular cloning and characterization of caffeic acid 3-O-methyltransferase from the rhizome of Ligusticum chuanxiong. Biotechnol. Lett. 2015, 37, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bugos, R.C.; Chiang, V.; Campbell, W.H. cDNA cloning, sequence analysis and seasonal expression of lignin-bispecific caffeic acid/5-hydroxyferulic acid O-methyltransferase of aspen. Plant Mol. Biol. 1991, 17, 1203–1215. [Google Scholar] [CrossRef]
- Carocha, V.; Soler, M.; Hefer, C.; Cassan-Wang, H.; Fevereiro, P.; Myburg, A.A.; Paiva, J.A.; Grima-Pettenati, J. Genome-wide analysis of the lignin toolbox of Eucalyptus grandis. New Phytol. 2015, 206, 1297–1313. [Google Scholar] [CrossRef]
- Lu, N.; Ma, W.; Han, D.; Liu, Y.; Wang, Z.; Wang, N.; Yang, G.; Qu, G.; Wang, Q.; Zhao, K.; et al. Genome-wide analysis of the Catalpa bungei caffeic acid O-methyltransferase (COMT) gene family: Identification and expression profiles in normal, tension, and opposite wood. PeerJ 2019, 7, 6520. [Google Scholar] [CrossRef] [PubMed]
- Chiang, V.L. Towards a systems approach for lignin biosynthesis in Populus trichocarpa: Transcript abundance and specificity of the monolignol biosynthetic genes. Plant Cell Physiol. 2010, 51, 144–163. [Google Scholar]
- Wei, L.; Lu, J.; Lu, K.; Yuan, J.; Li, J. Cloning and phylogenetic analysis of Brassica napus L. Caffeic acid O-methyltransferase 1 gene family and its expression pattern under drought stress. PLoS ONE 2016, 11, e0165975. [Google Scholar]
- Chen, S.; Zhao, Y.; Zhao, X.; Chen, S. Identification of putative lignin biosynthesis genes in Betula pendula. Trees 2020, 34, 1255–1265. [Google Scholar] [CrossRef]
- Lv, G.; Tang, D.; Chen, F.; Sun, Y.; Fang, W.; Guan, Z.; Liu, Z.; Chen, S. The anatomy and physiology of spray cut chrysanthemum pedicels, and expression of a caffeic acid 3-O-methyltransferase homologue. Postharvest Biol. Technol. 2011, 60, 244–250. [Google Scholar] [CrossRef]
- Oraby, H.F.; Ramadan, M.F. Impact of suppressing the caffeic acid O-methyltransferase (COMT) gene on lignin, fiber, and seed oil composition in Brassica napus transgenic plants. Eur. Food Res. Technol. 2015, 240, 931–938. [Google Scholar] [CrossRef]
- Lee, H.Y.; Byeon, Y.; Lee, K.; Lee, H.; Back, K. Cloning of Arabidopsis serotonin N-acetyltransferase and its role with caffeic acid O-methyltransferase in the biosynthesis of melatonin in vitro despite their different subcellular localizations. J. Pineal Res. 2015, 57, 418–426. [Google Scholar] [CrossRef]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef]
- Bailey, T.L.; Mikael, B.; Buske, F.A.; Martin, F.; Grant, C.E.; Luca, C.; Jingyuan, R.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Liu, Y.; Du, H.; Li, P.; Shen, Y.; Peng, H.; Liu, S.; Zhou, G.A.; Zhang, H.; Liu, Z.; Shi, M.; et al. Pan-genome of wild and cultivated soybeans. Cell 2020, 182, 162–176. [Google Scholar] [CrossRef]
- Lei, L.; Zhou, S.; Ma, H.; Zhang, L. Expansion and diversification of the SET domain gene family following whole-genome duplications in Populus trichocarpa. BMC Evol. Biol. 2012, 12, 51. [Google Scholar] [CrossRef]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin biosynthesis. Annu. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Liu, X.B.; Wang, C.; Yang, H.; Li, H.X.; Ruan, R.W.; Yuan, X.H.; Yi, Z.L. Expression analysis of key enzyme genes in lignin synthesis of culm among different lodging resistances of common buckwheat (Fagopyrum esculentum Moench.). Sci. Agric. Sin. 2015, 48, 1864–1872. [Google Scholar]
- Schuetz, M.; Benske, A.; Smith, R.A.; Watanabe, Y.; Tobimatsu, Y.; Ralph, J.; Demura, T.; Ellis, B.; Samuels, A.L. Laccases direct lignification in the discrete secondary cell wall domains of protoxylem. Plant Physiol. 2014, 166, 798–807. [Google Scholar] [CrossRef]
- Bellaloui, N. Soybean seed phenol, lignin, and isoflavones partitioning as affected by seed node position and genotype differences. Food Nutr. Sci. 2012, 3, 447–454. [Google Scholar] [CrossRef][Green Version]
- Casler, M. Breeding forage crops for increased nutritional value. Adv. Agron. 2001, 71, 51–107. [Google Scholar]
- Rogers, L.A.; Campbell, M.M. The genetic control of lignin deposition during plant growth and development. New Phytol. 2004, 164, 17–30. [Google Scholar] [CrossRef]
- Rinaldi, R.; Jastrzebski, R.; Clough, M.T.; Ralph, J.; Kennema, M.; Bruijnincx, P.; Weckhuysen, B.M. Paving the way for lignin valorisation: Recent advances in bioengineering, biorefining and catalysis. Angew. Chem. Int. Ed. Engl. 2016, 55, 8164–8215. [Google Scholar] [CrossRef]
- Vanholme, R.; Ralph, J.; Akiyama, T.; Lu, F.; Boerjan, W. Engineering traditional monolignols out of lignin by concomitant up-regulation of F5H1 and down-regulation of COMT in Arabidopsis. Plant J. Cell Mol. Biol. 2010, 64, 885–897. [Google Scholar] [CrossRef]
- Byeon, Y.; Lee, H.Y.; Lee, K.; Back, K. Caffeic acid O-methyltransferase is involved in the synthesis of melatonin by methylating N-acetylserotonin in Arabidopsis. J. Pineal Res. 2015, 57, 219–227. [Google Scholar] [CrossRef]
- Cipolla-Neto, J.; Amaral, F.G.; Afeche, S.C.; Tan, D.X.; Reiter, R.J. Melatonin, energy metabolism, and obesity: A review. J. Pineal Res. 2014, 56, 371–381. [Google Scholar] [CrossRef]
- Zhang, N.; Sun, Q.; Zhang, H.; Cao, Y.; Weeda, S.; Ren, S.; Guo, Y.D. Roles of melatonin in abiotic stress resistance in plants. J. Exp. Bot. 2015, 66, 647–656. [Google Scholar] [CrossRef]
- Barakat, A.; Choi, A.; Yassin, N.B.M.; Park, J.S.; Sun, Z.; Carlson, J.E. Comparative genomics and evolutionary analyses of the O-methyltransferase gene family in Populus. Gene 2011, 479, 37–46. [Google Scholar] [CrossRef]
- Robin, A.Y.; Giustini, C.; Graindorge, M.; Matringe, M.; Dumas, R. Crystal structure of norcoclaurine-6-O-methyltransferase, a key rate-limiting step in the synthesis of benzylisoquinoline alkaloids. Plant J. 2016, 87, 641–653. [Google Scholar] [CrossRef]
- Kim, J.; Choi, B.; Cho, B.K.; Lim, H.S.; Bae, H. Molecular cloning, characterization and expression of the caffeic acid O-methyltransferase (COMT) ortholog from kenaf (Hibiscus cannabinus). Plant Omics 2013, 6, 246–253. [Google Scholar]
- Zhang, K.; Cui, H.; Cao, S.; Yan, L.; Sun, Y. Overexpression of CrCOMT from Carex rigescens increases salt stress and modulates melatonin synthesis in Arabidopsis thaliana. Plant Cell Rep. 2019, 38, 1501–1514. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, 29–37. [Google Scholar] [CrossRef]
- Panu, A.; Manohar, J.; Konstantin, A.; Delphine, B.; Gabor, C.; Edouard, D.C.; Séverine, D.; Volker, F.; Arnaud, F.; Elisabeth, G. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, 597–603. [Google Scholar]
- Saitou, N. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Sudhir, K.; Glen, S.; Li, M.; Christina, K.; Koichiro, T. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Heredity 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Tae-Ho, L.; Jin, H.; Barry, M.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, 49. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Chen, C.; Li, C.; Liu, J.; Liu, C.; He, Y. Genome-wide investigation of WRKY gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genom. 2018, 19, 490. [Google Scholar] [CrossRef] [PubMed]
- Koch, M.; Al-Shehbaz, I.; Mummenhoff, K. Molecular systematics, evolution, and population biology in the mustard family (Brassicaceae). Ann. Mo. Bot. Gard. 2003, 1, 151–171. [Google Scholar] [CrossRef]
- Rombauts, S.; Déhais, P.; Van, M.; Rouzé, P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar] [CrossRef]
- Lescot, M. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Chen, B.; Wang, L.; Ali, S.; Guo, Y.; Liu, J.; Wang, J.; Xie, L.; Zhang, Q. Genome-Wide Identification and Characterization of Caffeic Acid O-Methyltransferase Gene Family in Soybean. Plants 2021, 10, 2816. https://doi.org/10.3390/plants10122816
Zhang X, Chen B, Wang L, Ali S, Guo Y, Liu J, Wang J, Xie L, Zhang Q. Genome-Wide Identification and Characterization of Caffeic Acid O-Methyltransferase Gene Family in Soybean. Plants. 2021; 10(12):2816. https://doi.org/10.3390/plants10122816
Chicago/Turabian StyleZhang, Xu, Bowei Chen, Lishan Wang, Shahid Ali, Yile Guo, Jiaxi Liu, Jiang Wang, Linan Xie, and Qingzhu Zhang. 2021. "Genome-Wide Identification and Characterization of Caffeic Acid O-Methyltransferase Gene Family in Soybean" Plants 10, no. 12: 2816. https://doi.org/10.3390/plants10122816
APA StyleZhang, X., Chen, B., Wang, L., Ali, S., Guo, Y., Liu, J., Wang, J., Xie, L., & Zhang, Q. (2021). Genome-Wide Identification and Characterization of Caffeic Acid O-Methyltransferase Gene Family in Soybean. Plants, 10(12), 2816. https://doi.org/10.3390/plants10122816





